Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in the Northwestern Mediterranean Sea - PubMed (original) (raw)
Comparative Study
Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in the Northwestern Mediterranean Sea
C J Phillips et al. Appl Environ Microbiol. 1999 Feb.
Abstract
The aim of this study was to determine if there were differences between the types of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria associated with particulate material and planktonic samples obtained from the northwestern Mediterranean Sea. A nested PCR procedure performed with ammonia oxidizer-selective primers was used to amplify 16S rRNA genes from extracted DNA. The results of partial and full-length sequence analyses of 16S rRNA genes suggested that different groups of ammonia-oxidizing bacteria were associated with the two sample types. The particle-associated sequences were predominantly related to Nitrosomonas eutropha, while the sequences obtained from the planktonic samples were related to a novel marine Nitrosospira group (cluster 1) for which there is no cultured representative yet. A number of oligonucleotide probes specific for different groups of ammonia oxidizers were used to estimate the relative abundance of sequence types in samples of clone libraries. The planktonic libraries contained lower proportions of ammonia oxidizer clones (0 to 26%) than the particulate material libraries (9 to 83%). Samples of the planktonic and particle-associated libraries showed that there were depth-related differences in the ammonia oxidizer populations, with the highest number of positive clones in the particle-associated sample occurring at a depth of 700 m. The greatest difference between planktonic and particle-associated populations occurred at a depth of 400 m, where only 4% of the clones in the planktonic library were identified as Nitrosomonas clones, while 96% of these clones were identified as clones that were related to the marine Nitrosospira species. Conversely, all ammonia oxidizer-positive clones obtained from the particle-associated library were members of the Nitrosomonas group. This is the first indication that Nitrosomonas species and Nitrosospira species may occupy at least two distinct environmental niches in marine environments. The occurrence of these groups in different niches may result from differences in physiological properties and, coupled with the different environmental conditions associated with these niches, may lead to significant differences in the nature and rates of nitrogen cycling in these environments.
Figures
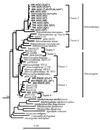
FIG. 1
Phylogenetic tree showing the positions, within the β-proteobacterial ammonia oxidizers, of partial sequences obtained from DNA extracted from planktonic samples and particulate material obtained from site 1 at depths of 100 and 400 m and from site 2 at a depth of 700 m. Clones were obtained by using primers designed to amplify sequences from the β-proteobacterial ammonia oxidizers (27). The tree was generated by using a 252-bp region of the 5′ region of the 16S rDNA and neighbor joining (34) with the Jukes-Cantor (20) correction in ARB. The cluster designations are the cluster designations described by Stephen et al. (39, 40). Asterisks indicate clones whose full-length sequences were used to construct the tree shown in Fig. 2. Environmental clones whose designations begin with Env and pH were obtained from the ammonia oxidizer database of Stephen et al. (39). Clone pH4.2A/23 is a representative of cluster 6a. The other members of cluster 6 are cluster 6b organisms (40). Scale bar = 0.1 estimated change per nucleotide position.
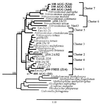
FIG. 2
Phylogenetic tree showing the positions, within the ammonia oxidizer group of the β-proteobacteria, of almost full-length sequences obtained from DNA extracted from planktonic samples and particulate material obtained from site 1 at depths of 100 and 400 m and from site 2 at a depth of 700 m. The tree was generated by using a 1,011-bp region of the 16S rDNA and neighbor joining (34) with the Jukes-Cantor (20) correction in ARB. The cluster designations are the cluster designations described by Stephen et al. (39, 40). For other details see the legend to Fig. 1.
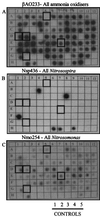
FIG. 3
Hybridization of members of the clone library obtained from aggregate samples collected at site 2 at a depth of 700 m (700 AGG clones) with ammonia oxidizer-specific probes. (A, B, and C) Autoradiographs showing the results of hybridizations with probes β-AO233, Nsp436, and Nmo254, respectively. Controls 1 through 5 were representatives of ammonia oxidizer cultures (control 1, Nitrosospira sp. strain Np 22-21 [cluster 3]; control 2, Nitrosospira sp. strain NPAV [cluster 3]; control 3, Nitrosospira multiformis [cluster 3]; control 4, Nitrosospira sp. strain C-141 [cluster 3]; control 5, Nitrosomonas europaea [cluster 7]). Partial sequences of the clones enclosed in boxes were determined to check the validity of the results.
References
- Acinas S G, Rodriguez-Valera F, Pedrós-Alió C. Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16S rDNA. FEMS Microbiol Ecol. 1997;24:27–40.
- Alldredge A L, Passow U, Logan B E. The abundance and significance of a class of large transparent organic particles in the ocean. Deep Sea Res Part I. 1993;40:1131–1140.
- Capone D G. Methane, nitrogen oxides and halomethanes. In: Rodgers J E, Whitman W B, editors. Microbial consumption of greenhouse gases. Washington, D.C: American Society for Microbiology; 1991. pp. 225–275.
- Cox D J, Bazin M J, Gull K. Distribution of bacteria in a continuous-flow nitrification column. Soil Biol Biochem. 1980;12:241–246.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases