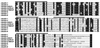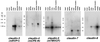Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands - PubMed (original) (raw)
Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands
K Morita et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Two related integral membrane proteins, claudin-1 and -2, recently were identified as novel components of tight junction (TJ) strands. Here, we report six more claudin-like proteins, indicating the existence of a claudin gene family. Three of these were reported previously as RVP1, Clostridium perfringens enterotoxin receptor, and TMVCF, but their physiological functions were not determined. Through similarity searches followed by PCR, we isolated full length cDNAs of mouse RVP1, Clostridium perfringens enterotoxin receptor, and TMVCF as well as three mouse claudin-like proteins and designated them as claudin-3 to -8, respectively. All of these claudin family members showed similar patterns on hydrophilicity plots, which predicted four transmembrane domains in each molecule. Northern blotting showed that the tissue distribution pattern varied significantly, depending on claudin species. Similarly to claudin-1 and -2, when these claudins were HA-tagged and introduced into cultured Madin-Darby canine kidney cells, all showed a tendency to concentrate at TJs. Immunofluorescence and immunoelectron microscopy with polyclonal antibodies specific for claudin-3, -4, or -8 revealed that these molecules were exclusively concentrated at TJs in the liver and/or kidney. These findings indicated that multiple claudin family members are involved in the formation of TJ strands in various tissues.
Figures
Figure 1
Multiple sequence alignments for the members of the claudin family. Putative membrane-spanning domains are represented by bars. Residues conserved in at least six of the eight sequences are highlighted with black backgrounds. The amino acid sequence was fairly conserved in the first and fourth transmembrane domains as well as in the first and second extracellular loops but was diversified in the second and third transmembrane domains. Note that all of the claudin family members end in -Y-V at their COOH-termini. The nucleotide sequence data of mouse claudin-1 to -8 are available from European Molecular Biology Laboratory/GenBank database/DNA Data Base in Japan under accession numbers AF072127, AF072128, AF087821, AF087822, AF087823, AF087824, AF087825, and AF087826, respectively.
Figure 2
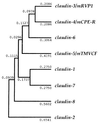
A potential phylogenetic tree of the claudin gene family. The tree was constructed by the unweighed pair group method (–41) using the calculated genetic distances (presented numerically) between pairs of members.
Figure 3
Northern blots of mouse claudin-3, -4, -5, -7, and -8 expression. Mouse Multiple Tissue Northern Blot (CLONTECH) was probed with DNA fragments of claudins (see Materials and Methods). Tissue distribution patterns were distinct between claudin species. The expression of claudin-6 was not detected by Northern blotting, although it was detected by reverse transcription–PCR at least in mouse kidney.
Figure 4
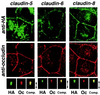
Concentration of HA-tagged claudins at the occludin-positive tight junctions in MDCK transfectants. Confluent cultures of MDCK transfectants expressing HA-tagged claudin-5, -6, or -8 (claudin-5, claudin-6, claudin-8) were doubly stained with anti-HA mAb (anti-HA) and anti-occludin mAb (anti-occludin). Images were obtained at the focal plane of the most apical region of lateral membranes by confocal microscopy. HA-tagged claudins were coconcentrated with occludin at the level of TJs. As shown in the bottom panels, in computer-generated cross-sectional views, HA-claudins (HA) and occludin (Oc) were highly concentrated at the most apical portion of lateral membranes of MDCK cells, and overlaid images showed that HA-claudins were precisely colocalized with occludin (Comp.). The thickness of each cellular sheet is indicated by arrows. The same results were obtained from MDCK transfectants expressing HA-tagged claudin-3, -4, and -7 (data not shown).
Figure 5
Subcellular distribution of endogenous claudin-3/mRVP-1, claudin-4/mCPE-R, and claudin-8. (A) Specificity of anticlaudin-3, -4, and -8 pAbs. Immunoblots of total lysates of E. coli expressing GST fusion proteins with cytoplasmic domains of all claudin family members (arrow) confirmed their specificities. CBB, Coomassie brilliant blue staining. (B) Frozen sections of mouse liver (a and b) and kidney (c_–_f) were stained with affinity-purified anti-claudin-3 pAb (a), anti-claudin-4 pAb (c), or anti-claudin-8 pAb (e). All sections were counter-stained with anti-occludin mAb (b, d, and f). Arrows_,_ distal tubules; asterisks, proximal tubules. (Bar = 10 μm.)
Figure 6
Immunoelectron microscopic images of freeze-fracture replicas of mouse liver with affinity-purified anti-claudin-3 pAb. Tight junction strands themselves (T) were specifically labeled with anticlaudin-3 pAb. (Bar = 0.2 μm.)
Comment in
- Plugging the leaks.
Goodenough DA. Goodenough DA. Proc Natl Acad Sci U S A. 1999 Jan 19;96(2):319-21. doi: 10.1073/pnas.96.2.319. Proc Natl Acad Sci U S A. 1999. PMID: 9892627 Free PMC article. No abstract available.
Similar articles
- Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier.
Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Sonoda N, et al. J Cell Biol. 1999 Oct 4;147(1):195-204. doi: 10.1083/jcb.147.1.195. J Cell Biol. 1999. PMID: 10508866 Free PMC article. - Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells.
Morita K, Sasaki H, Furuse M, Tsukita S. Morita K, et al. J Cell Biol. 1999 Oct 4;147(1):185-94. doi: 10.1083/jcb.147.1.185. J Cell Biol. 1999. PMID: 10508865 Free PMC article. - Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin.
Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Furuse M, et al. J Cell Biol. 1998 Jun 29;141(7):1539-50. doi: 10.1083/jcb.141.7.1539. J Cell Biol. 1998. PMID: 9647647 Free PMC article. - The structure and function of claudins, cell adhesion molecules at tight junctions.
Tsukita S, Furuse M. Tsukita S, et al. Ann N Y Acad Sci. 2000;915:129-35. doi: 10.1111/j.1749-6632.2000.tb05235.x. Ann N Y Acad Sci. 2000. PMID: 11193568 Review. - Crystal structures of claudins: insights into their intermolecular interactions.
Suzuki H, Tani K, Fujiyoshi Y. Suzuki H, et al. Ann N Y Acad Sci. 2017 Jun;1397(1):25-34. doi: 10.1111/nyas.13371. Epub 2017 Jun 12. Ann N Y Acad Sci. 2017. PMID: 28605828 Review.
Cited by
- Regulation of colon injury and improvement of exercise performance in exhausted running mice by Lactobacillus pentosus CQZC02.
Cai L, Wang B. Cai L, et al. Front Physiol. 2024 Sep 20;15:1475413. doi: 10.3389/fphys.2024.1475413. eCollection 2024. Front Physiol. 2024. PMID: 39371599 Free PMC article. - Expression and Targeted Application of Claudins Family in Hepatobiliary and Pancreatic Diseases.
Du F, Xie Y, Wu S, Ji M, Dong B, Zhu C. Du F, et al. J Hepatocell Carcinoma. 2024 Sep 25;11:1801-1821. doi: 10.2147/JHC.S483861. eCollection 2024. J Hepatocell Carcinoma. 2024. PMID: 39345937 Free PMC article. Review. - The Basic Requirement of Tight Junction Proteins in Blood-Brain Barrier Function and Their Role in Pathologies.
Dithmer S, Blasig IE, Fraser PA, Qin Z, Haseloff RF. Dithmer S, et al. Int J Mol Sci. 2024 May 21;25(11):5601. doi: 10.3390/ijms25115601. Int J Mol Sci. 2024. PMID: 38891789 Free PMC article. Review. - Biophysics of claudin proteins in tight junction architecture: Three decades of progress.
Marsch P, Rajagopal N, Nangia S. Marsch P, et al. Biophys J. 2024 Aug 20;123(16):2363-2378. doi: 10.1016/j.bpj.2024.06.010. Epub 2024 Jun 10. Biophys J. 2024. PMID: 38859584 Review. - The Gut Microbiota Contributes to the Development of LPS-Induced Orchitis by Disrupting the Blood-Testosterone Barrier in Mice.
Guo Q, Cheng Y, Li T, Huang J, Li J, Zhang Z, Qu Y. Guo Q, et al. Reprod Sci. 2024 Nov;31(11):3379-3390. doi: 10.1007/s43032-024-01613-9. Epub 2024 Jun 10. Reprod Sci. 2024. PMID: 38858330
References
- Schneeberger E E, Lynch R D. Am J Physiol. 1992;262:L647–L661. - PubMed
- Gumbiner B. Am J Physiol. 1987;253:C749–C758. - PubMed
- Anderson J M, van Itallie C M. Am J Physiol. 1995;269:G467–G475. - PubMed
- Staehelin L A. J Cell Sci. 1973;13:763–786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases