 |
 |
PDBsum entry 1kqg    Go to PDB code: Go to PDB code:                                                                Oxidoreductase PDB id Oxidoreductase PDB id   1kqg 1kqg                  Loading ... Loading ...              Contents Contents       Protein chains Protein chains          982 a.a.* 982 a.a.*          289 a.a.* 289 a.a.*          216 a.a.* 216 a.a.*       Ligands Ligands         SF4 ×5 SF4 ×5         MGD ×2 MGD ×2         CDL CDL         HEM ×2 HEM ×2         HQO HQO       Metals Metals          6MO 6MO       Waters ×1984 Waters ×1984    * Residue conservation analysis * Residue conservation analysis  PDB id: PDB id:  1kqg 1kqg  Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT  Name: Name:  Oxidoreductase Oxidoreductase  Title: Title:  Formate dehydrogenase n from e. Coli Structure: Formate dehydrogenase n from e. Coli Structure:  Formate dehydrogenase, nitrate-inducible, major subunit. Chain: a. Synonym: formate dehydrogenase-n alpha subunit. Fdh-n alpha subunit. Anaerobic formate dehydrogenase major subunit. Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit. Chain: b. Synonym: formate dehydrogenase-n beta subunit. Fdh-n beta subunit. Anaerobic formate dehydrogenase iron-sulfur subunit. Source: Formate dehydrogenase, nitrate-inducible, major subunit. Chain: a. Synonym: formate dehydrogenase-n alpha subunit. Fdh-n alpha subunit. Anaerobic formate dehydrogenase major subunit. Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit. Chain: b. Synonym: formate dehydrogenase-n beta subunit. Fdh-n beta subunit. Anaerobic formate dehydrogenase iron-sulfur subunit. Source:  Escherichia coli. Organism_taxid: 562. Strain: gl101. Strain: gl101 Biol. unit: Escherichia coli. Organism_taxid: 562. Strain: gl101. Strain: gl101 Biol. unit:   Nonamer (from PDB file) Resolution: Nonamer (from PDB file) Resolution:  2.80Å R-factor: 0.198 R-free: 0.239 Authors: 2.80Å R-factor: 0.198 R-free: 0.239 Authors:  M.Jormakka,S.Tornroth,B.Byrne,S.Iwata Key ref: M.Jormakka,S.Tornroth,B.Byrne,S.Iwata Key ref:   M.Jormakka et al. (2002). Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.Science,295, 1863-1868.PubMed id: 11884747 DOI: 10.1126/science.1068186 M.Jormakka et al. (2002). Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.Science,295, 1863-1868.PubMed id: 11884747 DOI: 10.1126/science.1068186   Date: Date:  05-Jan-02 Release date: 15-Mar-02 05-Jan-02 Release date: 15-Mar-02   PROCHECK PROCHECK    Headers Headers   References References       Protein chain Protein chain               ? ?       P24183 (FDNG_ECOLI) - Formate dehydrogenase, nitrate-inducible, major subunit from Escherichia coli (strain K12) P24183 (FDNG_ECOLI) - Formate dehydrogenase, nitrate-inducible, major subunit from Escherichia coli (strain K12)   Seq:Struc: Seq:Struc:       Seq:Struc: Seq:Struc:    1015 a.a. 982 a.a.* 1015 a.a. 982 a.a.*                 Protein chain Protein chain               ? ?       P0AAJ3 (FDNH_ECOLI) - Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit from Escherichia coli (strain K12) P0AAJ3 (FDNH_ECOLI) - Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit from Escherichia coli (strain K12)   Seq:Struc: Seq:Struc:    294 a.a. 289 a.a. 294 a.a. 289 a.a.                 Protein chain Protein chain               ? ?       P0AEK7 (FDNI_ECOLI) - Formate dehydrogenase, nitrate-inducible, cytochrome b556(Fdn) subunit from Escherichia coli (strain K12) P0AEK7 (FDNI_ECOLI) - Formate dehydrogenase, nitrate-inducible, cytochrome b556(Fdn) subunit from Escherichia coli (strain K12)   Seq:Struc: Seq:Struc:    217 a.a. 216 a.a. 217 a.a. 216 a.a.           Key: Key:  PfamA domain PfamA domain    Secondary structure Secondary structure   CATH domain CATH domain  * PDB and UniProt seqs differ at 1 residue position (black cross) * PDB and UniProt seqs differ at 1 residue position (black cross)         Enzyme reactions Enzyme reactions           Enzyme class 2: Enzyme class 2:  Chain A: E.C.1.17.5.3 - formate dehydrogenase-N. [IntEnz] [ExPASy] [KEGG] [BRENDA] Chain A: E.C.1.17.5.3 - formate dehydrogenase-N. [IntEnz] [ExPASy] [KEGG] [BRENDA]       Reaction: Reaction:  a quinone + formate + H+ = a quinol + CO2 a quinone + formate + H+ = a quinol + CO2       quinone + quinone +  formate + H(+) = quinol + formate + H(+) = quinol +  CO2 CO2         Cofactor: Cofactor:  Heme; Iron-sulfur; Molybdopterin guanine dinucleotide Heme; Iron-sulfur; Molybdopterin guanine dinucleotide      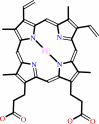 Heme Bound ligand (Het Group name = HEM) matches with 95.45% similarity Heme Bound ligand (Het Group name = HEM) matches with 95.45% similarity  Iron-sulfur Molybdopterin guanine dinucleotide Iron-sulfur Molybdopterin guanine dinucleotide   Enzyme class 3: Enzyme class 3:  Chains B, C: E.C.1.2.1.2 - Transferred entry: 1.17.1.9. [IntEnz] [ExPASy] [KEGG] [BRENDA] Chains B, C: E.C.1.2.1.2 - Transferred entry: 1.17.1.9. [IntEnz] [ExPASy] [KEGG] [BRENDA]       Reaction: Reaction:  Formate + NAD+ = CO2 + NADH Formate + NAD+ = CO2 + NADH       Formate + Formate +  NAD(+) Bound ligand (Het Group name = MGD) matches with 75.00% similarity = NAD(+) Bound ligand (Het Group name = MGD) matches with 75.00% similarity =  CO(2) + CO(2) +  NADH NADH         Cofactor: Cofactor:  Flavin; Iron-sulfur; Mo cation Flavin; Iron-sulfur; Mo cation       Flavin Flavin  Iron-sulfur Mo cation Iron-sulfur Mo cation     Note, where more than one E.C. class is given (as above), each may correspond to a different protein domain or, in the case of polyprotein precursors, to a different mature protein. Note, where more than one E.C. class is given (as above), each may correspond to a different protein domain or, in the case of polyprotein precursors, to a different mature protein.  Molecule diagrams generated from .mol files obtained from theKEGG ftp site Molecule diagrams generated from .mol files obtained from theKEGG ftp site                   reference DOI no: 10.1126/science.1068186 Science 295:1863-1868 (2002) PubMed id: 11884747 reference DOI no: 10.1126/science.1068186 Science 295:1863-1868 (2002) PubMed id: 11884747  Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. M.Jormakka, S.Törnroth, B.Byrne, S.Iwata. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. M.Jormakka, S.Törnroth, B.Byrne, S.Iwata.  ABSTRACT ABSTRACT   The structure of the membrane protein formate dehydrogenase-N (Fdn-N), a major component of Escherichia coli nitrate respiration, has been determined at 1.6 angstroms. The structure demonstrates 11 redox centers, including molybdopterin-guanine dinucleotides, five [4Fe-4S] clusters, two heme b groups, and a menaquinone analog. These redox centers are aligned in a single chain, which extends almost 90 angstroms through the enzyme. The menaquinone reduction site associated with a possible proton pathway was also characterized. This structure provides critical insights into the proton motive force generation by redox loop, a common mechanism among a wide range of respiratory enzymes. The structure of the membrane protein formate dehydrogenase-N (Fdn-N), a major component of Escherichia coli nitrate respiration, has been determined at 1.6 angstroms. The structure demonstrates 11 redox centers, including molybdopterin-guanine dinucleotides, five [4Fe-4S] clusters, two heme b groups, and a menaquinone analog. These redox centers are aligned in a single chain, which extends almost 90 angstroms through the enzyme. The menaquinone reduction site associated with a possible proton pathway was also characterized. This structure provides critical insights into the proton motive force generation by redox loop, a common mechanism among a wide range of respiratory enzymes.    Selected figure(s) Selected figure(s)        Figure 1. Fig. 1. A proposal for how proton motive force is generated by Fdh-N and nitrate reductase in Escherichia coli inner membrane. MK, menaquinone; MKH[2], reduced form of menaquinone; MGD, molybdopterin-guanine dinucleotide; b, heme b; FeS, iron-sulfur cluster. Figure 1. Fig. 1. A proposal for how proton motive force is generated by Fdh-N and nitrate reductase in Escherichia coli inner membrane. MK, menaquinone; MKH[2], reduced form of menaquinone; MGD, molybdopterin-guanine dinucleotide; b, heme b; FeS, iron-sulfur cluster.  Figure 2. Fig. 2. Overall structure of Fdh-N. The figures are based on the native Fdh-N structure except for the position of HQNO, which is determined in the Fdh-N and HQNO complex structure. The Figure 2. Fig. 2. Overall structure of Fdh-N. The figures are based on the native Fdh-N structure except for the position of HQNO, which is determined in the Fdh-N and HQNO complex structure. The , ,  , and , and  subunits are shown in brown, blue, and magenta, respectively. Heme groups and MGD cofactors are shown in green, and Mo, cardiolipin (CL), and HQNO are colored in magenta, yellow, and navy, respectively. Five [4Fe-4S] clusters are painted as red (Fe atoms) and yellow (S atoms). (A) Fdh-N trimer viewed parallel to the membrane. (B) Fdh-N trimer viewed from the periplasm along the membrane normal. (C) View of the Fdh-N monomer parallel to the membrane. Center-to-center and edge-to-edge (in parentheses) (12) distances in angstroms between each of the redox centers are also shown. The edge-to-edge distance for HQNO to heme b[C] is 0 because HQNO is directly binding to the histidine ligand of heme b[C]. All figures were made with MOLSCRIPT (28) and BOBSCRIPT (29) and rendered with RASTER3D (30). subunits are shown in brown, blue, and magenta, respectively. Heme groups and MGD cofactors are shown in green, and Mo, cardiolipin (CL), and HQNO are colored in magenta, yellow, and navy, respectively. Five [4Fe-4S] clusters are painted as red (Fe atoms) and yellow (S atoms). (A) Fdh-N trimer viewed parallel to the membrane. (B) Fdh-N trimer viewed from the periplasm along the membrane normal. (C) View of the Fdh-N monomer parallel to the membrane. Center-to-center and edge-to-edge (in parentheses) (12) distances in angstroms between each of the redox centers are also shown. The edge-to-edge distance for HQNO to heme b[C] is 0 because HQNO is directly binding to the histidine ligand of heme b[C]. All figures were made with MOLSCRIPT (28) and BOBSCRIPT (29) and rendered with RASTER3D (30).   The above figures are reprinted by permission from the AAAs: Science (2002,295, 1863-1868) copyright 2002. Figures were selected by an automated process. The above figures are reprinted by permission from the AAAs: Science (2002,295, 1863-1868) copyright 2002. Figures were selected by an automated process.              Literature references that cite this PDB file's key reference Literature references that cite this PDB file's key reference  PubMed id PubMed id  Reference Reference      22683878 22683878  T.Palmer, and B.C.Berks (2012). The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol,10, 483-496. T.Palmer, and B.C.Berks (2012). The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol,10, 483-496.      21289038 21289038  J.Speck, K.M.Arndt, and K.M.Müller (2011). Efficient phage display of intracellularly folded proteins mediated by the TAT pathway. Protein Eng Des Sel,24, 473-484. J.Speck, K.M.Arndt, and K.M.Müller (2011). Efficient phage display of intracellularly folded proteins mediated by the TAT pathway. Protein Eng Des Sel,24, 473-484.      20938980 20938980  K.Illergård, A.Kauko, and A.Elofsson (2011). Why are polar residues within the membrane core evolutionary conserved? Proteins,79, 79-91. K.Illergård, A.Kauko, and A.Elofsson (2011). Why are polar residues within the membrane core evolutionary conserved? Proteins,79, 79-91.      21518899 21518899  R.Arias-Cartin, S.Grimaldi, J.Pommier, P.Lanciano, C.Schaefer, P.Arnoux, G.Giordano, B.Guigliarelli, and A.Magalon (2011). Cardiolipin-based respiratory complex activation in bacteria. Proc Natl Acad Sci U S A,108, 7781-7786. R.Arias-Cartin, S.Grimaldi, J.Pommier, P.Lanciano, C.Schaefer, P.Arnoux, G.Giordano, B.Guigliarelli, and A.Magalon (2011). Cardiolipin-based respiratory complex activation in bacteria. Proc Natl Acad Sci U S A,108, 7781-7786.      20819103 20819103  X.Zhang, E.G.Matson, and J.R.Leadbetter (2011). Genes for selenium dependent and independent formate dehydrogenase in the gut microbial communities of three lower, wood-feeding termites and a wood-feeding roach. Environ Microbiol,13, 307-323. X.Zhang, E.G.Matson, and J.R.Leadbetter (2011). Genes for selenium dependent and independent formate dehydrogenase in the gut microbial communities of three lower, wood-feeding termites and a wood-feeding roach. Environ Microbiol,13, 307-323.      20645325 20645325  E.Cremades, J.Echeverría, and S.Alvarez (2010). The trigonal prism in coordination chemistry. Chemistry,16, 10380-10396. E.Cremades, J.Echeverría, and S.Alvarez (2010). The trigonal prism in coordination chemistry. Chemistry,16, 10380-10396.      19826804 19826804  K.McLuskey, A.W.Roszak, Y.Zhu, and N.W.Isaacs (2010). Crystal structures of all-alpha type membrane proteins. Eur Biophys J,39, 723-755. K.McLuskey, A.W.Roszak, Y.Zhu, and N.W.Isaacs (2010). Crystal structures of all-alpha type membrane proteins. Eur Biophys J,39, 723-755.      20667175 20667175  K.R.Vinothkumar, and R.Henderson (2010). Structures of membrane proteins. Q Rev Biophys,43, 65. K.R.Vinothkumar, and R.Henderson (2010). Structures of membrane proteins. Q Rev Biophys,43, 65.      20352202 20352202  M.Raja (2010). The role of phosphatidic acid and cardiolipin in stability of the tetrameric assembly of potassium channel KcsA. J Membr Biol,234, 235-240. M.Raja (2010). The role of phosphatidic acid and cardiolipin in stability of the tetrameric assembly of potassium channel KcsA. J Membr Biol,234, 235-240.      20406421 20406421  N.Borgese, and M.Righi (2010). Remote origins of tail-anchored proteins. Traffic,11, 877-885. N.Borgese, and M.Righi (2010). Remote origins of tail-anchored proteins. Traffic,11, 877-885.      20587053 20587053  T.E.Meyer, J.A.Kyndt, and M.A.Cusanovich (2010). Occurrence and sequence of Sphaeroides Heme Protein and diheme cytochrome C in purple photosynthetic bacteria in the family Rhodobacteraceae. BMC Biochem,11, 24. T.E.Meyer, J.A.Kyndt, and M.A.Cusanovich (2010). Occurrence and sequence of Sphaeroides Heme Protein and diheme cytochrome C in purple photosynthetic bacteria in the family Rhodobacteraceae. BMC Biochem,11, 24.      20479269 20479269  V.A.Gold, A.Robson, H.Bao, T.Romantsov, F.Duong, and I.Collinson (2010). The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci U S A,107, 10044-10049. V.A.Gold, A.Robson, H.Bao, T.Romantsov, F.Duong, and I.Collinson (2010). The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci U S A,107, 10044-10049.      19658738 19658738  A.Y.Smirnov, S.E.Savel'ev, and F.Nori (2009). Diffusion-controlled generation of a proton-motive force across a biomembrane. Phys Rev E Stat Nonlin Soft Matter Phys,80, 011916. A.Y.Smirnov, S.E.Savel'ev, and F.Nori (2009). Diffusion-controlled generation of a proton-motive force across a biomembrane. Phys Rev E Stat Nonlin Soft Matter Phys,80, 011916.      19371718 19371718  E.Mileykovskaya, and W.Dowhan (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta,1788, 2084-2091. E.Mileykovskaya, and W.Dowhan (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta,1788, 2084-2091.      19536822 19536822  G.Zoppellaro, K.L.Bren, A.A.Ensign, E.Harbitz, R.Kaur, H.P.Hersleth, U.Ryde, L.Hederstedt, and K.K.Andersson (2009). Review: studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers,91, 1064-1082. G.Zoppellaro, K.L.Bren, A.A.Ensign, E.Harbitz, R.Kaur, H.P.Hersleth, U.Ryde, L.Hederstedt, and K.K.Andersson (2009). Review: studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers,91, 1064-1082.      19452052 19452052  M.J.Romão (2009). Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans, (), 4053-4068. M.J.Romão (2009). Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans, (), 4053-4068.      19206188 19206188  S.Groysman, and R.H.Holm (2009). Biomimetic chemistry of iron, nickel, molybdenum, and tungsten in sulfur-ligated protein sites. Biochemistry,48, 2310-2320. S.Groysman, and R.H.Holm (2009). Biomimetic chemistry of iron, nickel, molybdenum, and tungsten in sulfur-ligated protein sites. Biochemistry,48, 2310-2320.      19416061 19416061  S.Raunser, and T.Walz (2009). Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys,38, 89. S.Raunser, and T.Walz (2009). Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys,38, 89.      19233927 19233927  X.Li, Q.Luo, N.Q.Wofford, K.L.Keller, M.J.McInerney, J.D.Wall, and L.R.Krumholz (2009). A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J Bacteriol,191, 2675-2682. X.Li, Q.Luo, N.Q.Wofford, K.L.Keller, M.J.McInerney, J.D.Wall, and L.R.Krumholz (2009). A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J Bacteriol,191, 2675-2682.      18658261 18658261  B.S.Pickering, and I.J.Oresnik (2008). Formate-dependent autotrophic growth in Sinorhizobium meliloti. J Bacteriol,190, 6409-6418. B.S.Pickering, and I.J.Oresnik (2008). Formate-dependent autotrophic growth in Sinorhizobium meliloti. J Bacteriol,190, 6409-6418.      18615097 18615097  C.F.Matos, C.Robinson, and A.Di Cola (2008). The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J,27, 2055-2063. C.F.Matos, C.Robinson, and A.Di Cola (2008). The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J,27, 2055-2063.      18247347 18247347  C.H.Lu, S.W.Huang, Y.L.Lai, C.P.Lin, C.H.Shih, C.C.Huang, W.L.Hsu, and J.K.Hwang (2008). On the relationship between the protein structure and protein dynamics. Proteins,72, 625-634. C.H.Lu, S.W.Huang, Y.L.Lai, C.P.Lin, C.H.Shih, C.C.Huang, W.L.Hsu, and J.K.Hwang (2008). On the relationship between the protein structure and protein dynamics. Proteins,72, 625-634.      18418633 18418633  E.A.Berry, and F.A.Walker (2008). Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem,13, 481-498. E.A.Berry, and F.A.Walker (2008). Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem,13, 481-498.      18535145 18535145  G.J.Workun, K.Moquin, R.A.Rothery, and J.H.Weiner (2008). Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold. Microbiol Mol Biol Rev,72, 228. G.J.Workun, K.Moquin, R.A.Rothery, and J.H.Weiner (2008). Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold. Microbiol Mol Biol Rev,72, 228.      18716757 18716757  I.Lüke, G.Butland, K.Moore, G.Buchanan, V.Lyall, S.A.Fairhurst, J.F.Greenblatt, A.Emili, T.Palmer, and F.Sargent (2008). Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch Microbiol,190, 685-696. I.Lüke, G.Butland, K.Moore, G.Buchanan, V.Lyall, S.A.Fairhurst, J.F.Greenblatt, A.Emili, T.Palmer, and F.Sargent (2008). Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch Microbiol,190, 685-696.  18751913 18751913  M.Bogdanov, E.Mileykovskaya, and W.Dowhan (2008). Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem,49, 197-239. M.Bogdanov, E.Mileykovskaya, and W.Dowhan (2008). Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem,49, 197-239.      18536726 18536726  M.Jormakka, K.Yokoyama, T.Yano, M.Tamakoshi, S.Akimoto, T.Shimamura, P.Curmi, and S.Iwata (2008). Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol,15, 730-737. M.Jormakka, K.Yokoyama, T.Yano, M.Tamakoshi, S.Akimoto, T.Shimamura, P.Curmi, and S.Iwata (2008). Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol,15, 730-737.  PDB codes: PDB codes:  2vpw 2vpx 2vpy 2vpz 2vpw 2vpx 2vpy 2vpz     18077827 18077827  M.Schlame (2008). Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res,49, 1607-1620. M.Schlame (2008). Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res,49, 1607-1620.      18500332 18500332  N.H.Joh, A.Min, S.Faham, J.P.Whitelegge, D.Yang, V.L.Woods, and J.U.Bowie (2008). Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature,453, 1266-1270. N.H.Joh, A.Min, S.Faham, J.P.Whitelegge, D.Yang, V.L.Woods, and J.U.Bowie (2008). Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature,453, 1266-1270.  PDB codes: PDB codes:  3coc 3cod 3coc 3cod     18218713 18218713  S.Newstead, S.Ferrandon, and S.Iwata (2008). Rationalizing alpha-helical membrane protein crystallization. Protein Sci,17, 466-472. S.Newstead, S.Ferrandon, and S.Iwata (2008). Rationalizing alpha-helical membrane protein crystallization. Protein Sci,17, 466-472.      18667702 18667702  T.Reda, C.M.Plugge, N.J.Abram, and J.Hirst (2008). Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc Natl Acad Sci U S A,105, 10654-10658. T.Reda, C.M.Plugge, N.J.Abram, and J.Hirst (2008). Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc Natl Acad Sci U S A,105, 10654-10658.      18501187 18501187  W.Liu, C.E.Rogge, G.F.da Silva, V.P.Shinkarev, A.L.Tsai, Y.Kamensky, G.Palmer, and R.J.Kulmacz (2008). His92 and His110 selectively affect different heme centers of adrenal cytochrome b(561). Biochim Biophys Acta,1777, 1218-1228. W.Liu, C.E.Rogge, G.F.da Silva, V.P.Shinkarev, A.L.Tsai, Y.Kamensky, G.Palmer, and R.J.Kulmacz (2008). His92 and His110 selectively affect different heme centers of adrenal cytochrome b(561). Biochim Biophys Acta,1777, 1218-1228.      17600788 17600788  K.Pal, P.K.Chaudhury, and S.Sarkar (2007). Structure of the Michaelis complex and function of the catalytic center in the reductive half-reaction of computational and synthetic models of sulfite oxidase. Chem Asian J,2, 956-964. K.Pal, P.K.Chaudhury, and S.Sarkar (2007). Structure of the Michaelis complex and function of the catalytic center in the reductive half-reaction of computational and synthetic models of sulfite oxidase. Chem Asian J,2, 956-964.      17557793 17557793  N.Buzhynskyy, P.Sens, V.Prima, J.N.Sturgis, and S.Scheuring (2007). Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J,93, 2870-2876. N.Buzhynskyy, P.Sens, V.Prima, J.N.Sturgis, and S.Scheuring (2007). Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J,93, 2870-2876.      16962969 16962969  D.P.Kloer, C.Hagel, J.Heider, and G.E.Schulz (2006). Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure,14, 1377-1388. D.P.Kloer, C.Hagel, J.Heider, and G.E.Schulz (2006). Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure,14, 1377-1388.  PDB code: PDB code:  2ivf 2ivf     16830149 16830149  H.C.Raaijmakers, and M.J.Romão (2006). Formate-reduced E. coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem,11, 849-854. H.C.Raaijmakers, and M.J.Romão (2006). Formate-reduced E. coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem,11, 849-854.  PDB code: PDB code:  2iv2 2iv2     16885262 16885262  H.Ridley, C.A.Watts, D.J.Richardson, and C.S.Butler (2006). Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl Environ Microbiol,72, 5173-5180. H.Ridley, C.A.Watts, D.J.Richardson, and C.S.Butler (2006). Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl Environ Microbiol,72, 5173-5180.      17024183 17024183  M.G.Madej, H.R.Nasiri, N.S.Hilgendorff, H.Schwalbe, and C.R.Lancaster (2006). Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J,25, 4963-4970. M.G.Madej, H.R.Nasiri, N.S.Hilgendorff, H.Schwalbe, and C.R.Lancaster (2006). Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J,25, 4963-4970.  PDB code: PDB code:  2bs2 2bs2     16802174 16802174  M.John, R.P.Schmitz, M.Westermann, W.Richter, and G.Diekert (2006). Growth substrate dependent localization of tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch Microbiol,186, 99. M.John, R.P.Schmitz, M.Westermann, W.Richter, and G.Diekert (2006). Growth substrate dependent localization of tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch Microbiol,186, 99.      17139260 17139260  M.L.Rodrigues, T.F.Oliveira, I.A.Pereira, and M.Archer (2006). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J,25, 5951-5960. M.L.Rodrigues, T.F.Oliveira, I.A.Pereira, and M.Archer (2006). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J,25, 5951-5960.  PDB code: PDB code:  2j7a 2j7a     16597999 16597999  S.S.Ruebush, S.L.Brantley, and M.Tien (2006). Reduction of soluble and insoluble iron forms by membrane fractions of Shewanella oneidensis grown under aerobic and anaerobic conditions. Appl Environ Microbiol,72, 2925-2935. S.S.Ruebush, S.L.Brantley, and M.Tien (2006). Reduction of soluble and insoluble iron forms by membrane fractions of Shewanella oneidensis grown under aerobic and anaerobic conditions. Appl Environ Microbiol,72, 2925-2935.      16433558 16433558  T.Teschner, L.Yatsunyk, V.Schünemann, H.Paulsen, H.Winkler, C.Hu, W.R.Scheidt, F.A.Walker, and A.X.Trautwein (2006). Models of the membrane-bound cytochromes: mössbauer spectra of crystalline low-spin ferriheme complexes having axial ligand plane dihedral angles ranging from 0 degree to 90 degrees. J Am Chem Soc,128, 1379-1389. T.Teschner, L.Yatsunyk, V.Schünemann, H.Paulsen, H.Winkler, C.Hu, W.R.Scheidt, F.A.Walker, and A.X.Trautwein (2006). Models of the membrane-bound cytochromes: mössbauer spectra of crystalline low-spin ferriheme complexes having axial ligand plane dihedral angles ranging from 0 degree to 90 degrees. J Am Chem Soc,128, 1379-1389.      16172928 16172928  A.J.Watson, A.V.Hughes, P.K.Fyfe, M.C.Wakeham, K.Holden-Dye, P.Heathcote, and M.R.Jones (2005). On the role of basic residues in adapting the reaction centre-LH1 complex for growth at elevated temperatures in purple bacteria. Photosynth Res,86, 81. A.J.Watson, A.V.Hughes, P.K.Fyfe, M.C.Wakeham, K.Holden-Dye, P.Heathcote, and M.R.Jones (2005). On the role of basic residues in adapting the reaction centre-LH1 complex for growth at elevated temperatures in purple bacteria. Photosynth Res,86, 81.      15802249 15802249  B.C.Berks, T.Palmer, and F.Sargent (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol,8, 174-181. B.C.Berks, T.Palmer, and F.Sargent (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol,8, 174-181.      16380425 16380425  C.R.Lancaster, U.S.Sauer, R.Gross, A.H.Haas, J.Graf, H.Schwalbe, W.Mäntele, J.Simon, and M.G.Madej (2005). Experimental support for the "E pathway hypothesis" of coupled transmembrane e- and H+ transfer in dihemic quinol:fumarate reductase. Proc Natl Acad Sci U S A,102, 18860-18865. C.R.Lancaster, U.S.Sauer, R.Gross, A.H.Haas, J.Graf, H.Schwalbe, W.Mäntele, J.Simon, and M.G.Madej (2005). Experimental support for the "E pathway hypothesis" of coupled transmembrane e- and H+ transfer in dihemic quinol:fumarate reductase. Proc Natl Acad Sci U S A,102, 18860-18865.  PDB codes: PDB codes:  2bs3 2bs4 2bs3 2bs4     15819632 15819632  M.L.Crouch, L.A.Becker, I.S.Bang, H.Tanabe, A.J.Ouellette, and F.C.Fang (2005). The alternative sigma factor sigma is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol Microbiol,56, 789-799. M.L.Crouch, L.A.Becker, I.S.Bang, H.Tanabe, A.J.Ouellette, and F.C.Fang (2005). The alternative sigma factor sigma is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol Microbiol,56, 789-799.      15949761 15949761  P.K.Fyfe, A.V.Hughes, P.Heathcote, and M.R.Jones (2005). Proteins, chlorophylls and lipids: X-ray analysis of a three-way relationship. Trends Plant Sci,10, 275-282. P.K.Fyfe, A.V.Hughes, P.Heathcote, and M.R.Jones (2005). Proteins, chlorophylls and lipids: X-ray analysis of a three-way relationship. Trends Plant Sci,10, 275-282.      15654871 15654871  R.A.Rothery, A.M.Seime, A.M.Spiers, E.Maklashina, I.Schröder, R.P.Gunsalus, G.Cecchini, and J.H.Weiner (2005). Defining the Q-site of Escherichia coli fumarate reductase by site-directed mutagenesis, fluorescence quench titrations and EPR spectroscopy. FEBS J,272, 313-326. R.A.Rothery, A.M.Seime, A.M.Spiers, E.Maklashina, I.Schröder, R.P.Gunsalus, G.Cecchini, and J.H.Weiner (2005). Defining the Q-site of Escherichia coli fumarate reductase by site-directed mutagenesis, fluorescence quench titrations and EPR spectroscopy. FEBS J,272, 313-326.      14681400 14681400  A.Andreeva, D.Howorth, S.E.Brenner, T.J.Hubbard, C.Chothia, and A.G.Murzin (2004). SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res,32, D226-D229. A.Andreeva, D.Howorth, S.E.Brenner, T.J.Hubbard, C.Chothia, and A.G.Murzin (2004). SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res,32, D226-D229.      15516557 15516557  A.C.Fisher, and M.P.DeLisa (2004). A little help from my friends: quality control of presecretory proteins in bacteria. J Bacteriol,186, 7467-7473. A.C.Fisher, and M.P.DeLisa (2004). A little help from my friends: quality control of presecretory proteins in bacteria. J Bacteriol,186, 7467-7473.      15284442 15284442  A.Messerschmidt, H.Niessen, D.Abt, O.Einsle, B.Schink, and P.M.Kroneck (2004). Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc Natl Acad Sci U S A,101, 11571-11576. A.Messerschmidt, H.Niessen, D.Abt, O.Einsle, B.Schink, and P.M.Kroneck (2004). Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc Natl Acad Sci U S A,101, 11571-11576.  PDB codes: PDB codes:  1ti2 1ti4 1ti6 1vld 1vle 1vlf 1ti2 1ti4 1ti6 1vld 1vle 1vlf     15342602 15342602  C.Punginelli, B.Ize, N.R.Stanley, V.Stewart, G.Sawers, B.C.Berks, and T.Palmer (2004). mRNA secondary structure modulates translation of Tat-dependent formate dehydrogenase N. J Bacteriol,186, 6311-6315. C.Punginelli, B.Ize, N.R.Stanley, V.Stewart, G.Sawers, B.C.Berks, and T.Palmer (2004). mRNA secondary structure modulates translation of Tat-dependent formate dehydrogenase N. J Bacteriol,186, 6311-6315.      15028701 15028701  F.Peters, M.Rother, and M.Boll (2004). Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J Bacteriol,186, 2156-2163. F.Peters, M.Rother, and M.Boll (2004). Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J Bacteriol,186, 2156-2163.      15311335 15311335  J.J.Moura, C.D.Brondino, J.Trincão, and M.J.Romão (2004). Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J Biol Inorg Chem,9, 791-799. J.J.Moura, C.D.Brondino, J.Trincão, and M.J.Romão (2004). Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J Biol Inorg Chem,9, 791-799.      15551861 15551861  O.Einsle, and P.M.Kroneck (2004). Structural basis of denitrification. Biol Chem,385, 875-883. O.Einsle, and P.M.Kroneck (2004). Structural basis of denitrification. Biol Chem,385, 875-883.      15321667 15321667  R.P.Schmitz, and G.Diekert (2004). The fdh operon of Sulfurospirillum multivorans. FEMS Microbiol Lett,237, 235-242. R.P.Schmitz, and G.Diekert (2004). The fdh operon of Sulfurospirillum multivorans. FEMS Microbiol Lett,237, 235-242.      15498942 15498942  T.J.Stevens, K.Mizuguchi, and I.T.Arkin (2004). Distinct protein interfaces in transmembrane domains suggest an in vivo folding model. Protein Sci,13, 3028-3037. T.J.Stevens, K.Mizuguchi, and I.T.Arkin (2004). Distinct protein interfaces in transmembrane domains suggest an in vivo folding model. Protein Sci,13, 3028-3037.      15486691 15486691  T.Tomiki, and N.Saitou (2004). Phylogenetic analysis of proteins associated in the four major energy metabolism systems: photosynthesis, aerobic respiration, denitrification, and sulfur respiration. J Mol Evol,59, 158-176. T.Tomiki, and N.Saitou (2004). Phylogenetic analysis of proteins associated in the four major energy metabolism systems: photosynthesis, aerobic respiration, denitrification, and sulfur respiration. J Mol Evol,59, 158-176.      12594934 12594934  F.Baymann, E.Lebrun, M.Brugna, B.Schoepp-Cothenet, M.T.Giudici-Orticoni, and W.Nitschke (2003). The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci,358, 267-274. F.Baymann, E.Lebrun, M.Brugna, B.Schoepp-Cothenet, M.T.Giudici-Orticoni, and W.Nitschke (2003). The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci,358, 267-274.      12823811 12823811  J.Simon, M.Sänger, S.C.Schuster, and R.Gross (2003). Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol Microbiol,49, 69-79. J.Simon, M.Sänger, S.C.Schuster, and R.Gross (2003). Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol Microbiol,49, 69-79.      12940994 12940994  K.Hatzixanthis, T.Palmer, and F.Sargent (2003). A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol,49, 1377-1390. K.Hatzixanthis, T.Palmer, and F.Sargent (2003). A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol,49, 1377-1390.      14511372 14511372  M.G.Almeida, S.Macieira, L.L.Gonçalves, R.Huber, C.A.Cunha, M.J.Romão, C.Costa, J.Lampreia, J.J.Moura, and I.Moura (2003). The isolation and characterization of cytochrome c nitrite reductase subunits (NrfA and NrfH) from Desulfovibrio desulfuricans ATCC 27774. Re-evaluation of the spectroscopic data and redox properties. Eur J Biochem,270, 3904-3915. M.G.Almeida, S.Macieira, L.L.Gonçalves, R.Huber, C.A.Cunha, M.J.Romão, C.Costa, J.Lampreia, J.J.Moura, and I.Moura (2003). The isolation and characterization of cytochrome c nitrite reductase subunits (NrfA and NrfH) from Desulfovibrio desulfuricans ATCC 27774. Re-evaluation of the spectroscopic data and redox properties. Eur J Biochem,270, 3904-3915.      12910261 12910261  M.G.Bertero, R.A.Rothery, M.Palak, C.Hou, D.Lim, F.Blasco, J.H.Weiner, and N.C.Strynadka (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol,10, 681-687. M.G.Bertero, R.A.Rothery, M.Palak, C.Hou, D.Lim, F.Blasco, J.H.Weiner, and N.C.Strynadka (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol,10, 681-687.  PDB code: PDB code:  1q16 1q16     12948771 12948771  M.Jormakka, B.Byrne, and S.Iwata (2003). Formate dehydrogenase--a versatile enzyme in changing environments. Curr Opin Struct Biol,13, 418-423. M.Jormakka, B.Byrne, and S.Iwata (2003). Formate dehydrogenase--a versatile enzyme in changing environments. Curr Opin Struct Biol,13, 418-423.      12815340 12815340  M.Machius (2003). Structural biology: a high-tech tool for biomedical research. Curr Opin Nephrol Hypertens,12, 431-438. M.Machius (2003). Structural biology: a high-tech tool for biomedical research. Curr Opin Nephrol Hypertens,12, 431-438.      12948779 12948779  P.D.Barker (2003). Designing redox metalloproteins from bottom-up and top-down perspectives. Curr Opin Struct Biol,13, 490-499. P.D.Barker (2003). Designing redox metalloproteins from bottom-up and top-down perspectives. Curr Opin Struct Biol,13, 490-499.      14640690 14640690  Z.Zhao, R.A.Rothery, and J.H.Weiner (2003). Effects of site-directed mutations on heme reduction in Escherichia coli nitrate reductase A by menaquinol: a stopped-flow study. Biochemistry,42, 14225-14233. Z.Zhao, R.A.Rothery, and J.H.Weiner (2003). Effects of site-directed mutations on heme reduction in Escherichia coli nitrate reductase A by menaquinol: a stopped-flow study. Biochemistry,42, 14225-14233.      11959503 11959503  B.Byrne, and S.Iwata (2002). Membrane protein complexes. Curr Opin Struct Biol,12, 239-243. B.Byrne, and S.Iwata (2002). Membrane protein complexes. Curr Opin Struct Biol,12, 239-243.      12220497 12220497  H.Raaijmakers, S.Macieira, J.M.Dias, S.Teixeira, S.Bursakov, R.Huber, J.J.Moura, I.Moura, and M.J.Romão (2002). Gene sequence and the 1.8 A crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure,10, 1261-1272. H.Raaijmakers, S.Macieira, J.M.Dias, S.Teixeira, S.Bursakov, R.Huber, J.J.Moura, I.Moura, and M.J.Romão (2002). Gene sequence and the 1.8 A crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure,10, 1261-1272.  PDB code: PDB code:  1h0h 1h0h     12165429 12165429  J.Simon (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev,26, 285-309. J.Simon (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev,26, 285-309.      11952800 11952800  S.Biel, J.Simon, R.Gross, T.Ruiz, M.Ruitenberg, and A.Kröger (2002). Reconstitution of coupled fumarate respiration in liposomes by incorporating the electron transport enzymes isolated from Wolinella succinogenes. Eur J Biochem,269, 1974-1983. S.Biel, J.Simon, R.Gross, T.Ruiz, M.Ruitenberg, and A.Kröger (2002). Reconstitution of coupled fumarate respiration in liposomes by incorporating the electron transport enzymes isolated from Wolinella succinogenes. Eur J Biochem,269, 1974-1983.    The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right. The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right.  |
 |




 Go to PDB code:
Go to PDB code: 





























































 Oxidoreductase PDB id
Oxidoreductase PDB id 
 1kqg
1kqg 















 Loading ...
Loading ... 











 Contents
Contents 




 Protein chains
Protein chains 







 982 a.a.*
982 a.a.* 







 289 a.a.*
289 a.a.* 







 216 a.a.*
216 a.a.* 




 Ligands
Ligands 






 SF4 ×5
SF4 ×5 






 MGD ×2
MGD ×2 






 CDL
CDL 






 HEM ×2
HEM ×2 






 HQO
HQO 




 Metals
Metals 







 6MO
6MO 




 Waters ×1984
Waters ×1984 

 * Residue conservation analysis
* Residue conservation analysis  PDB id:
PDB id:  1kqg
1kqg  Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT
Links PDBe RCSB MMDB JenaLib Proteopedia CATH SCOP PDBSWS PDBePISA CSA ProSAT  Name:
Name:  Oxidoreductase
Oxidoreductase  Title:
Title:  Formate dehydrogenase n from e. Coli Structure:
Formate dehydrogenase n from e. Coli Structure:  Formate dehydrogenase, nitrate-inducible, major subunit. Chain: a. Synonym: formate dehydrogenase-n alpha subunit. Fdh-n alpha subunit. Anaerobic formate dehydrogenase major subunit. Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit. Chain: b. Synonym: formate dehydrogenase-n beta subunit. Fdh-n beta subunit. Anaerobic formate dehydrogenase iron-sulfur subunit. Source:
Formate dehydrogenase, nitrate-inducible, major subunit. Chain: a. Synonym: formate dehydrogenase-n alpha subunit. Fdh-n alpha subunit. Anaerobic formate dehydrogenase major subunit. Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit. Chain: b. Synonym: formate dehydrogenase-n beta subunit. Fdh-n beta subunit. Anaerobic formate dehydrogenase iron-sulfur subunit. Source:  Escherichia coli. Organism_taxid: 562. Strain: gl101. Strain: gl101 Biol. unit:
Escherichia coli. Organism_taxid: 562. Strain: gl101. Strain: gl101 Biol. unit: 
 2.80Å R-factor: 0.198 R-free: 0.239 Authors:
2.80Å R-factor: 0.198 R-free: 0.239 Authors:  M.Jormakka,S.Tornroth,B.Byrne,S.Iwata Key ref:
M.Jormakka,S.Tornroth,B.Byrne,S.Iwata Key ref:  M.Jormakka et al. (2002). Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.Science,295, 1863-1868.PubMed id: 11884747 DOI: 10.1126/science.1068186
M.Jormakka et al. (2002). Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.Science,295, 1863-1868.PubMed id: 11884747 DOI: 10.1126/science.1068186 
 Date:
Date:  05-Jan-02 Release date: 15-Mar-02
05-Jan-02 Release date: 15-Mar-02 
 PROCHECK
PROCHECK 

 Headers
Headers 
 References
References 




 Protein chain
Protein chain 

















 P24183 (FDNG_ECOLI) - Formate dehydrogenase, nitrate-inducible, major subunit from Escherichia coli (strain K12)
P24183 (FDNG_ECOLI) - Formate dehydrogenase, nitrate-inducible, major subunit from Escherichia coli (strain K12) 
 Seq:Struc:
Seq:Struc: 




 Seq:Struc:
Seq:Struc: 

 1015 a.a. 982 a.a.*
1015 a.a. 982 a.a.* 














 Protein chain
Protein chain 

















 P0AAJ3 (FDNH_ECOLI) - Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit from Escherichia coli (strain K12)
P0AAJ3 (FDNH_ECOLI) - Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit from Escherichia coli (strain K12) 
 Seq:Struc:
Seq:Struc: 

 294 a.a. 289 a.a.
294 a.a. 289 a.a. 














 Protein chain
Protein chain 

















 P0AEK7 (FDNI_ECOLI) - Formate dehydrogenase, nitrate-inducible, cytochrome b556(Fdn) subunit from Escherichia coli (strain K12)
P0AEK7 (FDNI_ECOLI) - Formate dehydrogenase, nitrate-inducible, cytochrome b556(Fdn) subunit from Escherichia coli (strain K12) 
 Seq:Struc:
Seq:Struc: 

 217 a.a. 216 a.a.
217 a.a. 216 a.a. 








 Key:
Key:  PfamA domain
PfamA domain 

 Secondary structure
Secondary structure 
 CATH domain
CATH domain  * PDB and UniProt seqs differ at 1 residue position (black cross)
* PDB and UniProt seqs differ at 1 residue position (black cross) 






 Enzyme reactions
Enzyme reactions 








 Enzyme class 2:
Enzyme class 2:  Chain A: E.C.1.17.5.3 - formate dehydrogenase-N. [IntEnz] [ExPASy] [KEGG] [BRENDA]
Chain A: E.C.1.17.5.3 - formate dehydrogenase-N. [IntEnz] [ExPASy] [KEGG] [BRENDA] 




 Reaction:
Reaction:  a quinone + formate + H+ = a quinol + CO2
a quinone + formate + H+ = a quinol + CO2 











 Cofactor:
Cofactor:  Heme; Iron-sulfur; Molybdopterin guanine dinucleotide
Heme; Iron-sulfur; Molybdopterin guanine dinucleotide 





 Enzyme class 3:
Enzyme class 3:  Chains B, C: E.C.1.2.1.2 - Transferred entry: 1.17.1.9. [IntEnz] [ExPASy] [KEGG] [BRENDA]
Chains B, C: E.C.1.2.1.2 - Transferred entry: 1.17.1.9. [IntEnz] [ExPASy] [KEGG] [BRENDA] 




 Reaction:
Reaction:  Formate + NAD+ = CO2 + NADH
Formate + NAD+ = CO2 + NADH 











 Cofactor:
Cofactor:  Flavin; Iron-sulfur; Mo cation
Flavin; Iron-sulfur; Mo cation 







 Note, where more than one E.C. class is given (as above), each may correspond to a different protein domain or, in the case of polyprotein precursors, to a different mature protein.
Note, where more than one E.C. class is given (as above), each may correspond to a different protein domain or, in the case of polyprotein precursors, to a different mature protein.  Molecule diagrams generated from .mol files obtained from theKEGG ftp site
Molecule diagrams generated from .mol files obtained from theKEGG ftp site 
















 reference DOI no: 10.1126/science.1068186 Science 295:1863-1868 (2002) PubMed id: 11884747
reference DOI no: 10.1126/science.1068186 Science 295:1863-1868 (2002) PubMed id: 11884747  Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. M.Jormakka, S.Törnroth, B.Byrne, S.Iwata.
Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. M.Jormakka, S.Törnroth, B.Byrne, S.Iwata.  ABSTRACT
ABSTRACT 
 The structure of the membrane protein formate dehydrogenase-N (Fdn-N), a major component of Escherichia coli nitrate respiration, has been determined at 1.6 angstroms. The structure demonstrates 11 redox centers, including molybdopterin-guanine dinucleotides, five [4Fe-4S] clusters, two heme b groups, and a menaquinone analog. These redox centers are aligned in a single chain, which extends almost 90 angstroms through the enzyme. The menaquinone reduction site associated with a possible proton pathway was also characterized. This structure provides critical insights into the proton motive force generation by redox loop, a common mechanism among a wide range of respiratory enzymes.
The structure of the membrane protein formate dehydrogenase-N (Fdn-N), a major component of Escherichia coli nitrate respiration, has been determined at 1.6 angstroms. The structure demonstrates 11 redox centers, including molybdopterin-guanine dinucleotides, five [4Fe-4S] clusters, two heme b groups, and a menaquinone analog. These redox centers are aligned in a single chain, which extends almost 90 angstroms through the enzyme. The menaquinone reduction site associated with a possible proton pathway was also characterized. This structure provides critical insights into the proton motive force generation by redox loop, a common mechanism among a wide range of respiratory enzymes. 

 Selected figure(s)
Selected figure(s) 





 Figure 1. Fig. 1. A proposal for how proton motive force is generated by Fdh-N and nitrate reductase in Escherichia coli inner membrane. MK, menaquinone; MKH[2], reduced form of menaquinone; MGD, molybdopterin-guanine dinucleotide; b, heme b; FeS, iron-sulfur cluster.
Figure 1. Fig. 1. A proposal for how proton motive force is generated by Fdh-N and nitrate reductase in Escherichia coli inner membrane. MK, menaquinone; MKH[2], reduced form of menaquinone; MGD, molybdopterin-guanine dinucleotide; b, heme b; FeS, iron-sulfur cluster.  Figure 2. Fig. 2. Overall structure of Fdh-N. The figures are based on the native Fdh-N structure except for the position of HQNO, which is determined in the Fdh-N and HQNO complex structure. The
Figure 2. Fig. 2. Overall structure of Fdh-N. The figures are based on the native Fdh-N structure except for the position of HQNO, which is determined in the Fdh-N and HQNO complex structure. The ,
,  , and
, and  subunits are shown in brown, blue, and magenta, respectively. Heme groups and MGD cofactors are shown in green, and Mo, cardiolipin (CL), and HQNO are colored in magenta, yellow, and navy, respectively. Five [4Fe-4S] clusters are painted as red (Fe atoms) and yellow (S atoms). (A) Fdh-N trimer viewed parallel to the membrane. (B) Fdh-N trimer viewed from the periplasm along the membrane normal. (C) View of the Fdh-N monomer parallel to the membrane. Center-to-center and edge-to-edge (in parentheses) (12) distances in angstroms between each of the redox centers are also shown. The edge-to-edge distance for HQNO to heme b[C] is 0 because HQNO is directly binding to the histidine ligand of heme b[C]. All figures were made with MOLSCRIPT (28) and BOBSCRIPT (29) and rendered with RASTER3D (30).
subunits are shown in brown, blue, and magenta, respectively. Heme groups and MGD cofactors are shown in green, and Mo, cardiolipin (CL), and HQNO are colored in magenta, yellow, and navy, respectively. Five [4Fe-4S] clusters are painted as red (Fe atoms) and yellow (S atoms). (A) Fdh-N trimer viewed parallel to the membrane. (B) Fdh-N trimer viewed from the periplasm along the membrane normal. (C) View of the Fdh-N monomer parallel to the membrane. Center-to-center and edge-to-edge (in parentheses) (12) distances in angstroms between each of the redox centers are also shown. The edge-to-edge distance for HQNO to heme b[C] is 0 because HQNO is directly binding to the histidine ligand of heme b[C]. All figures were made with MOLSCRIPT (28) and BOBSCRIPT (29) and rendered with RASTER3D (30). 
 The above figures are reprinted by permission from the AAAs: Science (2002,295, 1863-1868) copyright 2002. Figures were selected by an automated process.
The above figures are reprinted by permission from the AAAs: Science (2002,295, 1863-1868) copyright 2002. Figures were selected by an automated process. 











 Literature references that cite this PDB file's key reference
Literature references that cite this PDB file's key reference  PubMed id
PubMed id  Reference
Reference 



 22683878
22683878  T.Palmer, and B.C.Berks (2012). The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol,10, 483-496.
T.Palmer, and B.C.Berks (2012). The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol,10, 483-496. 



 21289038
21289038  J.Speck, K.M.Arndt, and K.M.Müller (2011). Efficient phage display of intracellularly folded proteins mediated by the TAT pathway. Protein Eng Des Sel,24, 473-484.
J.Speck, K.M.Arndt, and K.M.Müller (2011). Efficient phage display of intracellularly folded proteins mediated by the TAT pathway. Protein Eng Des Sel,24, 473-484. 



 20938980
20938980  K.Illergård, A.Kauko, and A.Elofsson (2011). Why are polar residues within the membrane core evolutionary conserved? Proteins,79, 79-91.
K.Illergård, A.Kauko, and A.Elofsson (2011). Why are polar residues within the membrane core evolutionary conserved? Proteins,79, 79-91. 



 21518899
21518899  R.Arias-Cartin, S.Grimaldi, J.Pommier, P.Lanciano, C.Schaefer, P.Arnoux, G.Giordano, B.Guigliarelli, and A.Magalon (2011). Cardiolipin-based respiratory complex activation in bacteria. Proc Natl Acad Sci U S A,108, 7781-7786.
R.Arias-Cartin, S.Grimaldi, J.Pommier, P.Lanciano, C.Schaefer, P.Arnoux, G.Giordano, B.Guigliarelli, and A.Magalon (2011). Cardiolipin-based respiratory complex activation in bacteria. Proc Natl Acad Sci U S A,108, 7781-7786. 



 20819103
20819103  X.Zhang, E.G.Matson, and J.R.Leadbetter (2011). Genes for selenium dependent and independent formate dehydrogenase in the gut microbial communities of three lower, wood-feeding termites and a wood-feeding roach. Environ Microbiol,13, 307-323.
X.Zhang, E.G.Matson, and J.R.Leadbetter (2011). Genes for selenium dependent and independent formate dehydrogenase in the gut microbial communities of three lower, wood-feeding termites and a wood-feeding roach. Environ Microbiol,13, 307-323. 



 20645325
20645325  E.Cremades, J.Echeverría, and S.Alvarez (2010). The trigonal prism in coordination chemistry. Chemistry,16, 10380-10396.
E.Cremades, J.Echeverría, and S.Alvarez (2010). The trigonal prism in coordination chemistry. Chemistry,16, 10380-10396. 



 19826804
19826804  K.McLuskey, A.W.Roszak, Y.Zhu, and N.W.Isaacs (2010). Crystal structures of all-alpha type membrane proteins. Eur Biophys J,39, 723-755.
K.McLuskey, A.W.Roszak, Y.Zhu, and N.W.Isaacs (2010). Crystal structures of all-alpha type membrane proteins. Eur Biophys J,39, 723-755. 



 20667175
20667175  K.R.Vinothkumar, and R.Henderson (2010). Structures of membrane proteins. Q Rev Biophys,43, 65.
K.R.Vinothkumar, and R.Henderson (2010). Structures of membrane proteins. Q Rev Biophys,43, 65. 



 20352202
20352202  M.Raja (2010). The role of phosphatidic acid and cardiolipin in stability of the tetrameric assembly of potassium channel KcsA. J Membr Biol,234, 235-240.
M.Raja (2010). The role of phosphatidic acid and cardiolipin in stability of the tetrameric assembly of potassium channel KcsA. J Membr Biol,234, 235-240. 



 20406421
20406421  N.Borgese, and M.Righi (2010). Remote origins of tail-anchored proteins. Traffic,11, 877-885.
N.Borgese, and M.Righi (2010). Remote origins of tail-anchored proteins. Traffic,11, 877-885. 



 20587053
20587053  T.E.Meyer, J.A.Kyndt, and M.A.Cusanovich (2010). Occurrence and sequence of Sphaeroides Heme Protein and diheme cytochrome C in purple photosynthetic bacteria in the family Rhodobacteraceae. BMC Biochem,11, 24.
T.E.Meyer, J.A.Kyndt, and M.A.Cusanovich (2010). Occurrence and sequence of Sphaeroides Heme Protein and diheme cytochrome C in purple photosynthetic bacteria in the family Rhodobacteraceae. BMC Biochem,11, 24. 



 20479269
20479269  V.A.Gold, A.Robson, H.Bao, T.Romantsov, F.Duong, and I.Collinson (2010). The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci U S A,107, 10044-10049.
V.A.Gold, A.Robson, H.Bao, T.Romantsov, F.Duong, and I.Collinson (2010). The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci U S A,107, 10044-10049. 



