Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway - PubMed (original) (raw)
Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway
T Herman et al. Proc Natl Acad Sci U S A. 1999.
Abstract
We have molecularly analyzed three genes, sqv-3, sqv-7, and sqv-8, that are required for wild-type vulval invagination in Caenorhabditis elegans. The predicted SQV-8 protein is similar in sequence to two mammalian beta(1,3)-glucuronyltransferases, one of which adds glucuronic acid to protein-linked galactose-beta(1, 4)-N-acetylglucosamine. SQV-3 is similar to a family of glycosyltransferases that includes vertebrate beta(1, 4)-galactosyltransferases, which create galactose-beta(1, 4)-N-acetylglucosamine linkages. One model is therefore that SQV-8 uses a SQV-3 product as a substrate. SQV-7 is similar to members of a family of nucleotide-sugar transporters. The sqv genes therefore are likely to encode components of a conserved glycosylation pathway that assembles a C. elegans carbohydrate moiety, the absence of which perturbs vulval invagination.
Figures
Figure 1
SQV-8 is similar to a glucuronyltransferase. (A) Cosmid ZK1307 and plasmid clones assayed for sqv-8 rescue activity. + indicates that three or more independently transformed lines con tained rescued sqv-8 animals; − indicates that five or more independently transformed lines contained no rescued sqv-8 animals. Underlined restriction sites are unique in the region of the cosmid that is depicted in detail (i.e., the region from _Eag_I to _Sac_I). The open arrowhead indicates the site at which a frame-shifting insert was introduced, and the vertical line in the bottom clone indicates the site from which this insert was precisely removed. (B) Alignment of the SQV-8, GlcAT-P, and GlcAT-I protein sequences as well as protein sequences predicted from a schistosomal cDNA, and six C. elegans ORFs (W07G9.A, T09E11.1, T15D6.7, C54C8.5, C47F8.4, and E03H4.12) predicted by the Genome Sequencing Consortium. Identities between SQV-8 and any other protein are boxed in black. N-terminal identities are not indicated, because they seem likely to occur randomly in this region. Three or more identical residues that are not shared with SQV-8 are boxed in gray (in some cases two different sets of three identities are in gray within a single column). The underlined amino acids were predicted by the algorithm of (54) (using spans of seven) to have positive hydropathy and therefore may be contained within transmembrane domains. Other stretches of positive hydropathy occur within the regions of amino acid similarity but are not consistently conserved and so are not indicated. The predicted change in amino acid sequence caused by each mutant sqv-8 allele is indicated directly above the amino acid affected, and the DNA sequence of mutant stop codons is noted in parentheses. Cys-317 of GlcAT-P is indicated by a black arrowhead. The region of the alignment from SQV-8 amino acid positions 156–189 is bracketed underneath (see text).
Figure 2
SQV-3 is similar to members of a glycosyltransferase family. (A) Cosmid C47F11 and plasmid clones assayed for sqv-3 rescue activity. Symbols are used as in Fig. 1_A_. (B) Alignment of the SQV-3, GalT, GlcNAcT, and partial human N_-acetylgalactosaminyltransferase (GalNAcT) protein sequences and a C. elegans protein sequence, W02B12.11, predicted by the Genome Sequencing Consortium. Identities, transmembrane domains, and predicted changes in mutant alleles are indicated as in Fig. 1_B. Open arrowheads indicate Tyr and Trp residues in GalT implicated in binding GlcNAc and/or UDP-Gal, the black arrowhead indicates a Trp residue in GalT required for catalytic activity, and the gray arrowhead indicates a GalT Tyr residue that can be replaced by Phe without affecting catalytic activity (43). The long version of GalT is shown, and the initiator methionine of the short version is circled.
Figure 3
SQV-7 is similar to LPG2, a protein required for GDP-mannose transport. (A) Cosmid C52E12 and plasmid clones assayed for sqv-7 rescue activity. + indicates that two or more independently transformed lines contained rescued sqv-7 animals. Otherwise symbols are used as in Fig. 1_A_. (B) Alignment of the SQV-7 and Leishmania LPG2 protein sequences and protein sequences predicted from two human cDNAs. Human 1 is predicted from cDNA accession no. D87449, and human 2 from clone 132056 (see text). The human 2 cDNA is likely to be incomplete at its 5′ end, which is indicated by three dots at the human 2 N terminus. Identities and predicted changes in mutant alleles are indicated as in Fig. 1_B_. (C) Kyte-Doolittle hydropathy plots of SQV-7 and Leishmania LPG2. In each case, the amino acid sequence is plotted along the horizontal axis, and its corresponding hydropathy (54) is plotted along the vertical axis. Regions above the central horizontal line are of positive hydropathy and, if sufficient length, therefore may be contained within transmembrane domains. The two plots are drawn at the same scale.
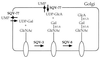
Figure 4
A model for sqv gene function. SQV-3, SQV-7, and SQV-8 may catalyze the formation of an oligosaccharide on one or more glycoproteins, glycolipids, or proteoglycans. The oval represents a glycoprotein, glycolipid, or proteoglycan, the longer straight line an oligosaccharide of unspecified structure, and the shorter straight lines sugar-sugar linkages. The biochemical activities proposed for SQV-3, SQV-7, and SQV-8 are based on a subset of those defined for the proteins related to them in amino acid sequence (see text). The terminal oligosaccharide is depicted as an unsulfated version of the HNK-1 epitope but instead may be less closely related to this epitope, may be further modified by the addition of sugars and/or sulfate groups, or may occur as a repeating rather than terminal unit (see text).
Similar articles
- sqv-3, -7, and -8, a set of genes affecting morphogenesis in Caenorhabditis elegans, encode enzymes required for glycosaminoglycan biosynthesis.
Bulik DA, Wei G, Toyoda H, Kinoshita-Toyoda A, Waldrip WR, Esko JD, Robbins PW, Selleck SB. Bulik DA, et al. Proc Natl Acad Sci U S A. 2000 Sep 26;97(20):10838-43. doi: 10.1073/pnas.97.20.10838. Proc Natl Acad Sci U S A. 2000. PMID: 11005858 Free PMC article. - sqv mutants of Caenorhabditis elegans are defective in vulval epithelial invagination.
Herman T, Hartwieg E, Horvitz HR. Herman T, et al. Proc Natl Acad Sci U S A. 1999 Feb 2;96(3):968-73. doi: 10.1073/pnas.96.3.968. Proc Natl Acad Sci U S A. 1999. PMID: 9927677 Free PMC article. - The Caenorhabditis elegans vulval morphogenesis gene sqv-4 encodes a UDP-glucose dehydrogenase that is temporally and spatially regulated.
Hwang HY, Horvitz HR. Hwang HY, et al. Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14224-9. doi: 10.1073/pnas.172522499. Epub 2002 Oct 21. Proc Natl Acad Sci U S A. 2002. PMID: 12391315 Free PMC article. - The Caenorhabditis elegans sqv genes and functions of proteoglycans in development.
Bulik DA, Robbins PW. Bulik DA, et al. Biochim Biophys Acta. 2002 Dec 19;1573(3):247-57. doi: 10.1016/s0304-4165(02)00391-4. Biochim Biophys Acta. 2002. PMID: 12417407 Review. - Mutations that perturb vulval invagination in C. elegans.
Herman T, Horvitz HR. Herman T, et al. Cold Spring Harb Symp Quant Biol. 1997;62:353-9. Cold Spring Harb Symp Quant Biol. 1997. PMID: 9598369 Review. No abstract available.
Cited by
- Glycosylation genes expressed in seam cells determine complex surface properties and bacterial adhesion to the cuticle of Caenorhabditis elegans.
Gravato-Nobre MJ, Stroud D, O'Rourke D, Darby C, Hodgkin J. Gravato-Nobre MJ, et al. Genetics. 2011 Jan;187(1):141-55. doi: 10.1534/genetics.110.122002. Epub 2010 Oct 26. Genetics. 2011. PMID: 20980242 Free PMC article. - Morphogenesis of the caenorhabditis elegans vulva.
Schindler AJ, Sherwood DR. Schindler AJ, et al. Wiley Interdiscip Rev Dev Biol. 2013 Jan-Feb;2(1):75-95. doi: 10.1002/wdev.87. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23418408 Free PMC article. Review. - Structural elucidation of zwitterionic carbohydrates derived from glycosphingolipids of the porcine parasitic nematode Ascaris suum.
Friedl CH, Lochnit G, Zähringer U, Bahr U, Geyer R. Friedl CH, et al. Biochem J. 2003 Jan 1;369(Pt 1):89-102. doi: 10.1042/BJ20021074. Biochem J. 2003. PMID: 12234251 Free PMC article. - Molecular physiology and pathology of the nucleotide sugar transporter family (SLC35).
Ishida N, Kawakita M. Ishida N, et al. Pflugers Arch. 2004 Feb;447(5):768-75. doi: 10.1007/s00424-003-1093-0. Epub 2003 May 21. Pflugers Arch. 2004. PMID: 12759756 Review.
References
- Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. - PubMed
- Kleene R, Berger E G. Biochim Biophys Acta. 1993;1154:283–325. - PubMed
- Stanley P, Ioffe E. FASEB J. 1995;9:1436–1444. - PubMed
- Stanley P. Annu Rev Genet. 1984;18:525–552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases