 |
 |
PDBsum entry 1b0p    Go to PDB code: Go to PDB code:                                                                Oxidoreductase PDB id Oxidoreductase PDB id   1b0p 1b0p                  Loading ... Loading ...              Contents Contents       Protein chains Protein chains           1231 a.a.* 1231 a.a.*       Ligands Ligands         SF4 ×6 SF4 ×6         TPP ×2 TPP ×2       Metals Metals          _MG×2 _MG×2          _CA×2 _CA×2       Waters ×543 Waters ×543    * Residue conservation analysis * Residue conservation analysis  PDB id: PDB id:  1b0p 1b0p  Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT  Name: Name:  Oxidoreductase Oxidoreductase  Title: Title:  Crystal structure of pyruvate-ferredoxin oxidoreductase from desulfovibrio africanus Structure: Crystal structure of pyruvate-ferredoxin oxidoreductase from desulfovibrio africanus Structure:  Protein (pyruvate-ferredoxin oxidoreductase). Chain: a, b. Ec: 1.2.7.1 Source: Protein (pyruvate-ferredoxin oxidoreductase). Chain: a, b. Ec: 1.2.7.1 Source:  Desulfovibrio africanus. Organism_taxid: 873. Strain: ncib 8401. Cellular_location: cytoplasm Biol. unit: Desulfovibrio africanus. Organism_taxid: 873. Strain: ncib 8401. Cellular_location: cytoplasm Biol. unit:  Homo-Dimer (from PDB file) Resolution: Homo-Dimer (from PDB file) Resolution:  2.31Å R-factor: 0.199 R-free: 0.271 Authors: 2.31Å R-factor: 0.199 R-free: 0.271 Authors:  E.Chabriere,M.H.Charon,A.Volbeda Key ref: E.Chabriere,M.H.Charon,A.Volbeda Key ref:   E.Chabrière et al. (1999). Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.Nat Struct Biol,6, 182-190.PubMed id: 10048931 DOI: 10.1038/5870 E.Chabrière et al. (1999). Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.Nat Struct Biol,6, 182-190.PubMed id: 10048931 DOI: 10.1038/5870   Date: Date:  12-Nov-98 Release date: 23-Apr-99 12-Nov-98 Release date: 23-Apr-99   PROCHECK PROCHECK    Headers Headers   References References       Protein chains Protein chains                ? ?       P94692 (PFOR_DESAF) - Pyruvate:ferredoxin oxidoreductase from Desulfocurvibacter africanus P94692 (PFOR_DESAF) - Pyruvate:ferredoxin oxidoreductase from Desulfocurvibacter africanus   Seq:Struc: Seq:Struc:       Seq:Struc: Seq:Struc:       Seq:Struc: Seq:Struc:    1232 a.a. 1231 a.a. 1232 a.a. 1231 a.a.           Key: Key:  PfamA domain PfamA domain    Secondary structure Secondary structure   CATH domain CATH domain          Enzyme reactions Enzyme reactions           Enzyme class: Enzyme class:  E.C.1.2.7.1 - pyruvate synthase. [IntEnz] [ExPASy] [KEGG] [BRENDA] E.C.1.2.7.1 - pyruvate synthase. [IntEnz] [ExPASy] [KEGG] [BRENDA]       Reaction: Reaction:  CoA + 2 oxidized [2Fe-2S]-[ferredoxin] + pyruvate = acetyl-CoA + CO2 + H+ + 2 reduced [2Fe-2S]-[ferredoxin] CoA + 2 oxidized [2Fe-2S]-[ferredoxin] + pyruvate = acetyl-CoA + CO2 + H+ + 2 reduced [2Fe-2S]-[ferredoxin]       CoA + 2 × oxidized [2Fe-2S]-[ferredoxin] + CoA + 2 × oxidized [2Fe-2S]-[ferredoxin] +  pyruvate = pyruvate =  acetyl-CoA + acetyl-CoA +  CO2 + H(+) + 2 × reduced [2Fe-2S]-[ferredoxin] CO2 + H(+) + 2 × reduced [2Fe-2S]-[ferredoxin]         Cofactor: Cofactor:  Iron-sulfur; Thiamine diphosphate Iron-sulfur; Thiamine diphosphate       Iron-sulfur Iron-sulfur 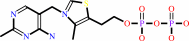 Thiamine diphosphate Bound ligand (Het Group name = TPP) corresponds exactly Thiamine diphosphate Bound ligand (Het Group name = TPP) corresponds exactly     Molecule diagrams generated from .mol files obtained from theKEGG ftp site Molecule diagrams generated from .mol files obtained from theKEGG ftp site                   reference DOI no: 10.1038/5870 Nat Struct Biol 6:182-190 (1999) PubMed id: 10048931 reference DOI no: 10.1038/5870 Nat Struct Biol 6:182-190 (1999) PubMed id: 10048931  Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. E.Chabrière, M.H.Charon, A.Volbeda, L.Pieulle, E.C.Hatchikian, J.C.Fontecilla-Camps. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. E.Chabrière, M.H.Charon, A.Volbeda, L.Pieulle, E.C.Hatchikian, J.C.Fontecilla-Camps.  ABSTRACT ABSTRACT   Oxidative decarboxylation of pyruvate to form acetyl-coenzyme A, a crucial step in many metabolic pathways, is carried out in most aerobic organisms by the multienzyme complex pyruvate dehydrogenase. In most anaerobes, the same reaction is usually catalyzed by a single enzyme, pyruvate:ferredoxin oxidoreductase (PFOR). Thus, PFOR is a potential target for drug design against certain anaerobic pathogens. Here, we report the crystal structures of the homodimeric Desulfovibrio africanus PFOR (data to 2.3 A resolution), and of its complex with pyruvate (3.0 A resolution). The structures show that each subunit consists of seven domains, one of which affords protection against oxygen. The thiamin pyrophosphate (TPP) cofactor and the three [4Fe-4S] clusters are suitably arranged to provide a plausible electron transfer pathway. In addition, the PFOR-pyruvate complex structure shows the noncovalent fixation of the substrate before the catalytic reaction. Oxidative decarboxylation of pyruvate to form acetyl-coenzyme A, a crucial step in many metabolic pathways, is carried out in most aerobic organisms by the multienzyme complex pyruvate dehydrogenase. In most anaerobes, the same reaction is usually catalyzed by a single enzyme, pyruvate:ferredoxin oxidoreductase (PFOR). Thus, PFOR is a potential target for drug design against certain anaerobic pathogens. Here, we report the crystal structures of the homodimeric Desulfovibrio africanus PFOR (data to 2.3 A resolution), and of its complex with pyruvate (3.0 A resolution). The structures show that each subunit consists of seven domains, one of which affords protection against oxygen. The thiamin pyrophosphate (TPP) cofactor and the three [4Fe-4S] clusters are suitably arranged to provide a plausible electron transfer pathway. In addition, the PFOR-pyruvate complex structure shows the noncovalent fixation of the substrate before the catalytic reaction.    Selected figure(s) Selected figure(s)        Figure 1. Figure 1. Ribbon drawings of Desulfovibrio africanus PFOR, made with MOLSCRIPT^48 and Raster3D^49. a,b, Two perpendicular views. Subunits are shown in light blue and dark blue. TPP cofactors are highlighted in bright red, Mg ions in green, iron atoms in brown and sulfur atoms in yellow. Figure 1. Figure 1. Ribbon drawings of Desulfovibrio africanus PFOR, made with MOLSCRIPT^48 and Raster3D^49. a,b, Two perpendicular views. Subunits are shown in light blue and dark blue. TPP cofactors are highlighted in bright red, Mg ions in green, iron atoms in brown and sulfur atoms in yellow.  Figure 3. Figure 3. Ribbon drawings of the seven structural domains of Desulfovibrio africanus PFOR. In the included topology diagrams, Figure 3. Figure 3. Ribbon drawings of the seven structural domains of Desulfovibrio africanus PFOR. In the included topology diagrams,  −helices are represented by circles and −helices are represented by circles and  −strands by triangles. A common folding motif in the core domains I, II and VI is indicated by the dots in their topology diagrams. For clarity, domains and topology diagrams are not represented in the same orientation. −strands by triangles. A common folding motif in the core domains I, II and VI is indicated by the dots in their topology diagrams. For clarity, domains and topology diagrams are not represented in the same orientation.   The above figures are reprinted by permission from Macmillan Publishers Ltd: Nat Struct Biol (1999,6, 182-190) copyright 1999. Figures were selected by an automated process. The above figures are reprinted by permission from Macmillan Publishers Ltd: Nat Struct Biol (1999,6, 182-190) copyright 1999. Figures were selected by an automated process.              Literature references that cite this PDB file's key reference Literature references that cite this PDB file's key reference  PubMed id PubMed id  Reference Reference      20884694 20884694  P.Worm, A.J.Stams, X.Cheng, and C.M.Plugge (2011). Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology,157, 280-289. P.Worm, A.J.Stams, X.Cheng, and C.M.Plugge (2011). Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology,157, 280-289.      20015072 20015072  T.Ikeda, M.Yamamoto, H.Arai, D.Ohmori, M.Ishii, and Y.Igarashi (2010). Enzymatic and electron paramagnetic resonance studies of anabolic pyruvate synthesis by pyruvate: ferredoxin oxidoreductase from Hydrogenobacter thermophilus. FEBS J,277, 501-510. T.Ikeda, M.Yamamoto, H.Arai, D.Ohmori, M.Ishii, and Y.Igarashi (2010). Enzymatic and electron paramagnetic resonance studies of anabolic pyruvate synthesis by pyruvate: ferredoxin oxidoreductase from Hydrogenobacter thermophilus. FEBS J,277, 501-510.      19590008 19590008  A.S.Eustáquio, R.P.McGlinchey, Y.Liu, C.Hazzard, L.L.Beer, G.Florova, M.M.Alhamadsheh, A.Lechner, A.J.Kale, Y.Kobayashi, K.A.Reynolds, and B.S.Moore (2009). Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U S A,106, 12295-12300. A.S.Eustáquio, R.P.McGlinchey, Y.Liu, C.Hazzard, L.L.Beer, G.Florova, M.M.Alhamadsheh, A.Lechner, A.J.Kale, Y.Kobayashi, K.A.Reynolds, and B.S.Moore (2009). Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U S A,106, 12295-12300.      19476486 19476486  B.Shaanan, and D.M.Chipman (2009). Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. FEBS J,276, 2447-2453. B.Shaanan, and D.M.Chipman (2009). Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. FEBS J,276, 2447-2453.      19476487 19476487  K.Tittmann (2009). Reaction mechanisms of thiamin diphosphate enzymes: redox reactions. FEBS J,276, 2454-2468. K.Tittmann (2009). Reaction mechanisms of thiamin diphosphate enzymes: redox reactions. FEBS J,276, 2454-2468.      18043855 18043855  S.J.Costelloe, J.M.Ward, and P.A.Dalby (2008). Evolutionary Analysis of the TPP-Dependent Enzyme Family. J Mol Evol,66, 36-49. S.J.Costelloe, J.M.Ward, and P.A.Dalby (2008). Evolutionary Analysis of the TPP-Dependent Enzyme Family. J Mol Evol,66, 36-49.      18801467 18801467  S.W.Ragsdale, and E.Pierce (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta,1784, 1873-1898. S.W.Ragsdale, and E.Pierce (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta,1784, 1873-1898.      18378591 18378591  S.W.Ragsdale (2008). Enzymology of the wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci,1125, 129-136. S.W.Ragsdale (2008). Enzymology of the wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci,1125, 129-136.      18004749 18004749  V.I.Bunik, and D.Degtyarev (2008). Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins,71, 874-890. V.I.Bunik, and D.Degtyarev (2008). Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins,71, 874-890.      17534532 17534532  A.W.Munro, H.M.Girvan, and K.J.McLean (2007). Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep,24, 585-609. A.W.Munro, H.M.Girvan, and K.J.McLean (2007). Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep,24, 585-609.      16430685 16430685  J.A.Imlay (2006). Iron-sulphur clusters and the problem with oxygen. Mol Microbiol,59, 1073-1082. J.A.Imlay (2006). Iron-sulphur clusters and the problem with oxygen. Mol Microbiol,59, 1073-1082.      16531404 16531404  P.Arjunan, M.Sax, A.Brunskill, K.Chandrasekhar, N.Nemeria, S.Zhang, F.Jordan, and W.Furey (2006). A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J Biol Chem,281, 15296-15303. P.Arjunan, M.Sax, A.Brunskill, K.Chandrasekhar, N.Nemeria, S.Zhang, F.Jordan, and W.Furey (2006). A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J Biol Chem,281, 15296-15303.  PDB codes: PDB codes:  2g25 2g28 2g25 2g28     16752902 16752902  S.O.Mansoorabadi, J.Seravalli, C.Furdui, V.Krymov, G.J.Gerfen, T.P.Begley, J.Melnick, S.W.Ragsdale, and G.H.Reed (2006). EPR spectroscopic and computational characterization of the hydroxyethylidene-thiamine pyrophosphate radical intermediate of pyruvate:ferredoxin oxidoreductase. Biochemistry,45, 7122-7131. S.O.Mansoorabadi, J.Seravalli, C.Furdui, V.Krymov, G.J.Gerfen, T.P.Begley, J.Melnick, S.W.Ragsdale, and G.H.Reed (2006). EPR spectroscopic and computational characterization of the hydroxyethylidene-thiamine pyrophosphate radical intermediate of pyruvate:ferredoxin oxidoreductase. Biochemistry,45, 7122-7131.      16603087 16603087  S.S.Krishna, R.I.Sadreyev, and N.V.Grishin (2006). A tale of two ferredoxins: sequence similarity and structural differences. BMC Struct Biol,6, 8. S.S.Krishna, R.I.Sadreyev, and N.V.Grishin (2006). A tale of two ferredoxins: sequence similarity and structural differences. BMC Struct Biol,6, 8.      15752351 15752351  R.Golbik, L.E.Meshalkina, T.Sandalova, K.Tittmann, E.Fiedler, H.Neef, S.König, R.Kluger, G.A.Kochetov, G.Schneider, and G.Hübner (2005). Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae. FEBS J,272, 1326-1342. R.Golbik, L.E.Meshalkina, T.Sandalova, K.Tittmann, E.Fiedler, H.Neef, S.König, R.Kluger, G.A.Kochetov, G.Schneider, and G.Hübner (2005). Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae. FEBS J,272, 1326-1342.      15305914 15305914  R.Rabus, A.Ruepp, T.Frickey, T.Rattei, B.Fartmann, M.Stark, M.Bauer, A.Zibat, T.Lombardot, I.Becker, J.Amann, K.Gellner, H.Teeling, W.D.Leuschner, F.O.Glöckner, A.N.Lupas, R.Amann, and H.P.Klenk (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol,6, 887-902. R.Rabus, A.Ruepp, T.Frickey, T.Rattei, B.Fartmann, M.Stark, M.Bauer, A.Zibat, T.Lombardot, I.Becker, J.Amann, K.Gellner, H.Teeling, W.D.Leuschner, F.O.Glöckner, A.N.Lupas, R.Amann, and H.P.Klenk (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol,6, 887-902.      14526024 14526024  C.Ebenau-Jehle, M.Boll, and G.Fuchs (2003). 2-Oxoglutarate:NADP(+) oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J Bacteriol,185, 6119-6129. C.Ebenau-Jehle, M.Boll, and G.Fuchs (2003). 2-Oxoglutarate:NADP(+) oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J Bacteriol,185, 6119-6129.      12594918 12594918  W.Martin, and M.J.Russell (2003). On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci,358, 59. W.Martin, and M.J.Russell (2003). On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci,358, 59.      12146957 12146957  C.Furdui, and S.W.Ragsdale (2002). The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry,41, 9921-9937. C.Furdui, and S.W.Ragsdale (2002). The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry,41, 9921-9937.      12081970 12081970  E.Dörner, and M.Boll (2002). Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J Bacteriol,184, 3975-3983. E.Dörner, and M.Boll (2002). Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J Bacteriol,184, 3975-3983.      12142484 12142484  P.J.Keeling, and N.M.Fast (2002). Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol,56, 93. P.J.Keeling, and N.M.Fast (2002). Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol,56, 93.      12167658 12167658  T.Iwasaki, A.Kounosu, M.Aoshima, D.Ohmori, T.Imai, A.Urushiyama, N.J.Cosper, and R.A.Scott (2002). Novel [2Fe-2S]-type redox center C in SdhC of archaeal respiratory complex II from Sulfolobus tokodaii strain 7. J Biol Chem,277, 39642-39648. T.Iwasaki, A.Kounosu, M.Aoshima, D.Ohmori, T.Imai, A.Urushiyama, N.J.Cosper, and R.A.Scott (2002). Novel [2Fe-2S]-type redox center C in SdhC of archaeal respiratory complex II from Sulfolobus tokodaii strain 7. J Biol Chem,277, 39642-39648.      12475211 12475211  V.L.Davidson (2002). Chemically gated electron transfer. A means of accelerating and regulating rates of biological electron transfer. Biochemistry,41, 14633-14636. V.L.Davidson (2002). Chemically gated electron transfer. A means of accelerating and regulating rates of biological electron transfer. Biochemistry,41, 14633-14636.      11422387 11422387  C.Y.Huang, A.K.Chang, P.F.Nixon, and R.G.Duggleby (2001). Site-directed mutagenesis of the ionizable groups in the active site of Zymomonas mobilis pyruvate decarboxylase: effect on activity and pH dependence. Eur J Biochem,268, 3558-3565. C.Y.Huang, A.K.Chang, P.F.Nixon, and R.G.Duggleby (2001). Site-directed mutagenesis of the ionizable groups in the active site of Zymomonas mobilis pyruvate decarboxylase: effect on activity and pH dependence. Eur J Biochem,268, 3558-3565.      11179210 11179210  D.Dobritzsch, G.Schneider, K.D.Schnackerz, and Y.Lindqvist (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J,20, 650-660. D.Dobritzsch, G.Schneider, K.D.Schnackerz, and Y.Lindqvist (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J,20, 650-660.  PDB codes: PDB codes:  1h7w 1h7x 1h7w 1h7x     11752578 11752578  E.Chabrière, X.Vernède, B.Guigliarelli, M.H.Charon, E.C.Hatchikian, and J.C.Fontecilla-Camps (2001). Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science,294, 2559-2563. E.Chabrière, X.Vernède, B.Guigliarelli, M.H.Charon, E.C.Hatchikian, and J.C.Fontecilla-Camps (2001). Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science,294, 2559-2563.  PDB code: PDB code:  1kek 1kek     11683888 11683888  E.Fukuda, H.Kino, H.Matsuzawa, and T.Wakagi (2001). Role of a highly conserved YPITP motif in 2-oxoacid:ferredoxin oxidoreductase: heterologous expression of the gene from Sulfolobus sp.strain 7, and characterization of the recombinant and variant enzymes. Eur J Biochem,268, 5639-5646. E.Fukuda, H.Kino, H.Matsuzawa, and T.Wakagi (2001). Role of a highly conserved YPITP motif in 2-oxoacid:ferredoxin oxidoreductase: heterologous expression of the gene from Sulfolobus sp.strain 7, and characterization of the recombinant and variant enzymes. Eur J Biochem,268, 5639-5646.      11568186 11568186  K.S.Yoon, C.Bobst, C.F.Hemann, R.Hille, and F.R.Tabita (2001). Spectroscopic and functional properties of novel 2[4Fe-4S] cluster-containing ferredoxins from the green sulfur bacterium Chlorobium tepidum. J Biol Chem,276, 44027-44036. K.S.Yoon, C.Bobst, C.F.Hemann, R.Hille, and F.R.Tabita (2001). Spectroscopic and functional properties of novel 2[4Fe-4S] cluster-containing ferredoxins from the green sulfur bacterium Chlorobium tepidum. J Biol Chem,276, 44027-44036.      10745006 10745006  A.AEvarsson, J.L.Chuang, R.M.Wynn, S.Turley, D.T.Chuang, and W.G.Hol (2000). Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure,8, 277-291. A.AEvarsson, J.L.Chuang, R.M.Wynn, S.Turley, D.T.Chuang, and W.G.Hol (2000). Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure,8, 277-291.  PDB code: PDB code:  1dtw 1dtw     10966480 10966480  R.N.Perham (2000). Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem,69, 961. R.N.Perham (2000). Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem,69, 961.      10848975 10848975  V.Bunik, A.H.Westphal, and A.de Kok (2000). Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii evidence for the formation of a precatalytic complex with 2-oxoglutarate. Eur J Biochem,267, 3583-3591. V.Bunik, A.H.Westphal, and A.de Kok (2000). Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii evidence for the formation of a precatalytic complex with 2-oxoglutarate. Eur J Biochem,267, 3583-3591.      10491097 10491097  L.Pieulle, M.H.Charon, P.Bianco, J.Bonicel, Y.Pétillot, and E.C.Hatchikian (1999). Structural and kinetic studies of the pyruvate-ferredoxin oxidoreductase/ferredoxin complex from Desulfovibrio africanus. Eur J Biochem,264, 500-508. L.Pieulle, M.H.Charon, P.Bianco, J.Bonicel, Y.Pétillot, and E.C.Hatchikian (1999). Structural and kinetic studies of the pyruvate-ferredoxin oxidoreductase/ferredoxin complex from Desulfovibrio africanus. Eur J Biochem,264, 500-508.      10607667 10607667  M.H.Charon, A.Volbeda, E.Chabriere, L.Pieulle, and J.C.Fontecilla-Camps (1999). Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol,9, 663-669. M.H.Charon, A.Volbeda, E.Chabriere, L.Pieulle, and J.C.Fontecilla-Camps (1999). Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol,9, 663-669.    The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right. The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right.  |
 |




 Go to PDB code:
Go to PDB code: 





























































 Oxidoreductase PDB id
Oxidoreductase PDB id 
 1b0p
1b0p 















 Loading ...
Loading ... 











 Contents
Contents 




 Protein chains
Protein chains 








 1231 a.a.*
1231 a.a.* 




 Ligands
Ligands 






 SF4 ×6
SF4 ×6 






 TPP ×2
TPP ×2 




 Metals
Metals 







 _MG×2
_MG×2 







 _CA×2
_CA×2 




 Waters ×543
Waters ×543 

 * Residue conservation analysis
* Residue conservation analysis  PDB id:
PDB id:  1b0p
1b0p  Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT
Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT  Name:
Name:  Oxidoreductase
Oxidoreductase  Title:
Title:  Crystal structure of pyruvate-ferredoxin oxidoreductase from desulfovibrio africanus Structure:
Crystal structure of pyruvate-ferredoxin oxidoreductase from desulfovibrio africanus Structure:  Protein (pyruvate-ferredoxin oxidoreductase). Chain: a, b. Ec: 1.2.7.1 Source:
Protein (pyruvate-ferredoxin oxidoreductase). Chain: a, b. Ec: 1.2.7.1 Source:  Desulfovibrio africanus. Organism_taxid: 873. Strain: ncib 8401. Cellular_location: cytoplasm Biol. unit:
Desulfovibrio africanus. Organism_taxid: 873. Strain: ncib 8401. Cellular_location: cytoplasm Biol. unit:  Homo-Dimer (from PDB file) Resolution:
Homo-Dimer (from PDB file) Resolution:  2.31Å R-factor: 0.199 R-free: 0.271 Authors:
2.31Å R-factor: 0.199 R-free: 0.271 Authors:  E.Chabriere,M.H.Charon,A.Volbeda Key ref:
E.Chabriere,M.H.Charon,A.Volbeda Key ref:  E.Chabrière et al. (1999). Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.Nat Struct Biol,6, 182-190.PubMed id: 10048931 DOI: 10.1038/5870
E.Chabrière et al. (1999). Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.Nat Struct Biol,6, 182-190.PubMed id: 10048931 DOI: 10.1038/5870 
 Date:
Date:  12-Nov-98 Release date: 23-Apr-99
12-Nov-98 Release date: 23-Apr-99 
 PROCHECK
PROCHECK 

 Headers
Headers 
 References
References 




 Protein chains
Protein chains 


















 P94692 (PFOR_DESAF) - Pyruvate:ferredoxin oxidoreductase from Desulfocurvibacter africanus
P94692 (PFOR_DESAF) - Pyruvate:ferredoxin oxidoreductase from Desulfocurvibacter africanus 
 Seq:Struc:
Seq:Struc: 




 Seq:Struc:
Seq:Struc: 




 Seq:Struc:
Seq:Struc: 

 1232 a.a. 1231 a.a.
1232 a.a. 1231 a.a. 








 Key:
Key:  PfamA domain
PfamA domain 

 Secondary structure
Secondary structure 
 CATH domain
CATH domain 







 Enzyme reactions
Enzyme reactions 








 Enzyme class:
Enzyme class:  E.C.1.2.7.1 - pyruvate synthase. [IntEnz] [ExPASy] [KEGG] [BRENDA]
E.C.1.2.7.1 - pyruvate synthase. [IntEnz] [ExPASy] [KEGG] [BRENDA] 




 Reaction:
Reaction:  CoA + 2 oxidized [2Fe-2S]-[ferredoxin] + pyruvate = acetyl-CoA + CO2 + H+ + 2 reduced [2Fe-2S]-[ferredoxin]
CoA + 2 oxidized [2Fe-2S]-[ferredoxin] + pyruvate = acetyl-CoA + CO2 + H+ + 2 reduced [2Fe-2S]-[ferredoxin] 











 Cofactor:
Cofactor:  Iron-sulfur; Thiamine diphosphate
Iron-sulfur; Thiamine diphosphate 







 Molecule diagrams generated from .mol files obtained from theKEGG ftp site
Molecule diagrams generated from .mol files obtained from theKEGG ftp site 
















 reference DOI no: 10.1038/5870 Nat Struct Biol 6:182-190 (1999) PubMed id: 10048931
reference DOI no: 10.1038/5870 Nat Struct Biol 6:182-190 (1999) PubMed id: 10048931  Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. E.Chabrière, M.H.Charon, A.Volbeda, L.Pieulle, E.C.Hatchikian, J.C.Fontecilla-Camps.
Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. E.Chabrière, M.H.Charon, A.Volbeda, L.Pieulle, E.C.Hatchikian, J.C.Fontecilla-Camps.  ABSTRACT
ABSTRACT 
 Oxidative decarboxylation of pyruvate to form acetyl-coenzyme A, a crucial step in many metabolic pathways, is carried out in most aerobic organisms by the multienzyme complex pyruvate dehydrogenase. In most anaerobes, the same reaction is usually catalyzed by a single enzyme, pyruvate:ferredoxin oxidoreductase (PFOR). Thus, PFOR is a potential target for drug design against certain anaerobic pathogens. Here, we report the crystal structures of the homodimeric Desulfovibrio africanus PFOR (data to 2.3 A resolution), and of its complex with pyruvate (3.0 A resolution). The structures show that each subunit consists of seven domains, one of which affords protection against oxygen. The thiamin pyrophosphate (TPP) cofactor and the three [4Fe-4S] clusters are suitably arranged to provide a plausible electron transfer pathway. In addition, the PFOR-pyruvate complex structure shows the noncovalent fixation of the substrate before the catalytic reaction.
Oxidative decarboxylation of pyruvate to form acetyl-coenzyme A, a crucial step in many metabolic pathways, is carried out in most aerobic organisms by the multienzyme complex pyruvate dehydrogenase. In most anaerobes, the same reaction is usually catalyzed by a single enzyme, pyruvate:ferredoxin oxidoreductase (PFOR). Thus, PFOR is a potential target for drug design against certain anaerobic pathogens. Here, we report the crystal structures of the homodimeric Desulfovibrio africanus PFOR (data to 2.3 A resolution), and of its complex with pyruvate (3.0 A resolution). The structures show that each subunit consists of seven domains, one of which affords protection against oxygen. The thiamin pyrophosphate (TPP) cofactor and the three [4Fe-4S] clusters are suitably arranged to provide a plausible electron transfer pathway. In addition, the PFOR-pyruvate complex structure shows the noncovalent fixation of the substrate before the catalytic reaction. 

 Selected figure(s)
Selected figure(s) 





 Figure 1. Figure 1. Ribbon drawings of Desulfovibrio africanus PFOR, made with MOLSCRIPT^48 and Raster3D^49. a,b, Two perpendicular views. Subunits are shown in light blue and dark blue. TPP cofactors are highlighted in bright red, Mg ions in green, iron atoms in brown and sulfur atoms in yellow.
Figure 1. Figure 1. Ribbon drawings of Desulfovibrio africanus PFOR, made with MOLSCRIPT^48 and Raster3D^49. a,b, Two perpendicular views. Subunits are shown in light blue and dark blue. TPP cofactors are highlighted in bright red, Mg ions in green, iron atoms in brown and sulfur atoms in yellow.  Figure 3. Figure 3. Ribbon drawings of the seven structural domains of Desulfovibrio africanus PFOR. In the included topology diagrams,
Figure 3. Figure 3. Ribbon drawings of the seven structural domains of Desulfovibrio africanus PFOR. In the included topology diagrams,  −helices are represented by circles and
−helices are represented by circles and  −strands by triangles. A common folding motif in the core domains I, II and VI is indicated by the dots in their topology diagrams. For clarity, domains and topology diagrams are not represented in the same orientation.
−strands by triangles. A common folding motif in the core domains I, II and VI is indicated by the dots in their topology diagrams. For clarity, domains and topology diagrams are not represented in the same orientation. 
 The above figures are reprinted by permission from Macmillan Publishers Ltd: Nat Struct Biol (1999,6, 182-190) copyright 1999. Figures were selected by an automated process.
The above figures are reprinted by permission from Macmillan Publishers Ltd: Nat Struct Biol (1999,6, 182-190) copyright 1999. Figures were selected by an automated process. 











 Literature references that cite this PDB file's key reference
Literature references that cite this PDB file's key reference  PubMed id
PubMed id  Reference
Reference 



 20884694
20884694  P.Worm, A.J.Stams, X.Cheng, and C.M.Plugge (2011). Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology,157, 280-289.
P.Worm, A.J.Stams, X.Cheng, and C.M.Plugge (2011). Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology,157, 280-289. 



 20015072
20015072  T.Ikeda, M.Yamamoto, H.Arai, D.Ohmori, M.Ishii, and Y.Igarashi (2010). Enzymatic and electron paramagnetic resonance studies of anabolic pyruvate synthesis by pyruvate: ferredoxin oxidoreductase from Hydrogenobacter thermophilus. FEBS J,277, 501-510.
T.Ikeda, M.Yamamoto, H.Arai, D.Ohmori, M.Ishii, and Y.Igarashi (2010). Enzymatic and electron paramagnetic resonance studies of anabolic pyruvate synthesis by pyruvate: ferredoxin oxidoreductase from Hydrogenobacter thermophilus. FEBS J,277, 501-510. 



 19590008
19590008  A.S.Eustáquio, R.P.McGlinchey, Y.Liu, C.Hazzard, L.L.Beer, G.Florova, M.M.Alhamadsheh, A.Lechner, A.J.Kale, Y.Kobayashi, K.A.Reynolds, and B.S.Moore (2009). Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U S A,106, 12295-12300.
A.S.Eustáquio, R.P.McGlinchey, Y.Liu, C.Hazzard, L.L.Beer, G.Florova, M.M.Alhamadsheh, A.Lechner, A.J.Kale, Y.Kobayashi, K.A.Reynolds, and B.S.Moore (2009). Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U S A,106, 12295-12300. 



 19476486
19476486  B.Shaanan, and D.M.Chipman (2009). Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. FEBS J,276, 2447-2453.
B.Shaanan, and D.M.Chipman (2009). Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. FEBS J,276, 2447-2453. 



 19476487
19476487  K.Tittmann (2009). Reaction mechanisms of thiamin diphosphate enzymes: redox reactions. FEBS J,276, 2454-2468.
K.Tittmann (2009). Reaction mechanisms of thiamin diphosphate enzymes: redox reactions. FEBS J,276, 2454-2468. 



 18043855
18043855  S.J.Costelloe, J.M.Ward, and P.A.Dalby (2008). Evolutionary Analysis of the TPP-Dependent Enzyme Family. J Mol Evol,66, 36-49.
S.J.Costelloe, J.M.Ward, and P.A.Dalby (2008). Evolutionary Analysis of the TPP-Dependent Enzyme Family. J Mol Evol,66, 36-49. 



 18801467
18801467  S.W.Ragsdale, and E.Pierce (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta,1784, 1873-1898.
S.W.Ragsdale, and E.Pierce (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta,1784, 1873-1898. 



 18378591
18378591  S.W.Ragsdale (2008). Enzymology of the wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci,1125, 129-136.
S.W.Ragsdale (2008). Enzymology of the wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci,1125, 129-136. 



 18004749
18004749  V.I.Bunik, and D.Degtyarev (2008). Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins,71, 874-890.
V.I.Bunik, and D.Degtyarev (2008). Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins,71, 874-890. 



 17534532
17534532  A.W.Munro, H.M.Girvan, and K.J.McLean (2007). Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep,24, 585-609.
A.W.Munro, H.M.Girvan, and K.J.McLean (2007). Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep,24, 585-609. 



 16430685
16430685  J.A.Imlay (2006). Iron-sulphur clusters and the problem with oxygen. Mol Microbiol,59, 1073-1082.
J.A.Imlay (2006). Iron-sulphur clusters and the problem with oxygen. Mol Microbiol,59, 1073-1082. 



 16531404
16531404  P.Arjunan, M.Sax, A.Brunskill, K.Chandrasekhar, N.Nemeria, S.Zhang, F.Jordan, and W.Furey (2006). A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J Biol Chem,281, 15296-15303.
P.Arjunan, M.Sax, A.Brunskill, K.Chandrasekhar, N.Nemeria, S.Zhang, F.Jordan, and W.Furey (2006). A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J Biol Chem,281, 15296-15303.  PDB codes:
PDB codes:  2g25 2g28
2g25 2g28 


 16752902
16752902  S.O.Mansoorabadi, J.Seravalli, C.Furdui, V.Krymov, G.J.Gerfen, T.P.Begley, J.Melnick, S.W.Ragsdale, and G.H.Reed (2006). EPR spectroscopic and computational characterization of the hydroxyethylidene-thiamine pyrophosphate radical intermediate of pyruvate:ferredoxin oxidoreductase. Biochemistry,45, 7122-7131.
S.O.Mansoorabadi, J.Seravalli, C.Furdui, V.Krymov, G.J.Gerfen, T.P.Begley, J.Melnick, S.W.Ragsdale, and G.H.Reed (2006). EPR spectroscopic and computational characterization of the hydroxyethylidene-thiamine pyrophosphate radical intermediate of pyruvate:ferredoxin oxidoreductase. Biochemistry,45, 7122-7131. 



 16603087
16603087  S.S.Krishna, R.I.Sadreyev, and N.V.Grishin (2006). A tale of two ferredoxins: sequence similarity and structural differences. BMC Struct Biol,6, 8.
S.S.Krishna, R.I.Sadreyev, and N.V.Grishin (2006). A tale of two ferredoxins: sequence similarity and structural differences. BMC Struct Biol,6, 8. 



 15752351
15752351  R.Golbik, L.E.Meshalkina, T.Sandalova, K.Tittmann, E.Fiedler, H.Neef, S.König, R.Kluger, G.A.Kochetov, G.Schneider, and G.Hübner (2005). Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae. FEBS J,272, 1326-1342.
R.Golbik, L.E.Meshalkina, T.Sandalova, K.Tittmann, E.Fiedler, H.Neef, S.König, R.Kluger, G.A.Kochetov, G.Schneider, and G.Hübner (2005). Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae. FEBS J,272, 1326-1342. 



 15305914
15305914  R.Rabus, A.Ruepp, T.Frickey, T.Rattei, B.Fartmann, M.Stark, M.Bauer, A.Zibat, T.Lombardot, I.Becker, J.Amann, K.Gellner, H.Teeling, W.D.Leuschner, F.O.Glöckner, A.N.Lupas, R.Amann, and H.P.Klenk (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol,6, 887-902.
R.Rabus, A.Ruepp, T.Frickey, T.Rattei, B.Fartmann, M.Stark, M.Bauer, A.Zibat, T.Lombardot, I.Becker, J.Amann, K.Gellner, H.Teeling, W.D.Leuschner, F.O.Glöckner, A.N.Lupas, R.Amann, and H.P.Klenk (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol,6, 887-902. 



 14526024
14526024  C.Ebenau-Jehle, M.Boll, and G.Fuchs (2003). 2-Oxoglutarate:NADP(+) oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J Bacteriol,185, 6119-6129.
C.Ebenau-Jehle, M.Boll, and G.Fuchs (2003). 2-Oxoglutarate:NADP(+) oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J Bacteriol,185, 6119-6129. 



 12594918
12594918  W.Martin, and M.J.Russell (2003). On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci,358, 59.
W.Martin, and M.J.Russell (2003). On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci,358, 59. 



 12146957
12146957  C.Furdui, and S.W.Ragsdale (2002). The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry,41, 9921-9937.
C.Furdui, and S.W.Ragsdale (2002). The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry,41, 9921-9937. 



 12081970
12081970  E.Dörner, and M.Boll (2002). Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J Bacteriol,184, 3975-3983.
E.Dörner, and M.Boll (2002). Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J Bacteriol,184, 3975-3983. 



 12142484
12142484  P.J.Keeling, and N.M.Fast (2002). Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol,56, 93.
P.J.Keeling, and N.M.Fast (2002). Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol,56, 93. 



 12167658
12167658  T.Iwasaki, A.Kounosu, M.Aoshima, D.Ohmori, T.Imai, A.Urushiyama, N.J.Cosper, and R.A.Scott (2002). Novel [2Fe-2S]-type redox center C in SdhC of archaeal respiratory complex II from Sulfolobus tokodaii strain 7. J Biol Chem,277, 39642-39648.
T.Iwasaki, A.Kounosu, M.Aoshima, D.Ohmori, T.Imai, A.Urushiyama, N.J.Cosper, and R.A.Scott (2002). Novel [2Fe-2S]-type redox center C in SdhC of archaeal respiratory complex II from Sulfolobus tokodaii strain 7. J Biol Chem,277, 39642-39648. 



 12475211
12475211  V.L.Davidson (2002). Chemically gated electron transfer. A means of accelerating and regulating rates of biological electron transfer. Biochemistry,41, 14633-14636.
V.L.Davidson (2002). Chemically gated electron transfer. A means of accelerating and regulating rates of biological electron transfer. Biochemistry,41, 14633-14636. 



 11422387
11422387  C.Y.Huang, A.K.Chang, P.F.Nixon, and R.G.Duggleby (2001). Site-directed mutagenesis of the ionizable groups in the active site of Zymomonas mobilis pyruvate decarboxylase: effect on activity and pH dependence. Eur J Biochem,268, 3558-3565.
C.Y.Huang, A.K.Chang, P.F.Nixon, and R.G.Duggleby (2001). Site-directed mutagenesis of the ionizable groups in the active site of Zymomonas mobilis pyruvate decarboxylase: effect on activity and pH dependence. Eur J Biochem,268, 3558-3565. 



 11179210
11179210  D.Dobritzsch, G.Schneider, K.D.Schnackerz, and Y.Lindqvist (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J,20, 650-660.
D.Dobritzsch, G.Schneider, K.D.Schnackerz, and Y.Lindqvist (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J,20, 650-660.  PDB codes:
PDB codes:  1h7w 1h7x
1h7w 1h7x 


 11752578
11752578  E.Chabrière, X.Vernède, B.Guigliarelli, M.H.Charon, E.C.Hatchikian, and J.C.Fontecilla-Camps (2001). Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science,294, 2559-2563.
E.Chabrière, X.Vernède, B.Guigliarelli, M.H.Charon, E.C.Hatchikian, and J.C.Fontecilla-Camps (2001). Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science,294, 2559-2563.  PDB code:
PDB code:  1kek
1kek 


 11683888
11683888  E.Fukuda, H.Kino, H.Matsuzawa, and T.Wakagi (2001). Role of a highly conserved YPITP motif in 2-oxoacid:ferredoxin oxidoreductase: heterologous expression of the gene from Sulfolobus sp.strain 7, and characterization of the recombinant and variant enzymes. Eur J Biochem,268, 5639-5646.
E.Fukuda, H.Kino, H.Matsuzawa, and T.Wakagi (2001). Role of a highly conserved YPITP motif in 2-oxoacid:ferredoxin oxidoreductase: heterologous expression of the gene from Sulfolobus sp.strain 7, and characterization of the recombinant and variant enzymes. Eur J Biochem,268, 5639-5646. 



 11568186
11568186  K.S.Yoon, C.Bobst, C.F.Hemann, R.Hille, and F.R.Tabita (2001). Spectroscopic and functional properties of novel 2[4Fe-4S] cluster-containing ferredoxins from the green sulfur bacterium Chlorobium tepidum. J Biol Chem,276, 44027-44036.
K.S.Yoon, C.Bobst, C.F.Hemann, R.Hille, and F.R.Tabita (2001). Spectroscopic and functional properties of novel 2[4Fe-4S] cluster-containing ferredoxins from the green sulfur bacterium Chlorobium tepidum. J Biol Chem,276, 44027-44036. 



 10745006
10745006  A.AEvarsson, J.L.Chuang, R.M.Wynn, S.Turley, D.T.Chuang, and W.G.Hol (2000). Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure,8, 277-291.
A.AEvarsson, J.L.Chuang, R.M.Wynn, S.Turley, D.T.Chuang, and W.G.Hol (2000). Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure,8, 277-291.  PDB code:
PDB code:  1dtw
1dtw 


 10966480
10966480  R.N.Perham (2000). Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem,69, 961.
R.N.Perham (2000). Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem,69, 961. 



 10848975
10848975  V.Bunik, A.H.Westphal, and A.de Kok (2000). Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii evidence for the formation of a precatalytic complex with 2-oxoglutarate. Eur J Biochem,267, 3583-3591.
V.Bunik, A.H.Westphal, and A.de Kok (2000). Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii evidence for the formation of a precatalytic complex with 2-oxoglutarate. Eur J Biochem,267, 3583-3591. 



 10491097
10491097  L.Pieulle, M.H.Charon, P.Bianco, J.Bonicel, Y.Pétillot, and E.C.Hatchikian (1999). Structural and kinetic studies of the pyruvate-ferredoxin oxidoreductase/ferredoxin complex from Desulfovibrio africanus. Eur J Biochem,264, 500-508.
L.Pieulle, M.H.Charon, P.Bianco, J.Bonicel, Y.Pétillot, and E.C.Hatchikian (1999). Structural and kinetic studies of the pyruvate-ferredoxin oxidoreductase/ferredoxin complex from Desulfovibrio africanus. Eur J Biochem,264, 500-508. 



 10607667
10607667  M.H.Charon, A.Volbeda, E.Chabriere, L.Pieulle, and J.C.Fontecilla-Camps (1999). Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol,9, 663-669.
M.H.Charon, A.Volbeda, E.Chabriere, L.Pieulle, and J.C.Fontecilla-Camps (1999). Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr Opin Struct Biol,9, 663-669. 

 The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right.
The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right. 
