Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats - PubMed (original) (raw)
Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats
U Nübel et al. Appl Environ Microbiol. 1999 Feb.
Abstract
We quantified the diversity of oxygenic phototrophic microorganisms present in eight hypersaline microbial mats on the basis of three cultivation-independent approaches. Morphological diversity was studied by microscopy. The diversity of carotenoids was examined by extraction from mat samples and high-pressure liquid chromatography analysis. The diversity of 16S rRNA genes from oxygenic phototrophic microorganisms was investigated by extraction of total DNA from mat samples, amplification of 16S rRNA gene segments from cyanobacteria and plastids of eukaryotic algae by phylum-specific PCR, and sequence-dependent separation of amplification products by denaturing-gradient gel electrophoresis. A numerical approach was introduced to correct for crowding the results of chromatographic and electrophoretic analyses. Diversity estimates typically varied up to twofold among mats. The congruence of richness estimates and Shannon-Weaver indices based on numbers and proportional abundances of unique morphotypes, 16S rRNA genes, and carotenoids unveiled the underlying diversity of oxygenic phototrophic microorganisms in the eight mat communities studied.
Figures
FIG. 1
Morphotypes of oxygenic-phototrophic microorganisms from eight mat communities as observed by phase-contrast light microscopy. Morphotypes 1 to 25 are cyanobacteria, of which 1 to 17 can be assigned to the order Oscillatoriales and 18 to 25 can be assigned to the order Chroococcales (7). Morphotypes 26 to 35 are diatoms, to which the following generic assignments can be made (49): Nitzschia, 26, 30, 33; Brachysira, 27; Navicula, 28; Amphora, 31; Mastogloia, 32; Entomoneis, 34; and Gyrosigma, 35. Morphotype 36 is a green alga (Dunaliella sp.).
FIG. 2
Proportional abundances of morphotypes based on cell counts. The morphotype numbers refer to Fig. 1. The data are from analyses of communities in mats P4 and NC3. Because sets of morphotypes found in triplicate subcores from the same mat do not necessarily completely coincide, the cumulative number of morphotypes observed in a community may exceed the mean richness, S M (Table 2).
FIG. 3
Relationship of S M (A) and H_′_M (B [based on cell counts]) to the number of cells encountered. The data are from analyses of communities in mats P4 and NC3.
FIG. 4
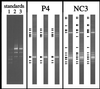
Composite figure of ethidium bromide-stained DGGE separation patterns of PCR-amplified segments of 16S rRNA genes. Mixtures of PCR products derived from five cyanobacterial strains were applied to each gel as standards (in lanes 1 to 3 [top to bottom] are Scytonema sp. strain B-77-Scy.jav., Synechococcus leopoliensis SAG 1402-1, Microcoleus chthonoplastes MPI-NDN-1, Geitlerinema sp. strain PCC 9452 [“_Microcoleus_” sp. strain 10 mfx], and Cyanothece sp. strain PCC 7418). The standard in lane 1 allows gel-to-gel comparisons. The DNA mass calibration standard in lanes 2 and 3 enables the transformation of measured band fluorescence values into amounts of DNA (in lane 2, the amounts of DNA in individual bands are (top to bottom) 528, 176, 59, 20, and 7 ng; in lane 3, half of the amount of the standard in lane 2 was applied). The gels labelled P4 and NC3 show separation patterns of PCR products derived from triplicate sampling cores of those microbial mats. The arrowheads indicate the bands included in the subsequent analyses.
FIG. 5
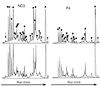
Duplicate carotenoid analysis by HPLC in two mats, P4 and NC3. Two independent analyses for each mat are shown. Each peak in the 470-nm chromatrogram was assigned to either a carotenoid (solid circles), a tetrapyrrol (chlorophylls and phaeophytin) (arrowheads), or scytonemin (arrow) on the basis of absorption spectra obtained on-line.
FIG. 6
Relationships among estimates of the richness of oxygenic-phototrophic microorganisms in eight microbial mats based on triplicate analyses of morphologies (S M), 16S rRNA genes (S R), and carotenoids (S C). The arithmetic means and standard errors from triplicate analyses are shown. The Pearson correlation coefficient, r, and its statistical significance, P, have been calculated.
FIG. 7
Relationships among Shannon-Weaver indices for communities of oxygenic-phototrophic microorganisms in eight microbial mats based on analyses of morphologies (H_′_M), 16S rRNA genes (H_′_R), and carotenoids (H_′_C). The arithmetic means and standard errors from triplicate analyses are shown. The Pearson correlation coefficient, r, and its statistical significance, P, have been calculated.
Similar articles
- Diversity of phototrophic bacteria in microbial mats from Arctic hot springs (Greenland).
Roeselers G, Norris TB, Castenholz RW, Rysgaard S, Glud RN, Kühl M, Muyzer G. Roeselers G, et al. Environ Microbiol. 2007 Jan;9(1):26-38. doi: 10.1111/j.1462-2920.2006.01103.x. Environ Microbiol. 2007. PMID: 17227409 - Matching molecular diversity and ecophysiology of benthic cyanobacteria and diatoms in communities along a salinity gradient.
Nübel U, Garcia-Pichel F, Clavero E, Muyzer G. Nübel U, et al. Environ Microbiol. 2000 Apr;2(2):217-26. doi: 10.1046/j.1462-2920.2000.00094.x. Environ Microbiol. 2000. PMID: 11220307 - Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: 'La Salada de Chiprana' (NE Spain).
Jonkers HM, Ludwig R, Wit R, Pringault O, Muyzer G, Niemann H, Finke N, Beer D. Jonkers HM, et al. FEMS Microbiol Ecol. 2003 May 1;44(2):175-89. doi: 10.1016/S0168-6496(02)00464-6. FEMS Microbiol Ecol. 2003. PMID: 19719635 - Molecular ecology of microbial mats.
Bolhuis H, Cretoiu MS, Stal LJ. Bolhuis H, et al. FEMS Microbiol Ecol. 2014 Nov;90(2):335-50. doi: 10.1111/1574-6941.12408. Epub 2014 Aug 28. FEMS Microbiol Ecol. 2014. PMID: 25109247 Review. - Bioremediation of oil by marine microbial mats.
Cohen Y. Cohen Y. Int Microbiol. 2002 Dec;5(4):189-93. doi: 10.1007/s10123-002-0089-5. Epub 2002 Nov 5. Int Microbiol. 2002. PMID: 12497184 Review.
Cited by
- Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input.
Cho JC, Kim SJ. Cho JC, et al. Appl Environ Microbiol. 2000 Mar;66(3):956-65. doi: 10.1128/AEM.66.3.956-965.2000. Appl Environ Microbiol. 2000. PMID: 10698758 Free PMC article. - Diazotrophic diversity and distribution in the tropical and subtropical Atlantic Ocean.
Langlois RJ, LaRoche J, Raab PA. Langlois RJ, et al. Appl Environ Microbiol. 2005 Dec;71(12):7910-9. doi: 10.1128/AEM.71.12.7910-7919.2005. Appl Environ Microbiol. 2005. PMID: 16332767 Free PMC article. - Determining Microeukaryotic Plankton Community around Xiamen Island, Southeast China, Using Illumina MiSeq and PCR-DGGE Techniques.
Yu L, Zhang W, Liu L, Yang J. Yu L, et al. PLoS One. 2015 May 28;10(5):e0127721. doi: 10.1371/journal.pone.0127721. eCollection 2015. PLoS One. 2015. PMID: 26020532 Free PMC article. - Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization.
Rudi K, Skulberg OM, Skulberg R, Jakobsen KS. Rudi K, et al. Appl Environ Microbiol. 2000 Sep;66(9):4004-11. doi: 10.1128/AEM.66.9.4004-4011.2000. Appl Environ Microbiol. 2000. PMID: 10966421 Free PMC article. - The Complete Genome and Physiological Analysis of the Microbialite-Dwelling Agrococcus pavilionensis sp. nov; Reveals Genetic Promiscuity and Predicted Adaptations to Environmental Stress.
White RA 3rd, Gavelis G, Soles SA, Gosselin E, Slater GF, Lim DSS, Leander B, Suttle CA. White RA 3rd, et al. Front Microbiol. 2018 Oct 15;9:2180. doi: 10.3389/fmicb.2018.02180. eCollection 2018. Front Microbiol. 2018. PMID: 30374333 Free PMC article.
References
- Anagnostidis K, Komárek J. Modern approach to the classification system of cyanophytes. 1. Introduction. Arch Hydrobiol (Suppl) 1985;71:291–302.
- Begon M, Harper J L, Townsend C R. Ecology—individuals, populations, communities. Oxford, United Kingdom: Blackwell Scientific Publications; 1990.
- Brosius M, Dull T, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources