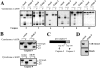Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner - PubMed (original) (raw)
. 1999 Jan 25;144(2):281-92.
doi: 10.1083/jcb.144.2.281.
M T Harte, R M Kluck, B B Wolf, C A Casiano, D D Newmeyer, H G Wang, J C Reed, D W Nicholson, E S Alnemri, D R Green, S J Martin
Affiliations
- PMID: 9922454
- PMCID: PMC2132895
- DOI: 10.1083/jcb.144.2.281
Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner
E A Slee et al. J Cell Biol. 1999.
Abstract
Exit of cytochrome c from mitochondria into the cytosol has been implicated as an important step in apoptosis. In the cytosol, cytochrome c binds to the CED-4 homologue, Apaf-1, thereby triggering Apaf-1-mediated activation of caspase-9. Caspase-9 is thought to propagate the death signal by triggering other caspase activation events, the details of which remain obscure. Here, we report that six additional caspases (caspases-2, -3, -6, -7, -8, and -10) are processed in cell-free extracts in response to cytochrome c, and that three others (caspases-1, -4, and -5) failed to be activated under the same conditions. In vitro association assays confirmed that caspase-9 selectively bound to Apaf-1, whereas caspases-1, -2, -3, -6, -7, -8, and -10 did not. Depletion of caspase-9 from cell extracts abrogated cytochrome c-inducible activation of caspases-2, -3, -6, -7, -8, and -10, suggesting that caspase-9 is required for all of these downstream caspase activation events. Immunodepletion of caspases-3, -6, and -7 from cell extracts enabled us to order the sequence of caspase activation events downstream of caspase-9 and reveal the presence of a branched caspase cascade. Caspase-3 is required for the activation of four other caspases (-2, -6, -8, and -10) in this pathway and also participates in a feedback amplification loop involving caspase-9.
Figures
Figure 1
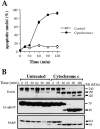
Cytochrome c initiates apoptotic changes in Jurkat cell–free extracts. Rat liver nuclei were incubated at 37°C in postnuclear extracts of Jurkat cells, prepared as described in Materials and Methods, in the presence or absence of 50 μg/ml of bovine heart cytochrome c. (A) At the indicated times, 2-μl aliquots of extract were removed for analysis of nuclear morphology by Hoescht 33342 staining. Nuclei were scored as apoptotic if they exhibited chromatin condensation and nuclear fragmentation characteristic of apoptosis. Each data point represents counts on 300 nuclei. Triplicate determinations (± SEM) are shown from a representative experiment. (B) At the indicated times, samples of each cell-free reaction were taken for SDS-PAGE, followed by Western blotting and probing for the indicated proteins.
Figure 2
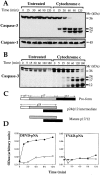
Cytochrome c initiates processing and activation of caspase-3 but not caspase-1 in Jurkat cell extracts. (A) Processing of caspase-3 (top) and caspase-1 (bottom) in Jurkat cell–free extracts incubated for the indicated times with or without cytochrome c (50 μg/ml). Caspase processing was assessed by Western blot. (B) Cell-free reactions were set up as in A except that [35S]methionine-labeled caspase-3, prepared by coupled in vitro transcription/translation, was added to the extracts. At the indicated times, portions of the extract were removed and were separated by SDS-PAGE followed by direct autoradiography. (C) Schematic representation of cytochrome c–initiated caspase-3 processing. The pattern of processing was deduced from the data shown in A and B. (D) At the indicated time points, extracts were assessed for DEVD-pNA or YVAD-pNA hydrolyzing activity, as described in Materials and Methods. Cell-free reactions were set up as described above and were incubated in the presence (closed circles) or absence (open circles) of cytochrome c (50 μg/ml). At the indicated times, samples of extract were removed and were assessed for their ability to hydrolyze the peptides DEVD-pNA and YVAD-pNA. Results shown are representative of three separate experiments.
Figure 3
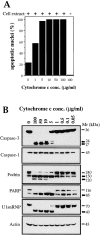
Titration of cytochrome c–initiated apoptosis in Jurkat cell extracts. (A) Rat liver nuclei were incubated in postnuclear extracts from Jurkat cells (75 μg protein in 20 μl) for 2 h at 37°C in the presence or absence of the indicated amounts of bovine heart cytochrome c. As a control, nuclei were also incubated in the highest concentration of cytochrome c in the absence of cell extract (CEB was substituted). Apoptosis was assessed by scoring nuclei with features of apoptosis, as described in Fig. 1. Results shown are from a representative experiment and were derived from counts performed on 300 nuclei from several fields. (B) Cell-free reactions, set up identically to those in A, were incubated for 2 h at 37°C followed by analysis by SDS-PAGE and Western blotting. Blots were then probed with antibodies directed against the indicated caspases and caspase substrates.
Figure 4
Cytochrome c–initiated activation of multiple caspases. [35S]Methionine-labeled caspases, prepared by coupled in vitro transcription/translation (∼0.25–0.5 μl of translation reaction in a total reaction volume of 10 μl), were incubated with cytochrome c (50 μg/ml) in the presence or absence of Jurkat postnuclear extract, as indicated. Where no cell extract was added to the reaction, an appropriate volume of extract dilution buffer was substituted. Cell-free reactions were incubated for 3 h at 37°C and were then analyzed by SDS-PAGE followed by fluorography.
Figure 5
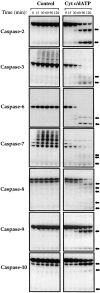
Kinetics of activation of caspases-2, -3, -6, -7, -8, -9, and -10 in response to cytochrome c. Cell-free reactions were assembled containing the indicated [35S]methionine-labeled caspases and were incubated with or without 50 μg/ml cytochrome c and 1 mM dATP, as indicated. Reactions were incubated at 37°C and samples were removed at the indicated times, followed by analysis of caspase processing by SDS-PAGE/fluorography. Cleavage products are denoted by arrows. To allow direct comparison, all reactions were set up in an identical manner. Results are representative of a minimum of three independent experiments.
Figure 6
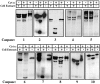
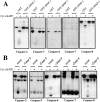
Selective binding of caspase-9 to Apaf-1. Glutathione Sepharose–immobilized GST-Apaf-11-97 fusion protein (CED-3 homology domain) was assessed for its ability to precipitate the indicated [35S]methionine-labeled caspases, as described in Materials and Methods. (A) The signal generated by 10% of the total input amount of each caspase is shown. (B) The total amount of each caspase precipitated by 6 μg of GST or GST-Apaf-11-97 fusion protein. Identical exposures of each gel are shown to enable direct comparison.
Figure 7
Depletion of caspase-9 from Jurkat extracts abolishes cytochrome c–initiated processing of all caspases. (A) Jurkat extracts were depleted of caspase-9 using glutathione Sepharose–immobilized GST-Apaf-11-97 fusion protein, or mock-depleted with GST, and were then assessed for their ability to support caspase processing, as indicated. Caspase processing was assessed in the presence of 50 μg/ml cytochrome c and 1 mM dATP. (B) Extracts were depleted of caspase-9 using protein A/G agarose– immobilized anti–caspase-9 antibody, or mock- depleted using a control antibody (anti–Rel A), as described in Materials and Methods. The same concentrations of cytochrome c/dATP were used as in A. Results are representative of three independent experiments.
Figure 8
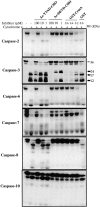
Differential inhibition of cytochrome c–initiated processing of caspases-2, -3, -6, -7, -8, and -10 by DEVD-CHO. The indicated [35S]methionine-labeled caspases were incubated in Jurkat cell extracts in the presence or absence of 10 μg/ml bovine heart cytochrome c and the indicated concentrations of peptides or recombinant proteins. Peptides and recombinant proteins were added to the extracts 30 min before the addition of cytochrome c. After incubation at 37°C for 3 h, reactions were stopped and caspase processing was subsequently assessed by SDS-PAGE/fluorography.
Figure 9
Depletion of caspase-3 from Jurkat extracts abolishes cytochrome c–initiated processing of caspases-2, -6, -8, and -10 and reveals a feedback amplification loop involving caspase-9. (A) Jurkat extracts were depleted of caspase-3 as described in Materials and Methods and were then assessed for their ability to support processing of the indicated 35S-labeled caspases. Caspase processing was assessed after incubation of extracts for 2 h at 37°C in the presence of 50 μg/ml cytochrome c and 1 mM dATP. (B) Caspase-9 processing is impaired in the absence of caspase-3. (Top) Jurkat extracts were depleted of caspase-3, or were mock depleted using a control antibody, and incubated under similar conditions to A. (Bottom) Cell extracts were prepared from Jurkat or MCF-7 cells and were compared for their ability to process caspase-9 to its p37 and p35 forms in the presence of cytochrome c/dATP. Caspase-9 was detected by Western blot. (C) Schematic representation of the sites at which caspase-9 and -3 are likely to cleave the caspase-9 pro-form to produce the p35 and p37 processed forms of caspase-9. (D) Verification of caspase-3 depletion from Jurkat extracts. Equal amounts of mock-depleted or caspase-3–depleted extract (top) or antibody-coupled protein A/G agarose beads used in the immunodepletions (bottom) were loaded, followed by probing for caspase-3.
Figure 10
Depletion of caspase-6 from Jurkat extracts abolishes cytochrome c/dATP-initiated processing of caspases-8 and -10, whereas depletion of caspase-7 has no effect. Jurkat extracts were depleted of caspase-6 (A) or caspase-7 (B) as described in Materials and Methods and were then assessed for their ability to support processing of the indicated 35S-labeled caspases. Caspase processing was assessed after incubation of extracts for 2 h at 37°C in the presence of 50 μg/ml cytochrome c and 1 mM dATP. (C) To confirm that caspases were depleted, equal amounts of either mock- depleted, caspase-6–depleted, or caspase-7–depleted Jurkat extracts were loaded, followed by probing for caspase-6 or caspase-7 by Western blot, as indicated.
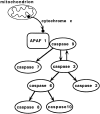
Figure 11
Schematic representation of events that take place distal to the entry of cytochrome c to the cytoplasm. The mitochondrial apoptosome is formed upon exit of cytochrome c from the mitochondrial intermembrane space, possibly triggered by either Bax or Bid. Upon activation, caspase-9 may disengage from the cytochrome c/Apaf-1 complex in order to activate downstream caspases, alternatively, downstream caspases may also become recruited to this apoptosome via suitable adaptor proteins.
Similar articles
- The adapter protein apoptotic protease-activating factor-1 (Apaf-1) is proteolytically processed during apoptosis.
Lauber K, Appel HA, Schlosser SF, Gregor M, Schulze-Osthoff K, Wesselborg S. Lauber K, et al. J Biol Chem. 2001 Aug 10;276(32):29772-81. doi: 10.1074/jbc.M101524200. Epub 2001 May 31. J Biol Chem. 2001. PMID: 11387322 - Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90.
Pandey P, Saleh A, Nakazawa A, Kumar S, Srinivasula SM, Kumar V, Weichselbaum R, Nalin C, Alnemri ES, Kufe D, Kharbanda S. Pandey P, et al. EMBO J. 2000 Aug 15;19(16):4310-22. doi: 10.1093/emboj/19.16.4310. EMBO J. 2000. PMID: 10944114 Free PMC article. - Activation of caspases triggered by cytochrome c in vitro.
Pan G, Humke EW, Dixit VM. Pan G, et al. FEBS Lett. 1998 Apr 10;426(1):151-4. doi: 10.1016/s0014-5793(98)00330-5. FEBS Lett. 1998. PMID: 9598997 - Caspase-8 in apoptosis: the beginning of "the end"?
Kruidering M, Evan GI. Kruidering M, et al. IUBMB Life. 2000 Aug;50(2):85-90. doi: 10.1080/713803693. IUBMB Life. 2000. PMID: 11185963 Review. - Apoptosis: checkpoint at the mitochondrial frontier.
Bossy-Wetzel E, Green DR. Bossy-Wetzel E, et al. Mutat Res. 1999 Jul 30;434(3):243-51. doi: 10.1016/s0921-8777(99)00032-4. Mutat Res. 1999. PMID: 10486595 Review.
Cited by
- Mitophagy in Cell Death Regulation: Insights into Mechanisms and Disease Implications.
Lin J, Chen X, Du Y, Li J, Guo T, Luo S. Lin J, et al. Biomolecules. 2024 Oct 9;14(10):1270. doi: 10.3390/biom14101270. Biomolecules. 2024. PMID: 39456203 Free PMC article. Review. - Role of layilin in regulating mitochondria-mediated apoptosis: a study on B cell lymphoma (BCL)-2 family proteins.
Arito M, Tsutiya A, Sato M, Omoteyama K, Sato T, Motonaga Y, Suematsu N, Kurokawa MS, Kato T. Arito M, et al. BMC Mol Cell Biol. 2024 Oct 25;25(1):24. doi: 10.1186/s12860-024-00521-9. BMC Mol Cell Biol. 2024. PMID: 39455917 Free PMC article. - Tocotrienol-rich fraction (TRF) protects against retinal cell apoptosis and preserves visual behavior in rats with streptozotocin-induced diabetic retinopathy.
Goh Y, Sadikan MZ, Jaiprakash H, Nasir NAA, Agarwal R, Iezhitsa I, Ismail NM. Goh Y, et al. BMC Complement Med Ther. 2024 Aug 30;24(1):322. doi: 10.1186/s12906-024-04614-y. BMC Complement Med Ther. 2024. PMID: 39215295 Free PMC article. - Hypoimmunogenic human iPSCs expressing HLA-G, PD-L1, and PD-L2 evade innate and adaptive immunity.
Tsuneyoshi N, Hosoya T, Takeno Y, Saitoh K, Murai H, Amimoto N, Tatsumi R, Watanabe S, Hasegawa Y, Kikkawa E, Goto K, Nishigaki F, Tamura K, Kimura H. Tsuneyoshi N, et al. Stem Cell Res Ther. 2024 Jul 2;15(1):193. doi: 10.1186/s13287-024-03810-4. Stem Cell Res Ther. 2024. PMID: 38956724 Free PMC article. - Intracerebroventricular BDNF infusion may reduce cerebral ischemia/reperfusion injury by promoting autophagy and suppressing apoptosis.
Yilmaz U, Tanbek K, Gul S, Koc A, Gul M, Sandal S. Yilmaz U, et al. J Cell Mol Med. 2024 Apr;28(8):e18246. doi: 10.1111/jcmm.18246. J Cell Mol Med. 2024. PMID: 38520223 Free PMC article.
References
- Ahmad M, Srinivasula SM, Wang L, Talanian RV, Litwack G, Fernandes-Alnemri T, Alnemri ES. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57:615–619. - PubMed
- Alnemri ES. Mammalian cell death proteases: a family of highly conserved aspartate specific cysteine proteases. J Cell Biochem. 1997;64:33–42. - PubMed
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. - PubMed
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1 and TNF receptor-induced cell death. Cell. 1996;85:803–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous