ROLE OF BACTERIA IN BIO SORPTION OF HEAVY METALS (original) (raw)

RESEARCH ARTICLE
ROLE OF BACTERIA IN BIO SORPTION OF HEAVY METALS.
Anu Bala and Namita Joshi.
Department of Environmental Science, Kanya Gurukul Campus, Haridwar, India.
Manuscript Info
Manuscript History
Received: 12 August 2016
Final Accepted: 22 September 2016
Published: October 2016
Key words:-
Heavy metals, Biosorption, Bacterial strains,
Abstract
Presented study was focussed on isolation and screening the heavy metal resistant microbes and to evaluate the biosorption potential of heavy metals from contaminated soil of Bhagwanpur industrial area. Soil samples for the study were randomly collected from Bhagwanpur industrial area. The soil sample was analysed for different physicochemical properties such as pH , moisture content, temperature, water holding capacity, carbon, organic matter, total nitrogen and available phosphorous. Heavy metal resistant bacteria were isolated and screened for their biosorption potential. The minimum inhibitory concentration (MIC) of Pb,Cr,Ni\mathrm{Pb}, \mathrm{Cr}, \mathrm{Ni} and Zn was determined by agar diffusion method. The selected bacterial isolates were identified by gram staining and biochemical parameters. Isolates were further characterized molecularly with 16 S rRNA gene sequence analysis. Furthermore, biosorption capacity i.e. amount of metal ion (mg) bioabsorbed/g of dried biomass was calculated for identified strains. Carbon and organic matter content was found to be 0.23±0.1%0.23 \pm 0.1 \% and 0.33±0.08%0.33 \pm 0.08 \% respectively, while nitrogen in the sample was 0.34±0.34 \pm 0.07%0.07 \%.
Among ten isolates only four bacteria showed resistance against metals ( Ni,Cr,Pb\mathrm{Ni}, \mathrm{Cr}, \mathrm{Pb} and Zn ). MIC range of isolates against various metal concentrations was in the range of 25 PPM to 400 PPM. Molecular characterization of the isolates revealed 99%99 \% similarities of S44, S3B, S12 and S13 with Staphylococcus gallinarum, Acinetobacter pitti and Pantoea agglomerans and Enterobactor spp. respectively. Biosorption experiments indicated that, Acinetobactor spp. (S3B) and Enterobactor spp. (S13) could bioadsorb metals in the order Zn>Pb>Ni>Cr\mathrm{Zn}>\mathrm{Pb}>\mathrm{Ni}>\mathrm{Cr}; Staphylococcus spp. (S44) showed in Ni>Zn>Cr>Pb\mathrm{Ni}>\mathrm{Zn}>\mathrm{Cr}>\mathrm{Pb}; Pantoea spp. (S12) in order Zn>Ni>Pb>Cr\mathrm{Zn}>\mathrm{Ni}>\mathrm{Pb}>\mathrm{Cr}. Thus, Biosorption was influenced by the initial metal concentration. Among the isolated bacterial strains, biosorption capacity was found in order as Acinetobacter calcoaceticus >> Enterobacter spp. >> Staphylococcus gallinarum >> Pantoea agglomerans.
In this study Cr,Zn,Pb\mathrm{Cr}, \mathrm{Zn}, \mathrm{Pb} and Ni resistant bacteria were isolated from heavy metal contaminated soil. Tolerance data with extremely high range of heavy metal concentrations, revelaed that heavy metal resistant bacterial isolates can tolerate metal toxicity up to 400 . Thus paving a new way for safe, reliable and cost-effective treatment of heavy metal contaminated soil around the world.
Introduction:-
Heavy metal contamination of soils constitutes an ill-defined group of inorganic chemical hazards such as, Lead (Pb)(\mathrm{Pb}), Chromium (Cr), Arsenic (As), Zinc (Zn), Cadmium (Cd), Copper (Cu), Mercury (Hg) and Nickel (Ni) 1{ }^{1}. Heavy metal contamination of soil has become a serious environmental threat around the world due to industrial activities, soil waste disposal, fertilizers and sludge application, irrigation, wastewater and automobile exhausts 2−5{ }^{2-5}. In contaminated soil, the physical and chemical form of contaminants strongly influences the selection of appropriate treatment approach. Conventional techniques used for removing dissolved heavy metals include chemical precipitation, carbon adsorption, electrolytic recovery, ion-exchange, chelation and solvent extraction or liquid membrane separation 6{ }^{6}. Disadvantages associated with above procedures like high cost, incomplete removal, low selectivity, high energy consumption end generation of toxic slurries limit its application at large scale. Microbes offer an alternative to traditional adsorbents, as they are cheap, readily available and are ecofriendly. Microorganisms are the natural decomposers and can be used in heavy metal removal process. Bioremediation, which is removal of heavy metals by microorganisms is readily used as a strategy to remove pollutants 7{ }^{7}.
Biosorption can be defined as the selective sequestering of metal soluble species, resulting in the mobilization of metals by microbes. Metal sequestering takes place via complex formation, chelation, co-ordination, ion-exchange, precipitation and reduction 8{ }^{8}. Bioremediation uses mainly microorganisms such as, yeasts, fungi or bacteria to clean up contaminated soil and water. Heavy metal resistant microbes present in the contaminated sites use the contaminants ( Pb,Cr\mathrm{Pb}, \mathrm{Cr}, etc) as a source of nutrient and energy 9−11{ }^{9-11}. Biosorption of heavy metals by microbes can follow either metabolism independent or dependent pathways. Independent metabolism can be physical or chemical sorption onto the microbial cell walls whereas, dependent metabolism comprises transport, internal compartmentalization and extracellular precipitation by metabolism. Interestingly, biosorption can also be carried out by metabolically active or inactive cells 12{ }^{12}. Different species of Aspergillus, Pseudomona, Saprophytic, Bacillus, Phanerochaete, etc. have been reported as efficient chromium and nickel reducers. The response of microorganisms towards toxic heavy metals is very important for reclamation of polluted sites 13{ }^{13}. Our study was focussed on isolating and screening the heavy metal resistant microbes and to evaluate the biosorption potential of heavy metals from contaminated soil of Bhagwanpur industrial area.
Materials and Methods:-
Sampling:-
Soil samples were randomly collected from the industrial area of Bhagwanpur and composite sample was formed by mixing the collected samples, Sample was stored in polythene bags 4∘C4^{\circ} \mathrm{C} for further analysis.
Physico-chemical and heavy metals analysis of soil sample:-
The soil sample was analysed for different physico-chemical properties such as pH , moisture content, temperature, water holding capacity, carbon, organic matter, total nitrogen and available phosphorous 14{ }^{14}. Samples was digested by taking 1 g of sample in 250 mL glass beaker with 8 mL of aqua regia on a sand bath for 2 h . After evaporation to near dryness the samples were dissolved with 10 mL nitric acid and diluted to 50 mL using distilled water. Total metal concentration ( Cr,Ni,Co,Cu,Fe,Cd,Pb\mathrm{Cr}, \mathrm{Ni}, \mathrm{Co}, \mathrm{Cu}, \mathrm{Fe}, \mathrm{Cd}, \mathrm{Pb}, and Zn ) of digested soil samples was analysed by using AAS.
Isolation and screening of heavy metal resistant bacterial strains:-
Soil samples were serially diluted upto 10−710^{-7} and were spread on nutrient agar plates. Plates were incubated at 37∘C37^{\circ} \mathrm{C} for 24 h1524 \mathrm{~h}^{15}. . After 24 h of incubation, colony forming unit of each plate was calculated and according to morphology, bacterial isolates were purified further by streaking on nutrient agar plate. Bacterial isolates were maintained on agar slants. Isolates were screened on nutrient agar enriched with 25 ppm of different heavy metals and incubated at 37∘C37^{\circ} \mathrm{C} for 24 h . After incubation plates were observed and four bacterial isolates were used for biosorption potential.
Minimum inhibitory concentration:-
The minimum inhibitory concentration (MIC) of Pb,Cr,Ni\mathrm{Pb}, \mathrm{Cr}, \mathrm{Ni} and Zn at which no growth occurred was determined by agar diffusion method 16{ }^{16}. Nutrient agar containing bacterial strain was poured in petri-plates and allowed to solidify. After solidification five partitions were formed with bore well depth of 2 mm . Metal solution (10μl)(10 \mu \mathrm{l}) of Pb,Cr,Ni\mathrm{Pb}, \mathrm{Cr}, \mathrm{Ni}
and Zn having concentration 25,50,100,20025,50,100,200 and 400 ppm were poured in wells separately. These petri-plates were incubated at 37∘C37^{\circ} \mathrm{C} for 24 h , and after incubation plates were observed.
Identification of bacterial isolates:-
The selected bacterial isolates were identified by Gram staining and some biochemical tests such as Indole test, Methyl red (MR) and Voges-Proskauer (VP) test, Starch hydrolysis test, Urease production test, Citrate utilization test, Catalase activity and Oxidase test.
The molecular characterization of selected isolates was done by 16 S sequence analysis. The bacterial genomic DNA was extracted using standard protocol 17{ }^{17}. The strains were amplified by PCR using two bacterial 16 S primers, 5’AGAGTTTGATCMTGGCTCA-3’ (27F) for forward sequencing and 5’-CGGTTACCTTGTTACGACTT-3’ (1492R) for reverse sequencing. 16S rRNA sequences were deposited to GenBank database to get the accession number and most similar sequence alignment using www.ncbi.nlm.nih.gov/BLAST 18{ }^{18} was identified. The nucleotide sequences were aligned with MUSCLE and phylogenetic tree was constructed with the help of MEGA 5.2 software.
Biosorbent Preparation:-
All the strains were inoculated separately into 100 mL nutrient broth in 500 mL conical flasks and incubated on a shaker at 150 rpm for 24 h at 28∘C28^{\circ} \mathrm{C}. The cells were grown to late exponential phase, harvested by centrifugation (REMI, India) at 10,000rpm10,000 \mathrm{rpm} for 30 min at 4∘C4^{\circ} \mathrm{C} and washed three times with deionized water. Cell suspensions for assay of biosorption potential of live bacteria were prepared by resuspending the cell pellet in deionized water. Biomass concentration in cell suspensions were determined by drying an aliquot in a preweighed aluminum foil container to a constant weight at 80∘C1980^{\circ} \mathrm{C}^{19}.
Biosorption experiment:-
Dried and powdered living biomass ( 10.0mg±0.110.0 \mathrm{mg} \pm 0.1 ) of four isolates was inoculated separately into 100 mL of metal solution containing 25 to 400 ppm of Ni,Cr,Zn\mathrm{Ni}, \mathrm{Cr}, \mathrm{Zn} and Pb . Metal-free and biosorbent-free solutions were prepared as controls. The flasks (250 mL)(250 \mathrm{~mL}) were kept on rotatory shaker for 24 h at 30∘C30^{\circ} \mathrm{C} and 150 rpm . After 24 h , cells were harvested from the medium and content of supernatant were analyzed after proper digestion and dilution by AAS. The optimum pH and temperature were maintained for the growth of microorganisms in batch culture 20{ }^{20}.
Biosorption capacity i.e. amount of metal ion (mg) bioabsorbed/g of dried biomass was calculated using the following equation:
Q=((Ci−Ci)V)/m\mathrm{Q}=\left(\left(\mathrm{C}_{\mathrm{i}}-\mathrm{C}_{\mathrm{i}}\right) \mathrm{V}\right) / \mathrm{m}
Whereas Q=mg\mathrm{Q}=\mathrm{mg} of metal ions uptake per gram biomass ( mg/g\mathrm{mg} / \mathrm{g} ), Ci\mathrm{C}_{\mathrm{i}} is initial concentration of metallic ions ( mg/L\mathrm{mg} / \mathrm{L} ); m is dried mass of biosorbent in the reaction mixture ( g ) and V is volume of reaction mixture (mL).
Results and Discussion:-
Physico-chemical analysis of soil sample:-
The physico-chemical parameters such as, pH , temperature, moisture content, water holding capacity, carbon, organic matter and nitrogen are listed in Table 1. pH of soil quality is an important index of acidity or alkalinity. The pH of soil sample analysed was found to be 6.3±0.46.3 \pm 0.4 at temperature 35.58±2.5∘C35.58 \pm 2.5^{\circ} \mathrm{C}. Water holding capacity is the total amount of water that a given type of soil can hold and is an important indicator for crop production. Average water holding capacity and moisture content was found to be 35.58±2.5%35.58 \pm 2.5 \% and 1.23±0.7%1.23 \pm 0.7 \% respectively. Soil carbon content improves the physical properties of soil whereas, organic matter is important for plant growth as it provides nutrients and trace elements. Carbon and organic matter content was found to be 0.23±0.1%0.23 \pm 0.1 \% and 0.33±0.33 \pm 0.08%0.08 \% respectively, while nitrogen in the sample was 0.34±0.07%0.34 \pm 0.07 \%. This indicates that due to industrialization there is a decrease in nitrogen content availability for crop or plant growth.
Heavy metal analysis of soil samples:-
The total metal analysis was done by ICP-MS and concentration of Zinc ( Zn ), Nickel ( Ni ), Chromium (Cr), Cobalt (Co)(\mathrm{Co}), Lead (Pb)(\mathrm{Pb}), Cadmium (Cd)(\mathrm{Cd}), Iron (Fe)(\mathrm{Fe}) and Copper (Cu)(\mathrm{Cu}) were determined in soil sample (Table 2). High contamination of Zn in the sample was found due to higher input of Zn in the industrial products of Bhagwanpur industrial area. Contamination of soil with zinc is associated with the haematological disorders. Concentration of Pb,Ni\mathrm{Pb}, \mathrm{Ni} and Cr were also high in soil sample. Higher doses of chromium is known to cause liver and kidney damage, while chromate dust is reported to be carcinogenic. Nickel is highly corrosion-resistant metal and commonly used
for pure nickel plating. According to Indian standards, desirable limit for Cr,Pb,Zn\mathrm{Cr}, \mathrm{Pb}, \mathrm{Zn} and Ni is 0.05mg/g,0.05mg/g0.05 \mathrm{mg} / \mathrm{g}, 0.05 \mathrm{mg} / \mathrm{g}, 5.0mg/g5.0 \mathrm{mg} / \mathrm{g} and 0.01mg/g0.01 \mathrm{mg} / \mathrm{g} respectively. These results indicate that soil is contaminated with many heavy metals, and urgent action is required for its remediation.
Isolation and screening of resistant bacterial strains:-
After serial dilution of soil sample, bacterial isolates were selected based on colony morphology and colour. These isolates were further purified by streaking separately on nutrient agar plates. Isolated bacteria were further screened for resistance against heavy metals. As soil was highly contaminated with Ni,Cr,Pb\mathrm{Ni}, \mathrm{Cr}, \mathrm{Pb} and Zn , resistivity of four isolates against 25mg/g25 \mathrm{mg} / \mathrm{g} concentration of these four metals was tested. Among ten isolates only four bacteria showed resistance against metals ( Ni,Cr,Pb\mathrm{Ni}, \mathrm{Cr}, \mathrm{Pb} and Zn ). These isolates were named as S12, S13, S3B and S44.
Minimum Inhibitory concentration:-
The minimum inhibitory concentration (MIC) of bacterial isolates has been depicted in Table 3. MIC range of isolates against various metal concentrations was in the range of 25mg/g25 \mathrm{mg} / \mathrm{g} to 400mg/g400 \mathrm{mg} / \mathrm{g}. MIC was found species specific and metal dependent.
Identification:-
The selected bacterial isolates listed in Table 4 were positive to several biochemical tests. Based on these biochemical tests and Bergey’s manual of bacteriology 21{ }^{21}, isolates were identified as Staphylococcus spp (S44), Acinetobacter spp. (S3B), and Enterobacter spp. (S13) and Pantoea spp.(S12).
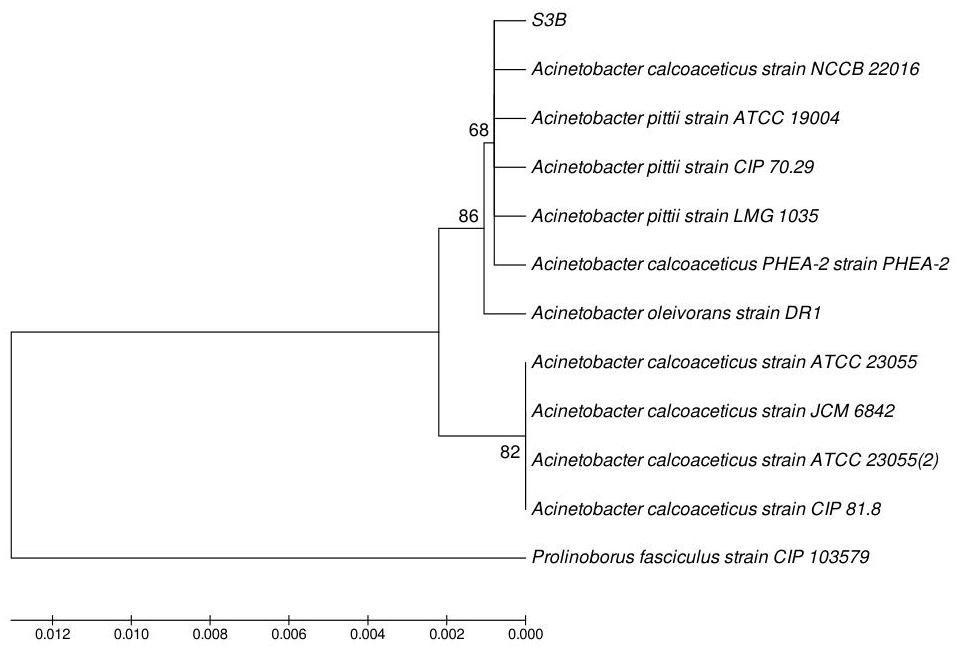
The molecular characterization of selected bacterial isolates was done by 16 S rRNA sequence analysis, which revealed the bacterial strains. After sequencing 99%99 \% similarities were found between S44, S3B and S12 with Staphylococcus gallinarum, Acinetobacter calcoaceticus and Pantoea agglomerans respectively by BLAST (www.ncbi.nlm.nih.gov/BLAST). One of the isolates was identified as new species of Enterobacter genera (S13) as it did not match with any sequence in BLAST. Phylogenetic tree was prepared by using neighbour joining tool, which showed the relation between the isolates and their respective neighbour type strains.(Figure 1, 2, 3 and 4).
Biosorption Experiment:-
On the basis of minimum inhibitory concentration to various metals, four isolates were evaluated for metal biosorption potential. Biosorption of heavy metals is the passive accumulation by micro-organisms like bacteria, algae and fungi. Dried biomass ( 0.1 g ) of four isolated species (S12, S13, S44 and S3B) was used for biosorption of Ni,Cr,Pb\mathrm{Ni}, \mathrm{Cr}, \mathrm{Pb} and Zn at five initial concentrations of 25,50,100,20025,50,100,200 and 400mg/g400 \mathrm{mg} / \mathrm{g}. The pH of the working solution was kept constant as the biosorption capacity of cell is sensitive to pH22\mathrm{pH}^{22}. The biosorption capacities of metal by four isolates are described in Table 5. Biosorption of Pb and Zn was maximum at all initial concentrations both in Acinetobacter spp. (S3B) and Enterobacter spp. (S13). Biosorption capacity increases with the metal concentration. Biosorption of Zn and Cr was maximum at 100mg/g100 \mathrm{mg} / \mathrm{g} initial concentration for Staphylococcus spp. (S44) and Enterobacter spp. (S13). Minimum biosorption capacity was showed by Pantoea agglomerans (S12) at all initial concentration of four metals individually. Biosorption of Zn was recorded maximum at 400 ppm initial metal concentration. Acinetobacter spp. (S3B) and Enterobacter spp. (S13) is 229.0±2.0229.0 \pm 2.0 and 216.8±0.8216.8 \pm 0.8 respectively. Thus, Acinetobactor spp. (S3B) and Enterobactor spp. (S13) could bioadsorb metals in the order Zn>Pb>Ni>Cr\mathrm{Zn}>\mathrm{Pb}>\mathrm{Ni}>\mathrm{Cr}; Staphylococcus spp. (S44) showed in Ni>Zn>Cr>Pb\mathrm{Ni}>\mathrm{Zn}>\mathrm{Cr}>\mathrm{Pb}; Pantoea agglomerans. (S12) in order Zn>Ni>Pb>Cr\mathrm{Zn}>\mathrm{Ni}>\mathrm{Pb}>\mathrm{Cr}. Thus, Biosorption was influenced by the initial metal concentration. Among the isolated bacterial strains, biosorption capacity was found in order as Acinetobacter calcoaceticus> Enterobacter spp.> Staphylococcus gallinarum> Pantoea agglomerans. The data on biosorption of various metal varied due to difference in the biomass, biosorption condition include pH and method employed 23−24{ }^{23-24}.
Conclusion:-
In this study Cr,Zn,Pb\mathrm{Cr}, \mathrm{Zn}, \mathrm{Pb} and Ni resistant bacteria were isolated from heavy metal contaminated soil. Tolerance data with extremely high range of heavy metal concentrations, revelaed that heavy metal resistant bacterial isolates can tolerate metal toxicity up to 400 , thus paving a new way for safe, reliable and cost-effective treatment of heavy metal contaminated soil around the world.
Table 1:- Physico-chemical properties of soil sample.
| S.No. | Parameter | Test value |
|---|---|---|
| 1. | pH | 6.3±0.266.3 \pm 0.26 |
| 2. | Temperature | 30∘C±0.530^{\circ} \mathrm{C} \pm 0.5 |
| 3. | Moisture content(%) | 1.23±0.71.23 \pm 0.7 |
| 4. | Water holding capacity (%) | 35.58±2.535.58 \pm 2.5 |
| 5. | Organic carbon (%) | 0.23±0.10.23 \pm 0.1 |
| 6. | Organic matter (%) | 0.33±0.080.33 \pm 0.08 |
| 7. | Nitrogen (%) | 0.34±0.070.34 \pm 0.07 |
Table 2:- Heavy meal concentration of soil sample.
| S.No. | Metals | Concentration (ppm) |
|---|---|---|
| 1. | Chromium (Cr)(\mathrm{Cr}) | 11.29±3.511.29 \pm 3.5 |
| 2. | Nickel (Ni)(\mathrm{Ni}) | 6.27±1.16.27 \pm 1.1 |
| 3. | Copper (Cu)(\mathrm{Cu}) | 3.21±0.63.21 \pm 0.6 |
| 4. | Cobalt (Co)(\mathrm{Co}) | 1.34±0.11.34 \pm 0.1 |
| 5. | Zinc (Zn)(\mathrm{Zn}) | 30.11±0.730.11 \pm 0.7 |
| 6. | Lead (Pb)(\mathrm{Pb}) | 3.50±0.53.50 \pm 0.5 |
| 7. | Cadmium (Cd)(\mathrm{Cd}) | 0.027±0.0040.027 \pm 0.004 |
| 8. | Iron (Fe)(\mathrm{Fe}) | 0.58±0.10.58 \pm 0.1 |
Table 3:- Minimum inhibition concentration of four isolates (‘+++’ is 75%75 \% inhibition, ‘++’ is 50%50 \% inhibition, ’ + ’ is 25%25 \% inhibition, ‘-’ is no inhibition).
| Metals | Initial concentration | S3B | S44 | S12 | S13 |
|---|---|---|---|---|---|
| 25 | + | + | ++ | + | |
| Pb | 50 | + | ++ | + | + |
| 100 | + | + | + | - | |
| 200 | ++ | + | ++ | - | |
| 500 | + | - | + | - | |
| 25 | +++ | +++ | ++ | ++ | |
| 50 | +++ | ++ | +++ | + | |
| Zn | 100 | +++ | + | +++ | + |
| 200 | +++ | + | +++ | + | |
| 500 | +++ | + | +++ | - | |
| 25 | ++ | ++ | + | + | |
| 50 | ++ | + | + | + | |
| Cr | 100 | + | + | + | + |
| 200 | + | +++ | + | - | |
| 500 | - | +++ | + | - | |
| Ni | 25 | ++ | +++ | + | +++ |
| 50 | + | +++ | + | + | |
| 100 | + | ++ | - | + | |
| 200 | + | ++ | - | - | |
| 500 | - | + | - | - |
Table 4:- Identification of Isolates.
| Characteristics | S3B | S44 | S12 | S13 |
|---|---|---|---|---|
| Gram staining | - | + | - | - |
| Methyl red | - | - | + | + |
| Voges proskeur | - | - | + | + |
| Urease production | - | - | + | + |
| Catalase activity | + | + | + | + |
| Oxidase test | - | - | - | - |
| Nitrate reduction | - | + | + | + |
|---|---|---|---|---|
| Alkaline phosphatase | - | + | - | - |
| Phenylalanine deaminase | + | - | - | - |
| Acid from glucose | + | - | + | + |
Table 5:- Biosorption of heavy metals by bacterial isolates [values are mean ±\pm SE of 3 replicates]
| Metals | Initial concentration | S3B | S44 | S13 | S12 |
|---|---|---|---|---|---|
| Pb | 25 | 19.06±0.6519.06 \pm 0.65 | 18.2±0.5118.2 \pm 0.51 | 20.6±0.520.6 \pm 0.5 | 12.8±0.0312.8 \pm 0.03 |
| 50 | 31.7±0.7631.7 \pm 0.76 | 30.3±0.3830.3 \pm 0.38 | 29.5±1.829.5 \pm 1.8 | 24.7±0.1524.7 \pm 0.15 | |
| 100 | 68.5±1.0168.5 \pm 1.01 | 67.2±1.4667.2 \pm 1.46 | 59.2±3.059.2 \pm 3.0 | - | |
| 200 | 96.29±3.0196.29 \pm 3.01 | 77.3±1.3577.3 \pm 1.35 | 87.6±0.0887.6 \pm 0.08 | - | |
| 500 | 123.6±3.12123.6 \pm 3.12 | - | 105.4±2.7105.4 \pm 2.7 | - | |
| Zn | 25 | 13.4±0.7913.4 \pm 0.79 | 19.12±0.3319.12 \pm 0.33 | 14±0.1914 \pm 0.19 | 13.6±0.113.6 \pm 0.1 |
| 50 | 42.2±1.1542.2 \pm 1.15 | 39.2±1.139.2 \pm 1.1 | 43.6±1.243.6 \pm 1.2 | 32.46±0.0632.46 \pm 0.06 | |
| 100 | 91.6±0.0391.6 \pm 0.03 | 73.1±0.873.1 \pm 0.8 | 92.01±0.392.01 \pm 0.3 | 64.9±0.2664.9 \pm 0.26 | |
| 200 | 107.39±1.27107.39 \pm 1.27 | 101.6±0.79101.6 \pm 0.79 | 115.4±1.8115.4 \pm 1.8 | 99.7±1.0199.7 \pm 1.01 | |
| 500 | 229.07±2.0229.07 \pm 2.0 | 208.9±0.6208.9 \pm 0.6 | 216.86±0.8216.86 \pm 0.8 | - | |
| Cr | |||||
| 25 | 15.23±0.3815.23 \pm 0.38 | 16.9±0.516.9 \pm 0.5 | 9±0.79 \pm 0.7 | - | |
| 50 | 27.8±2.0127.8 \pm 2.01 | 32.2±1.332.2 \pm 1.3 | 19.4±0.3819.4 \pm 0.38 | 26.7±0.1826.7 \pm 0.18 | |
| 100 | 61.8±0.7761.8 \pm 0.77 | 72.9±0.572.9 \pm 0.5 | 67.6±0.367.6 \pm 0.3 | 3.6±0.113.6 \pm 0.11 | |
| 200 | - | 109.9±0.51109.9 \pm 0.51 | 99.03±0.399.03 \pm 0.3 | - | |
| 500 | - | 258.8±0.67258.8 \pm 0.67 | 157.03±0.6157.03 \pm 0.6 | - | |
| Ni | 25 | 16.5±1.2316.5 \pm 1.23 | 22.8±0.6422.8 \pm 0.64 | 14.81±0.0714.81 \pm 0.07 | 15.2±0.0815.2 \pm 0.08 |
| 50 | 34.4±0.2534.4 \pm 0.25 | 40.6±0.5940.6 \pm 0.59 | 23.9±0.5223.9 \pm 0.52 | 30.7±0.1530.7 \pm 0.15 | |
| 100 | 61.6±0.361.6 \pm 0.3 | 79.1±0.2679.1 \pm 0.26 | - | 43.7±0.0643.7 \pm 0.06 | |
| 200 | 89.5±0.2989.5 \pm 0.29 | 119.3±0.49119.3 \pm 0.49 | - | - | |
| 500 | - | 236±2.66236 \pm 2.66 | - | - |
Figure 1:- S44 - Staphylococcus gallinarum.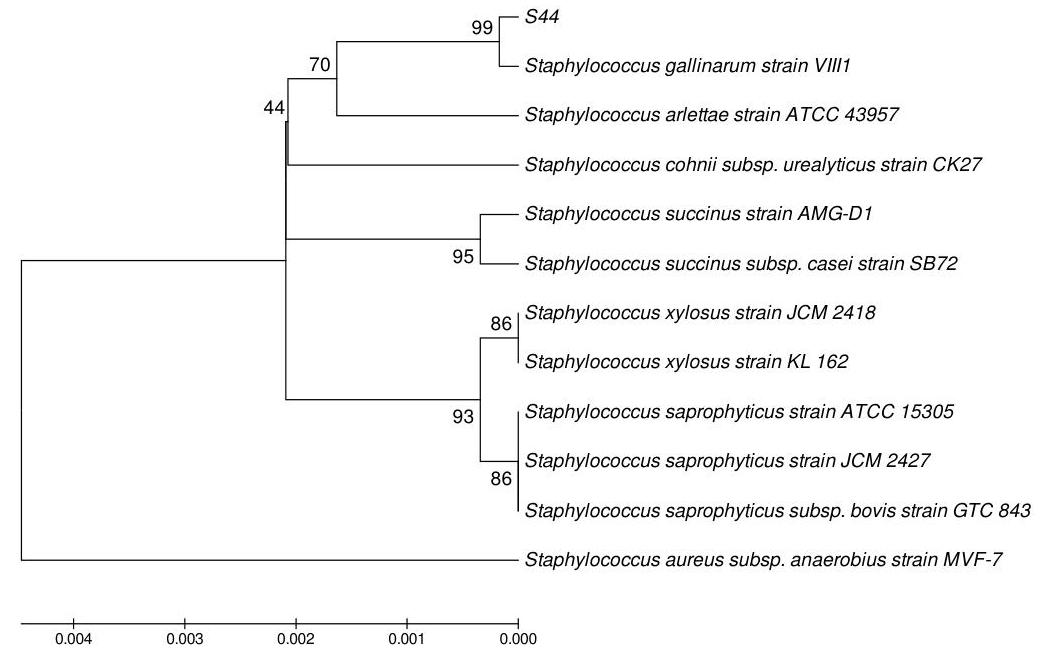
Figure 2:- S12- Pantoea agglomerans.
Inti. I. Adv. Res. 4(10), 416-424
Figure 3: S3B-Acinetobacter pitti.
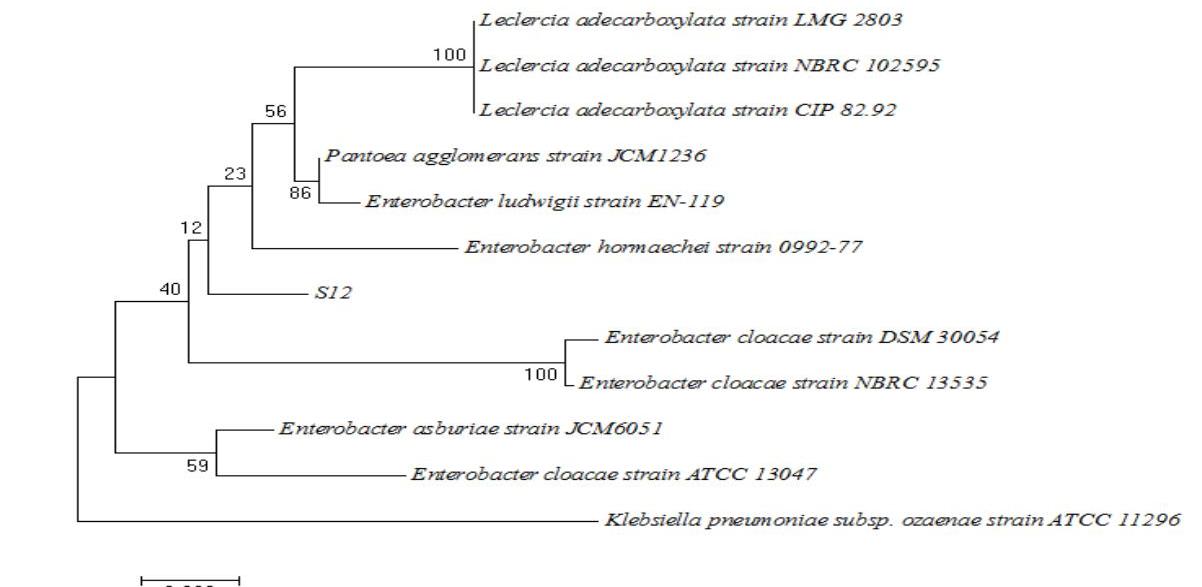
Figure 4: S13-Enterobacter spp.
References:-
GWRTAC, “Remediation of metals - contaminated soils and ground water”.
Karaca A, Naseby D, Lynch J (2002). Effects of cadmium contamination with sewage sludge and phosphate fertilizers amendments on soil enzyme activities. Microbial structure and available cadmium. Bio. Fertilizer soil 35:435-440.
Karaca (2004). Effects of organic wastes on the extractability of cadmium, copper, nickel and zinc in soil. Geoderma 122:297-305.
Khan S, Cao Q, Hesham AEL, Xia Y, He J (2007). Soil enzymatic activities and microbial community structure with difference application rates of Cd and Pb . Journal of environment Science. 19:834-840.
Yang Z, Liu S, Zheng D, Feng S(2006). Effects of Cadmium, Zinc and lead on soil enzymatic activities. Journal of Environment Science.18: 1135-1141.
M Malakootian; J Nouri; H Hossaini.(2009). International Journal of Environment Science and Technology. 6: 183-190.
S.P. Kamaludeen; K. R. Arun kumar; S R Avadainayagam.(2003). International Journal of Environment Science & Technology. 41:972-985.
K. Ashok ; B.S. Balwant and J D Vishnu. (2010). Biosorption of heavy metal by four acclimated microbial species, bacillus spp. Pseudomonas spp, staphylococcus spp. and Aspergillus spp.Journal of biology and environment science. 4(12) :97-108.
Hess a, Zarda B, Hahn D; Hanner A, Stax D. (1997). In-situ analysis of denitrifying toluene and m-xylene degrading bacteria in disel fuel contaminated laboratory aquifer column. Applied Environment microbes. 63:2136-2141.
Aggarwal SK (1998). Environment Biotechnology, Ist edition, APHA publishing corporation, New Delhi, India, 267-289.
Tang C Y, Criddle QS Fu Cs, Leckie JQ. (2007). Effect of flux (transmembranne pressue) and membrane properties on fouling and rejection of reverse osmoisis and nanofilteration membrane treating.
B Y Chen; V P Utgikar; S M Harmon; H H Tabak; D F Bishop; R Govind.(2000) International Biodeterioration Biodegradation.46:11-18.
Congeevaram S; Dhanarani S; Park J Dexilin H and Thamaraiselvi K (2007). Biosorption of Chromium and nickel by heavy metal resistant fungal & bacterial isolates. J. hazards Mat. 146: 270-277.
Trivedi, R.K. and Goel,P.K…(1986).Chemicals ansd biological method for water pollution studies. Environment Publications, Karad.
Aneja, K. R.(2010).In: Experiments in microbiology, plant pathology and biotechnology ( 4th 4^{\text {th }} edition ). New age international (pvt) ltd., New Delhi.
Hansson, P.J.; Edwards, N.T. and Andrews, J.A. (2003).Effect of different tree species on soil properties.Journal of Applied Ecology.23: 657-666.
Sam brook J, Fritsch EF, Maniatis T(1989) Molecular cloning: a laboratory manual (2nd edn), cold spring harbour, New York.
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731-2739.
Puranik, P. R. and Paknikar, K. M. (1999): Biosorption of lead, cadmium and zinc by Citrobacter strain MCM B-181: characterization studies. Biotechnol. Prog., 15: 228-237.
Cybulski Z, Dzuirla E, Kaczorek E, Olszanowski A, (2003). The influence of emulsifiers on hydrocarbon biodegradation by pseudomonadacea and Bacillacea strains. Spill Science and technology bulletin 8:503-507.
John G H; Noel R K; Peter H A S; James T S and Stanley T W.(1994) Bergey’s manual of Determinative Bacteriology ( 9th 9^{\text {th }} edition). Lippincott Williams & Wilkins, New York.
D DSimie; C Finoli; A Vecchio; V Ancheoni. Journal industrial Microbiology biotechnology, 19898, 20, 116120 .
Gupta R, Ahuja P, Khan S and Sexena R K. (2000). Microbial biosorbent: Meeting challenges of heavy metal pollution in aqueous solutions. Current Science. 78(8). 25.
Kapoor A and Viraraghavan T. (1995). Fungal-biosorption- An alternative treatment option for heavy metal bearing wastewater. A review, Bioresource Technology. 53:19553: 195.