Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation - PubMed (original) (raw)
Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation
L Misquitta et al. Proc Natl Acad Sci U S A. 1999.
Abstract
The expression of the MyoD gene homolog, nautilus (nau), in the Drosophila embryo defines a subset of mesodermal cells known as the muscle "pioneer" or "founder" cells. These cells are thought to establish the future muscle pattern in each hemisegment. Founders appear to recruit fusion-competent mesodermal cells to establish a particular muscle fiber type. In support of this concept every somatic muscle in the embryo is associated with one or more nautilus-positive cells. However, because of the lack of known (isolated) nautilus mutations, no direct test of the founder cell hypothesis has been possible. We now have utilized toxin ablation and genetic interference by double-stranded RNA (RNA interference or RNA-i) to determine both the role of the nautilus-expressing cells and the nautilus gene, respectively, in embryonic muscle formation. In the absence of nautilus-expressing cells muscle formation is severely disrupted or absent. A similar phenotype is observed with the elimination of the nautilus gene product by genetic interference upon injection of nautilus double-stranded RNA. These results define a crucial role for nautilus in embryonic muscle formation. The application of RNA interference to a variety of known Drosophila mutations as controls gave phenotypes essentially indistinguishable from the original mutation. RNA-i provides a powerful approach for the targeted disruption of a given genetic function in Drosophila.
Figures
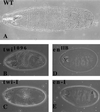
Figure 1
Muscle formation and gene expression patterns in the Drosophila embryo as modulated by specific cell ablation, antisense expression, and RNA interference by the injection of dsRNA. Ablation of the nautilus_-positive muscle founder cells by ricin toxin disrupts muscle formation. (A) Ricin not induced. (B and C) Ricin induced in nautilus_-positive cells. Antisense expression of nautilus RNA disrupts muscle formation in three different UAS antisense (AS) nautilus lines (D_– F). Injection of_nautilus dsRNA blocks muscle formation (G and H) and does not depend on the bHLH domain for the disruption [dsRNA for the C terminus (I), dsRNA for the bHLH domain (J), and dsRNA for the amino terminus of_nautilus (K)]. Injection of β-galactosidase dsRNA does not disrupt the muscle pattern but eliminates normal lacZ expression (shown in M) without affecting muscle pattern (similar to A), whereas injection of nautilus dsRNA into a_nautilus lacZ line 14.1 disrupts the lacZ muscle pattern (compare M and N) and reduces lacZ expression. Injection of dsRNA for DMEF2 [uninjected (L) and injected (O)], S59 [uninjected (P) and injected (Q)],daughterless [uninjected (R) and injected (S); C, CNS; P, PNS], and white [uninjected (T) (w+) and injected (U)] results in the disruption of gene function for these genes.A_–_L and_O_–Q were stained with antimyosin;M and N were stained with 5-bromo-4-chloro-3-indolyl β-
d
-galactoside;R and S were stained with monoclonal 22C10 and horseradish peroxide for CNS and PNS. AS in the upper left-hand corner marks the nautilus antisense lines, -U indicates the uninjected phenotype for the designated gene, and -I indicates dsRNA injection for the indicated gene.
Figure 2
Cuticular patterns in early larvae induced by injection of twist and engrailed dsRNAs. (A) Wild-type cuticular pattern for early larva. (B) Twist phenotype of the known_twist_ mutation twi_1096. (C) Embryos injected with twist dsRNA show the same phenotype as in B. (D) Fused cuticular band phenotype seen for enIIB86 null mutants. (E) Embryos injected with the engrailed dsRNA show the same_en null phenotype as in D.
Similar articles
- Stereotypic founder cell patterning and embryonic muscle formation in Drosophila require nautilus (MyoD) gene function.
Wei Q, Rong Y, Paterson BM. Wei Q, et al. Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5461-6. doi: 10.1073/pnas.0608739104. Epub 2007 Mar 21. Proc Natl Acad Sci U S A. 2007. PMID: 17376873 Free PMC article. - Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability.
Balagopalan L, Keller CA, Abmayr SM. Balagopalan L, et al. Dev Biol. 2001 Mar 15;231(2):374-82. doi: 10.1006/dbio.2001.0162. Dev Biol. 2001. PMID: 11237466 - Misexpression of nautilus induces myogenesis in cardioblasts and alters the pattern of somatic muscle fibers.
Keller CA, Erickson MS, Abmayr SM. Keller CA, et al. Dev Biol. 1997 Jan 15;181(2):197-212. doi: 10.1006/dbio.1996.8434. Dev Biol. 1997. PMID: 9013930 - Drosophila myogenesis and insights into the role of nautilus.
Abmayr SM, Keller CA. Abmayr SM, et al. Curr Top Dev Biol. 1998;38:35-80. doi: 10.1016/s0070-2153(08)60244-6. Curr Top Dev Biol. 1998. PMID: 9399076 Review. - Muscle differentiation: how two cells become one.
Taylor MV. Taylor MV. Curr Biol. 2002 Mar 19;12(6):R224-8. doi: 10.1016/s0960-9822(02)00757-1. Curr Biol. 2002. PMID: 11909553 Review.
Cited by
- Anion-sensitive fluorophore identifies the Drosophila swell-activated chloride channel in a genome-wide RNA interference screen.
Stotz SC, Clapham DE. Stotz SC, et al. PLoS One. 2012;7(10):e46865. doi: 10.1371/journal.pone.0046865. Epub 2012 Oct 4. PLoS One. 2012. PMID: 23056495 Free PMC article. - Targeted mRNA degradation by double-stranded RNA in vitro.
Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Tuschl T, et al. Genes Dev. 1999 Dec 15;13(24):3191-7. doi: 10.1101/gad.13.24.3191. Genes Dev. 1999. PMID: 10617568 Free PMC article. - Differential gene silencing induced by short interfering RNA in cultured pine cells associates with the cell cycle phase.
Tang W, Newton RJ, Weidner DA. Tang W, et al. Planta. 2006 Jun;224(1):53-60. doi: 10.1007/s00425-005-0190-z. Epub 2005 Dec 9. Planta. 2006. PMID: 16341704 - Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways.
Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Clemens JC, et al. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6499-503. doi: 10.1073/pnas.110149597. Proc Natl Acad Sci U S A. 2000. PMID: 10823906 Free PMC article. - In Vivo RNAi-Based Screens: Studies in Model Organisms.
Yamamoto-Hino M, Goto S. Yamamoto-Hino M, et al. Genes (Basel). 2013 Nov 25;4(4):646-65. doi: 10.3390/genes4040646. Genes (Basel). 2013. PMID: 24705267 Free PMC article.
References
- Rushton E, Drysdale R, Abmayr S M, Michelson A M, Bate M. Development. 1995;121:1979–1988. - PubMed
- Bate M. Development. 1990;110:791–804. - PubMed
- Ho R K, Ball E E, Goodman C S. Nature (London) 1983;301:66–69. - PubMed
- Keller C A, Grill M A, Abmayr S M. Dev Biol. 1998;202:153–156. - PubMed
- Abmayr S M, Erickson M S, Bour B A. Trends Genet. 1995;11:153–159. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases