 |
 |
PDBsum entry 1gt8    Go to PDB code: Go to PDB code:                                                          Oxidoreductase PDB id Oxidoreductase PDB id   1gt8 1gt8                  Loading ... Loading ...              Contents Contents       Protein chains Protein chains             1017 a.a.* 1017 a.a.*       Ligands Ligands         SF4 ×16 SF4 ×16         FMN ×4 FMN ×4         FAD ×4 FAD ×4         NDP ×4 NDP ×4         UAA ×4 UAA ×4    * Residue conservation analysis * Residue conservation analysis  PDB id: PDB id:  1gt8 1gt8  Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA PROCOGNATE ProSAT Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA PROCOGNATE ProSAT  Name: Name:  Oxidoreductase Oxidoreductase  Title: Title:  Dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with NADPH and uracil-4-acetic acid Structure: Dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with NADPH and uracil-4-acetic acid Structure:  Dihydropyrimidine dehydrogenase. Chain: a, b, c, d. Synonym: dpd, dhpdhase, dihydrouracil dehydrogenase, dihydrothymine dehydrogenase. Engineered: yes Source: Dihydropyrimidine dehydrogenase. Chain: a, b, c, d. Synonym: dpd, dhpdhase, dihydrouracil dehydrogenase, dihydrothymine dehydrogenase. Engineered: yes Source:  Sus scrofa. Pig. Organism_taxid: 9823. Organ: liver. Expressed in: escherichia coli dh5[alpha]. Expression_system_taxid: 668369. Biol. unit: Sus scrofa. Pig. Organism_taxid: 9823. Organ: liver. Expressed in: escherichia coli dh5[alpha]. Expression_system_taxid: 668369. Biol. unit:   Dimer (from PDB file) Resolution: Dimer (from PDB file) Resolution:  3.30Å R-factor: 0.214 R-free: 0.269 Authors: 3.30Å R-factor: 0.214 R-free: 0.269 Authors:  D.Dobritzsch,S.Ricagno,G.Schneider,K.D.Schnackerz,Y.Lindqvist Key ref: D.Dobritzsch,S.Ricagno,G.Schneider,K.D.Schnackerz,Y.Lindqvist Key ref:   D.Dobritzsch et al. (2002). Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.J Biol Chem,277, 13155-13166.PubMed id: 11796730 DOI: 10.1074/jbc.M111877200 D.Dobritzsch et al. (2002). Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.J Biol Chem,277, 13155-13166.PubMed id: 11796730 DOI: 10.1074/jbc.M111877200   Date: Date:  14-Jan-02 Release date: 11-Apr-02 14-Jan-02 Release date: 11-Apr-02   PROCHECK PROCHECK    Headers Headers   References References       Protein chains Protein chains                  ? ?       Q28943 (DPYD_PIG) - Dihydropyrimidine dehydrogenase [NADP(+)] from Sus scrofa Q28943 (DPYD_PIG) - Dihydropyrimidine dehydrogenase [NADP(+)] from Sus scrofa   Seq:Struc: Seq:Struc:       Seq:Struc: Seq:Struc:       Seq:Struc: Seq:Struc:    1025 a.a. 1017 a.a.* 1025 a.a. 1017 a.a.*           Key: Key:  PfamA domain PfamA domain    Secondary structure Secondary structure   CATH domain CATH domain  * PDB and UniProt seqs differ at 1 residue position (black cross) * PDB and UniProt seqs differ at 1 residue position (black cross)         Enzyme reactions Enzyme reactions           Enzyme class: Enzyme class:  E.C.1.3.1.2 - dihydropyrimidine dehydrogenase (NADP(+)). [IntEnz] [ExPASy] [KEGG] [BRENDA] E.C.1.3.1.2 - dihydropyrimidine dehydrogenase (NADP(+)). [IntEnz] [ExPASy] [KEGG] [BRENDA]       Reaction: Reaction:  5,6-dihydrouracil + NADP+ = H+ + NADPH + uracil 5,6-dihydrouracil + NADP+ = H+ + NADPH + uracil       5,6-dihydrouracil Bound ligand (Het Group name = UAA) matches with 66.67% similarity + 5,6-dihydrouracil Bound ligand (Het Group name = UAA) matches with 66.67% similarity +  NADP(+) Bound ligand (Het Group name = NDP) corresponds exactly = H(+) + NADP(+) Bound ligand (Het Group name = NDP) corresponds exactly = H(+) +  NADPH + NADPH + 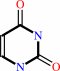 uracil uracil           Molecule diagrams generated from .mol files obtained from theKEGG ftp site Molecule diagrams generated from .mol files obtained from theKEGG ftp site                   reference DOI no: 10.1074/jbc.M111877200 J Biol Chem 277:13155-13166 (2002) PubMed id: 11796730 reference DOI no: 10.1074/jbc.M111877200 J Biol Chem 277:13155-13166 (2002) PubMed id: 11796730  Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. D.Dobritzsch, S.Ricagno, G.Schneider, K.D.Schnackerz, Y.Lindqvist. Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. D.Dobritzsch, S.Ricagno, G.Schneider, K.D.Schnackerz, Y.Lindqvist.  ABSTRACT ABSTRACT   Dihydroprymidine dehydrogenase catalyzes the first and rate-limiting step in pyrimidine degradation by converting pyrimidines to the corresponding 5,6- dihydro compounds. The three-dimensional structures of a binary complex with the inhibitor 5-iodouracil and two ternary complexes with NADPH and the inhibitors 5-iodouracil and uracil-4-acetic acid were determined by x-ray crystallography. In the ternary complexes, NADPH is bound in a catalytically competent fashion, with the nicotinamide ring in a position suitable for hydride transfer to FAD. The structures provide a complete picture of the electron transfer chain from NADPH to the substrate, 5-iodouracil, spanning a distance of 56 A and involving clusters, and FMN as cofactors. The crystallographic analysis further reveals that pyrimidine binding triggers a conformational change of a flexible active-site loop in the alpha/beta-barrel domain, resulting in placement of a catalytically crucial cysteine close to the bound substrate. Loop closure requires physiological pH, which is also necessary for correct binding of NADPH. Binding of the voluminous competitive inhibitor uracil-4-acetic acid prevents loop closure due to steric hindrance. The three-dimensional structure of the ternary complex enzyme-NADPH-5-iodouracil supports the proposal that this compound acts as a mechanism-based inhibitor, covalently modifying the active-site residue Cys-671, resulting in S-(hexahydro-2,4-dioxo-5-pyrimidinyl)cysteine. Dihydroprymidine dehydrogenase catalyzes the first and rate-limiting step in pyrimidine degradation by converting pyrimidines to the corresponding 5,6- dihydro compounds. The three-dimensional structures of a binary complex with the inhibitor 5-iodouracil and two ternary complexes with NADPH and the inhibitors 5-iodouracil and uracil-4-acetic acid were determined by x-ray crystallography. In the ternary complexes, NADPH is bound in a catalytically competent fashion, with the nicotinamide ring in a position suitable for hydride transfer to FAD. The structures provide a complete picture of the electron transfer chain from NADPH to the substrate, 5-iodouracil, spanning a distance of 56 A and involving clusters, and FMN as cofactors. The crystallographic analysis further reveals that pyrimidine binding triggers a conformational change of a flexible active-site loop in the alpha/beta-barrel domain, resulting in placement of a catalytically crucial cysteine close to the bound substrate. Loop closure requires physiological pH, which is also necessary for correct binding of NADPH. Binding of the voluminous competitive inhibitor uracil-4-acetic acid prevents loop closure due to steric hindrance. The three-dimensional structure of the ternary complex enzyme-NADPH-5-iodouracil supports the proposal that this compound acts as a mechanism-based inhibitor, covalently modifying the active-site residue Cys-671, resulting in S-(hexahydro-2,4-dioxo-5-pyrimidinyl)cysteine.    Selected figure(s) Selected figure(s)        Figure 5. Fig. 5. Conformational change of the active-site loop upon ligand binding and pH shift. The figure shows the pyrimidine-binding site in chain A of complex DPD·5IU·NADPH (pH 7.5). The active-site loop (comprising residues 670-682) and the adjacent residues Leu-669 and Ala-683 in the closed conformation (DPD·5IU·NADPH, pH 7.5) are shown in pink, with the loop residues 670, 671, and 673 given as ball-and-stick models (the carbon atoms of the modified Cys-671 (C671*) originating from 5IU are shown in brown; all others in pink). The same loop as observed in ligand-free DPD and DPD·5IU (pH 4.7) after superposition of domain IV with that of complex DPD·5IU·NADPH is shown in cyan. Here side chains are shown only for residues Ser-670 and Cys-671 (carbon atoms in cyan) to indicate their position in the open conformation. Ball-and-stick models of the cofactor FMN, substrate/inhibitor binding residues, and residue Lys-709 are given with carbon atoms in yellow. These residues do not change position or conformation upon ligand binding and pH shift. Hydrogen bond interactions of ligand binding residues Lys-709 and Ser-670 (as observed in DPD·5IU·NADPH (pH 7.5) and DPD·5IU (pH 4.7), respectively) are indicated by dotted lines. Labels in black mark residues placed at identical positions in both structures, and labels in cyan indicate the active-site-loop residues in ligand-free DPD. Active-site-loop residues of complex DPD·5IU·NADPH (pH 7.5) are labeled in pink. Figure 5. Fig. 5. Conformational change of the active-site loop upon ligand binding and pH shift. The figure shows the pyrimidine-binding site in chain A of complex DPD·5IU·NADPH (pH 7.5). The active-site loop (comprising residues 670-682) and the adjacent residues Leu-669 and Ala-683 in the closed conformation (DPD·5IU·NADPH, pH 7.5) are shown in pink, with the loop residues 670, 671, and 673 given as ball-and-stick models (the carbon atoms of the modified Cys-671 (C671*) originating from 5IU are shown in brown; all others in pink). The same loop as observed in ligand-free DPD and DPD·5IU (pH 4.7) after superposition of domain IV with that of complex DPD·5IU·NADPH is shown in cyan. Here side chains are shown only for residues Ser-670 and Cys-671 (carbon atoms in cyan) to indicate their position in the open conformation. Ball-and-stick models of the cofactor FMN, substrate/inhibitor binding residues, and residue Lys-709 are given with carbon atoms in yellow. These residues do not change position or conformation upon ligand binding and pH shift. Hydrogen bond interactions of ligand binding residues Lys-709 and Ser-670 (as observed in DPD·5IU·NADPH (pH 7.5) and DPD·5IU (pH 4.7), respectively) are indicated by dotted lines. Labels in black mark residues placed at identical positions in both structures, and labels in cyan indicate the active-site-loop residues in ligand-free DPD. Active-site-loop residues of complex DPD·5IU·NADPH (pH 7.5) are labeled in pink.  Figure 6. Fig. 6. The electron transfer pathway between the ligand-binding sites. The cofactors FAD and FMN (carbon atoms in cyan) and the four [4Fe-4S] clusters creating the electron transfer chain between both flavins (iron atoms in magenta, sulfur atoms in green) as well as all amino acids located in van der Waals distance (3.8 Å) to the [4Fe-4S] clusters to atoms N5, N1, and C7M of FMN or to atoms N5, N1, and O[2] of FAD are shown as ball-and-stick models. Residues mentioned in the text are labeled. Amino acids with carbon atoms in yellow originate from the same subunit as FAD and FMN. Orange carbon atoms mark residues originating from the other subunit in the dimer. Hydrogen bond interactions mentioned in the text are indicated by dotted lines. For clarity, NADPH and the pyrimidine substrate are not shown. Figure 6. Fig. 6. The electron transfer pathway between the ligand-binding sites. The cofactors FAD and FMN (carbon atoms in cyan) and the four [4Fe-4S] clusters creating the electron transfer chain between both flavins (iron atoms in magenta, sulfur atoms in green) as well as all amino acids located in van der Waals distance (3.8 Å) to the [4Fe-4S] clusters to atoms N5, N1, and C7M of FMN or to atoms N5, N1, and O[2] of FAD are shown as ball-and-stick models. Residues mentioned in the text are labeled. Amino acids with carbon atoms in yellow originate from the same subunit as FAD and FMN. Orange carbon atoms mark residues originating from the other subunit in the dimer. Hydrogen bond interactions mentioned in the text are indicated by dotted lines. For clarity, NADPH and the pyrimidine substrate are not shown.   The above figures are reprinted by permission from the ASBMB: J Biol Chem (2002,277, 13155-13166) copyright 2002. Figures were selected by an automated process. The above figures are reprinted by permission from the ASBMB: J Biol Chem (2002,277, 13155-13166) copyright 2002. Figures were selected by an automated process.              Literature references that cite this PDB file's key reference Literature references that cite this PDB file's key reference  PubMed id PubMed id  Reference Reference      17324113 17324113  X.Zhang, and R.B.Diasio (2007). Regulation of human dihydropyrimidine dehydrogenase: implications in the pharmacogenetics of 5-FU-based chemotherapy. Pharmacogenomics,8, 257-265. X.Zhang, and R.B.Diasio (2007). Regulation of human dihydropyrimidine dehydrogenase: implications in the pharmacogenetics of 5-FU-based chemotherapy. Pharmacogenomics,8, 257-265.      16595627 16595627  A.Osterman (2006). A hidden metabolic pathway exposed. Proc Natl Acad Sci U S A,103, 5637-5638. A.Osterman (2006). A hidden metabolic pathway exposed. Proc Natl Acad Sci U S A,103, 5637-5638.      12703798 12703798  F.Peyrane, J.L.Fourrey, and P.Clivio (2003). Thiation of 2'-deoxy-5,6-dihydropyrimidine nucleosides with Lawesson's reagent: characterisation of oxathiaphosphepane intermediates. Chem Commun (Camb), (), 736-737. F.Peyrane, J.L.Fourrey, and P.Clivio (2003). Thiation of 2'-deoxy-5,6-dihydropyrimidine nucleosides with Lawesson's reagent: characterisation of oxathiaphosphepane intermediates. Chem Commun (Camb), (), 736-737.      12493833 12493833  M.S.Yousef, S.A.Clark, P.K.Pruett, T.Somasundaram, W.R.Ellington, and M.S.Chapman (2003). Induced fit in guanidino kinases--comparison of substrate-free and transition state analog structures of arginine kinase. Protein Sci,12, 103-111. M.S.Yousef, S.A.Clark, P.K.Pruett, T.Somasundaram, W.R.Ellington, and M.S.Chapman (2003). Induced fit in guanidino kinases--comparison of substrate-free and transition state analog structures of arginine kinase. Protein Sci,12, 103-111.  PDB code: PDB code:  1m80 1m80   The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB code is shown on the right. The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB code is shown on the right.  |
 |




 Go to PDB code:
Go to PDB code: 























































 Oxidoreductase PDB id
Oxidoreductase PDB id 
 1gt8
1gt8 















 Loading ...
Loading ... 











 Contents
Contents 




 Protein chains
Protein chains 










 1017 a.a.*
1017 a.a.* 




 Ligands
Ligands 






 SF4 ×16
SF4 ×16 






 FMN ×4
FMN ×4 






 FAD ×4
FAD ×4 






 NDP ×4
NDP ×4 






 UAA ×4
UAA ×4 

 * Residue conservation analysis
* Residue conservation analysis  PDB id:
PDB id:  1gt8
1gt8  Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA PROCOGNATE ProSAT
Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA PROCOGNATE ProSAT  Name:
Name:  Oxidoreductase
Oxidoreductase  Title:
Title:  Dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with NADPH and uracil-4-acetic acid Structure:
Dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with NADPH and uracil-4-acetic acid Structure:  Dihydropyrimidine dehydrogenase. Chain: a, b, c, d. Synonym: dpd, dhpdhase, dihydrouracil dehydrogenase, dihydrothymine dehydrogenase. Engineered: yes Source:
Dihydropyrimidine dehydrogenase. Chain: a, b, c, d. Synonym: dpd, dhpdhase, dihydrouracil dehydrogenase, dihydrothymine dehydrogenase. Engineered: yes Source:  Sus scrofa. Pig. Organism_taxid: 9823. Organ: liver. Expressed in: escherichia coli dh5[alpha]. Expression_system_taxid: 668369. Biol. unit:
Sus scrofa. Pig. Organism_taxid: 9823. Organ: liver. Expressed in: escherichia coli dh5[alpha]. Expression_system_taxid: 668369. Biol. unit: 
 3.30Å R-factor: 0.214 R-free: 0.269 Authors:
3.30Å R-factor: 0.214 R-free: 0.269 Authors:  D.Dobritzsch,S.Ricagno,G.Schneider,K.D.Schnackerz,Y.Lindqvist Key ref:
D.Dobritzsch,S.Ricagno,G.Schneider,K.D.Schnackerz,Y.Lindqvist Key ref:  D.Dobritzsch et al. (2002). Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.J Biol Chem,277, 13155-13166.PubMed id: 11796730 DOI: 10.1074/jbc.M111877200
D.Dobritzsch et al. (2002). Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.J Biol Chem,277, 13155-13166.PubMed id: 11796730 DOI: 10.1074/jbc.M111877200 
 Date:
Date:  14-Jan-02 Release date: 11-Apr-02
14-Jan-02 Release date: 11-Apr-02 
 PROCHECK
PROCHECK 

 Headers
Headers 
 References
References 




 Protein chains
Protein chains 




















 Q28943 (DPYD_PIG) - Dihydropyrimidine dehydrogenase [NADP(+)] from Sus scrofa
Q28943 (DPYD_PIG) - Dihydropyrimidine dehydrogenase [NADP(+)] from Sus scrofa 
 Seq:Struc:
Seq:Struc: 




 Seq:Struc:
Seq:Struc: 




 Seq:Struc:
Seq:Struc: 

 1025 a.a. 1017 a.a.*
1025 a.a. 1017 a.a.* 








 Key:
Key:  PfamA domain
PfamA domain 

 Secondary structure
Secondary structure 
 CATH domain
CATH domain  * PDB and UniProt seqs differ at 1 residue position (black cross)
* PDB and UniProt seqs differ at 1 residue position (black cross) 






 Enzyme reactions
Enzyme reactions 








 Enzyme class:
Enzyme class:  E.C.1.3.1.2 - dihydropyrimidine dehydrogenase (NADP(+)). [IntEnz] [ExPASy] [KEGG] [BRENDA]
E.C.1.3.1.2 - dihydropyrimidine dehydrogenase (NADP(+)). [IntEnz] [ExPASy] [KEGG] [BRENDA] 




 Reaction:
Reaction:  5,6-dihydrouracil + NADP+ = H+ + NADPH + uracil
5,6-dihydrouracil + NADP+ = H+ + NADPH + uracil 













 Molecule diagrams generated from .mol files obtained from theKEGG ftp site
Molecule diagrams generated from .mol files obtained from theKEGG ftp site 
















 reference DOI no: 10.1074/jbc.M111877200 J Biol Chem 277:13155-13166 (2002) PubMed id: 11796730
reference DOI no: 10.1074/jbc.M111877200 J Biol Chem 277:13155-13166 (2002) PubMed id: 11796730  Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. D.Dobritzsch, S.Ricagno, G.Schneider, K.D.Schnackerz, Y.Lindqvist.
Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. D.Dobritzsch, S.Ricagno, G.Schneider, K.D.Schnackerz, Y.Lindqvist.  ABSTRACT
ABSTRACT 
 Dihydroprymidine dehydrogenase catalyzes the first and rate-limiting step in pyrimidine degradation by converting pyrimidines to the corresponding 5,6- dihydro compounds. The three-dimensional structures of a binary complex with the inhibitor 5-iodouracil and two ternary complexes with NADPH and the inhibitors 5-iodouracil and uracil-4-acetic acid were determined by x-ray crystallography. In the ternary complexes, NADPH is bound in a catalytically competent fashion, with the nicotinamide ring in a position suitable for hydride transfer to FAD. The structures provide a complete picture of the electron transfer chain from NADPH to the substrate, 5-iodouracil, spanning a distance of 56 A and involving clusters, and FMN as cofactors. The crystallographic analysis further reveals that pyrimidine binding triggers a conformational change of a flexible active-site loop in the alpha/beta-barrel domain, resulting in placement of a catalytically crucial cysteine close to the bound substrate. Loop closure requires physiological pH, which is also necessary for correct binding of NADPH. Binding of the voluminous competitive inhibitor uracil-4-acetic acid prevents loop closure due to steric hindrance. The three-dimensional structure of the ternary complex enzyme-NADPH-5-iodouracil supports the proposal that this compound acts as a mechanism-based inhibitor, covalently modifying the active-site residue Cys-671, resulting in S-(hexahydro-2,4-dioxo-5-pyrimidinyl)cysteine.
Dihydroprymidine dehydrogenase catalyzes the first and rate-limiting step in pyrimidine degradation by converting pyrimidines to the corresponding 5,6- dihydro compounds. The three-dimensional structures of a binary complex with the inhibitor 5-iodouracil and two ternary complexes with NADPH and the inhibitors 5-iodouracil and uracil-4-acetic acid were determined by x-ray crystallography. In the ternary complexes, NADPH is bound in a catalytically competent fashion, with the nicotinamide ring in a position suitable for hydride transfer to FAD. The structures provide a complete picture of the electron transfer chain from NADPH to the substrate, 5-iodouracil, spanning a distance of 56 A and involving clusters, and FMN as cofactors. The crystallographic analysis further reveals that pyrimidine binding triggers a conformational change of a flexible active-site loop in the alpha/beta-barrel domain, resulting in placement of a catalytically crucial cysteine close to the bound substrate. Loop closure requires physiological pH, which is also necessary for correct binding of NADPH. Binding of the voluminous competitive inhibitor uracil-4-acetic acid prevents loop closure due to steric hindrance. The three-dimensional structure of the ternary complex enzyme-NADPH-5-iodouracil supports the proposal that this compound acts as a mechanism-based inhibitor, covalently modifying the active-site residue Cys-671, resulting in S-(hexahydro-2,4-dioxo-5-pyrimidinyl)cysteine. 

 Selected figure(s)
Selected figure(s) 





 Figure 5. Fig. 5. Conformational change of the active-site loop upon ligand binding and pH shift. The figure shows the pyrimidine-binding site in chain A of complex DPD·5IU·NADPH (pH 7.5). The active-site loop (comprising residues 670-682) and the adjacent residues Leu-669 and Ala-683 in the closed conformation (DPD·5IU·NADPH, pH 7.5) are shown in pink, with the loop residues 670, 671, and 673 given as ball-and-stick models (the carbon atoms of the modified Cys-671 (C671*) originating from 5IU are shown in brown; all others in pink). The same loop as observed in ligand-free DPD and DPD·5IU (pH 4.7) after superposition of domain IV with that of complex DPD·5IU·NADPH is shown in cyan. Here side chains are shown only for residues Ser-670 and Cys-671 (carbon atoms in cyan) to indicate their position in the open conformation. Ball-and-stick models of the cofactor FMN, substrate/inhibitor binding residues, and residue Lys-709 are given with carbon atoms in yellow. These residues do not change position or conformation upon ligand binding and pH shift. Hydrogen bond interactions of ligand binding residues Lys-709 and Ser-670 (as observed in DPD·5IU·NADPH (pH 7.5) and DPD·5IU (pH 4.7), respectively) are indicated by dotted lines. Labels in black mark residues placed at identical positions in both structures, and labels in cyan indicate the active-site-loop residues in ligand-free DPD. Active-site-loop residues of complex DPD·5IU·NADPH (pH 7.5) are labeled in pink.
Figure 5. Fig. 5. Conformational change of the active-site loop upon ligand binding and pH shift. The figure shows the pyrimidine-binding site in chain A of complex DPD·5IU·NADPH (pH 7.5). The active-site loop (comprising residues 670-682) and the adjacent residues Leu-669 and Ala-683 in the closed conformation (DPD·5IU·NADPH, pH 7.5) are shown in pink, with the loop residues 670, 671, and 673 given as ball-and-stick models (the carbon atoms of the modified Cys-671 (C671*) originating from 5IU are shown in brown; all others in pink). The same loop as observed in ligand-free DPD and DPD·5IU (pH 4.7) after superposition of domain IV with that of complex DPD·5IU·NADPH is shown in cyan. Here side chains are shown only for residues Ser-670 and Cys-671 (carbon atoms in cyan) to indicate their position in the open conformation. Ball-and-stick models of the cofactor FMN, substrate/inhibitor binding residues, and residue Lys-709 are given with carbon atoms in yellow. These residues do not change position or conformation upon ligand binding and pH shift. Hydrogen bond interactions of ligand binding residues Lys-709 and Ser-670 (as observed in DPD·5IU·NADPH (pH 7.5) and DPD·5IU (pH 4.7), respectively) are indicated by dotted lines. Labels in black mark residues placed at identical positions in both structures, and labels in cyan indicate the active-site-loop residues in ligand-free DPD. Active-site-loop residues of complex DPD·5IU·NADPH (pH 7.5) are labeled in pink.  Figure 6. Fig. 6. The electron transfer pathway between the ligand-binding sites. The cofactors FAD and FMN (carbon atoms in cyan) and the four [4Fe-4S] clusters creating the electron transfer chain between both flavins (iron atoms in magenta, sulfur atoms in green) as well as all amino acids located in van der Waals distance (3.8 Å) to the [4Fe-4S] clusters to atoms N5, N1, and C7M of FMN or to atoms N5, N1, and O[2] of FAD are shown as ball-and-stick models. Residues mentioned in the text are labeled. Amino acids with carbon atoms in yellow originate from the same subunit as FAD and FMN. Orange carbon atoms mark residues originating from the other subunit in the dimer. Hydrogen bond interactions mentioned in the text are indicated by dotted lines. For clarity, NADPH and the pyrimidine substrate are not shown.
Figure 6. Fig. 6. The electron transfer pathway between the ligand-binding sites. The cofactors FAD and FMN (carbon atoms in cyan) and the four [4Fe-4S] clusters creating the electron transfer chain between both flavins (iron atoms in magenta, sulfur atoms in green) as well as all amino acids located in van der Waals distance (3.8 Å) to the [4Fe-4S] clusters to atoms N5, N1, and C7M of FMN or to atoms N5, N1, and O[2] of FAD are shown as ball-and-stick models. Residues mentioned in the text are labeled. Amino acids with carbon atoms in yellow originate from the same subunit as FAD and FMN. Orange carbon atoms mark residues originating from the other subunit in the dimer. Hydrogen bond interactions mentioned in the text are indicated by dotted lines. For clarity, NADPH and the pyrimidine substrate are not shown. 
 The above figures are reprinted by permission from the ASBMB: J Biol Chem (2002,277, 13155-13166) copyright 2002. Figures were selected by an automated process.
The above figures are reprinted by permission from the ASBMB: J Biol Chem (2002,277, 13155-13166) copyright 2002. Figures were selected by an automated process. 











 Literature references that cite this PDB file's key reference
Literature references that cite this PDB file's key reference  PubMed id
PubMed id  Reference
Reference 



 17324113
17324113  X.Zhang, and R.B.Diasio (2007). Regulation of human dihydropyrimidine dehydrogenase: implications in the pharmacogenetics of 5-FU-based chemotherapy. Pharmacogenomics,8, 257-265.
X.Zhang, and R.B.Diasio (2007). Regulation of human dihydropyrimidine dehydrogenase: implications in the pharmacogenetics of 5-FU-based chemotherapy. Pharmacogenomics,8, 257-265. 



 16595627
16595627  A.Osterman (2006). A hidden metabolic pathway exposed. Proc Natl Acad Sci U S A,103, 5637-5638.
A.Osterman (2006). A hidden metabolic pathway exposed. Proc Natl Acad Sci U S A,103, 5637-5638. 



 12703798
12703798  F.Peyrane, J.L.Fourrey, and P.Clivio (2003). Thiation of 2'-deoxy-5,6-dihydropyrimidine nucleosides with Lawesson's reagent: characterisation of oxathiaphosphepane intermediates. Chem Commun (Camb), (), 736-737.
F.Peyrane, J.L.Fourrey, and P.Clivio (2003). Thiation of 2'-deoxy-5,6-dihydropyrimidine nucleosides with Lawesson's reagent: characterisation of oxathiaphosphepane intermediates. Chem Commun (Camb), (), 736-737. 



 12493833
12493833  M.S.Yousef, S.A.Clark, P.K.Pruett, T.Somasundaram, W.R.Ellington, and M.S.Chapman (2003). Induced fit in guanidino kinases--comparison of substrate-free and transition state analog structures of arginine kinase. Protein Sci,12, 103-111.
M.S.Yousef, S.A.Clark, P.K.Pruett, T.Somasundaram, W.R.Ellington, and M.S.Chapman (2003). Induced fit in guanidino kinases--comparison of substrate-free and transition state analog structures of arginine kinase. Protein Sci,12, 103-111.  PDB code:
PDB code:  1m80
1m80 
 The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB code is shown on the right.
The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB code is shown on the right. 