 19658738
19658738  A.Y.Smirnov, S.E.Savel'ev, and F.Nori (2009). Diffusion-controlled generation of a proton-motive force across a biomembrane. Phys Rev E Stat Nonlin Soft Matter Phys,80, 011916.
A.Y.Smirnov, S.E.Savel'ev, and F.Nori (2009). Diffusion-controlled generation of a proton-motive force across a biomembrane. Phys Rev E Stat Nonlin Soft Matter Phys,80, 011916. 



 19371718
19371718  E.Mileykovskaya, and W.Dowhan (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta,1788, 2084-2091.
E.Mileykovskaya, and W.Dowhan (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta,1788, 2084-2091. 



 19536822
19536822  G.Zoppellaro, K.L.Bren, A.A.Ensign, E.Harbitz, R.Kaur, H.P.Hersleth, U.Ryde, L.Hederstedt, and K.K.Andersson (2009). Review: studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers,91, 1064-1082.
G.Zoppellaro, K.L.Bren, A.A.Ensign, E.Harbitz, R.Kaur, H.P.Hersleth, U.Ryde, L.Hederstedt, and K.K.Andersson (2009). Review: studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers,91, 1064-1082. 



 19452052
19452052  M.J.Romão (2009). Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans, (), 4053-4068.
M.J.Romão (2009). Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans, (), 4053-4068. 



 19206188
19206188  S.Groysman, and R.H.Holm (2009). Biomimetic chemistry of iron, nickel, molybdenum, and tungsten in sulfur-ligated protein sites. Biochemistry,48, 2310-2320.
S.Groysman, and R.H.Holm (2009). Biomimetic chemistry of iron, nickel, molybdenum, and tungsten in sulfur-ligated protein sites. Biochemistry,48, 2310-2320. 



 19416061
19416061  S.Raunser, and T.Walz (2009). Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys,38, 89.
S.Raunser, and T.Walz (2009). Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys,38, 89. 



 19233927
19233927  X.Li, Q.Luo, N.Q.Wofford, K.L.Keller, M.J.McInerney, J.D.Wall, and L.R.Krumholz (2009). A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J Bacteriol,191, 2675-2682.
X.Li, Q.Luo, N.Q.Wofford, K.L.Keller, M.J.McInerney, J.D.Wall, and L.R.Krumholz (2009). A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J Bacteriol,191, 2675-2682. 



 18658261
18658261  B.S.Pickering, and I.J.Oresnik (2008). Formate-dependent autotrophic growth in Sinorhizobium meliloti. J Bacteriol,190, 6409-6418.
B.S.Pickering, and I.J.Oresnik (2008). Formate-dependent autotrophic growth in Sinorhizobium meliloti. J Bacteriol,190, 6409-6418. 



 18615097
18615097  C.F.Matos, C.Robinson, and A.Di Cola (2008). The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J,27, 2055-2063.
C.F.Matos, C.Robinson, and A.Di Cola (2008). The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J,27, 2055-2063. 



 18247347
18247347  C.H.Lu, S.W.Huang, Y.L.Lai, C.P.Lin, C.H.Shih, C.C.Huang, W.L.Hsu, and J.K.Hwang (2008). On the relationship between the protein structure and protein dynamics. Proteins,72, 625-634.
C.H.Lu, S.W.Huang, Y.L.Lai, C.P.Lin, C.H.Shih, C.C.Huang, W.L.Hsu, and J.K.Hwang (2008). On the relationship between the protein structure and protein dynamics. Proteins,72, 625-634. 



 18418633
18418633  E.A.Berry, and F.A.Walker (2008). Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem,13, 481-498.
E.A.Berry, and F.A.Walker (2008). Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem,13, 481-498. 



 18535145
18535145  G.J.Workun, K.Moquin, R.A.Rothery, and J.H.Weiner (2008). Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold. Microbiol Mol Biol Rev,72, 228.
G.J.Workun, K.Moquin, R.A.Rothery, and J.H.Weiner (2008). Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold. Microbiol Mol Biol Rev,72, 228. 



 18716757
18716757  I.Lüke, G.Butland, K.Moore, G.Buchanan, V.Lyall, S.A.Fairhurst, J.F.Greenblatt, A.Emili, T.Palmer, and F.Sargent (2008). Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch Microbiol,190, 685-696.
I.Lüke, G.Butland, K.Moore, G.Buchanan, V.Lyall, S.A.Fairhurst, J.F.Greenblatt, A.Emili, T.Palmer, and F.Sargent (2008). Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch Microbiol,190, 685-696.  18751913
18751913  M.Bogdanov, E.Mileykovskaya, and W.Dowhan (2008). Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem,49, 197-239.
M.Bogdanov, E.Mileykovskaya, and W.Dowhan (2008). Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem,49, 197-239. 



 18536726
18536726  M.Jormakka, K.Yokoyama, T.Yano, M.Tamakoshi, S.Akimoto, T.Shimamura, P.Curmi, and S.Iwata (2008). Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol,15, 730-737.
M.Jormakka, K.Yokoyama, T.Yano, M.Tamakoshi, S.Akimoto, T.Shimamura, P.Curmi, and S.Iwata (2008). Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol,15, 730-737.  PDB codes:
PDB codes:  2vpw 2vpx 2vpy 2vpz
2vpw 2vpx 2vpy 2vpz 


 18077827
18077827  M.Schlame (2008). Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res,49, 1607-1620.
M.Schlame (2008). Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res,49, 1607-1620. 



 18500332
18500332  N.H.Joh, A.Min, S.Faham, J.P.Whitelegge, D.Yang, V.L.Woods, and J.U.Bowie (2008). Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature,453, 1266-1270.
N.H.Joh, A.Min, S.Faham, J.P.Whitelegge, D.Yang, V.L.Woods, and J.U.Bowie (2008). Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature,453, 1266-1270.  PDB codes:
PDB codes:  3coc 3cod
3coc 3cod 


 18218713
18218713  S.Newstead, S.Ferrandon, and S.Iwata (2008). Rationalizing alpha-helical membrane protein crystallization. Protein Sci,17, 466-472.
S.Newstead, S.Ferrandon, and S.Iwata (2008). Rationalizing alpha-helical membrane protein crystallization. Protein Sci,17, 466-472. 



 18667702
18667702  T.Reda, C.M.Plugge, N.J.Abram, and J.Hirst (2008). Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc Natl Acad Sci U S A,105, 10654-10658.
T.Reda, C.M.Plugge, N.J.Abram, and J.Hirst (2008). Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc Natl Acad Sci U S A,105, 10654-10658. 



 18501187
18501187  W.Liu, C.E.Rogge, G.F.da Silva, V.P.Shinkarev, A.L.Tsai, Y.Kamensky, G.Palmer, and R.J.Kulmacz (2008). His92 and His110 selectively affect different heme centers of adrenal cytochrome b(561). Biochim Biophys Acta,1777, 1218-1228.
W.Liu, C.E.Rogge, G.F.da Silva, V.P.Shinkarev, A.L.Tsai, Y.Kamensky, G.Palmer, and R.J.Kulmacz (2008). His92 and His110 selectively affect different heme centers of adrenal cytochrome b(561). Biochim Biophys Acta,1777, 1218-1228. 



 17600788
17600788  K.Pal, P.K.Chaudhury, and S.Sarkar (2007). Structure of the Michaelis complex and function of the catalytic center in the reductive half-reaction of computational and synthetic models of sulfite oxidase. Chem Asian J,2, 956-964.
K.Pal, P.K.Chaudhury, and S.Sarkar (2007). Structure of the Michaelis complex and function of the catalytic center in the reductive half-reaction of computational and synthetic models of sulfite oxidase. Chem Asian J,2, 956-964. 



 17557793
17557793  N.Buzhynskyy, P.Sens, V.Prima, J.N.Sturgis, and S.Scheuring (2007). Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J,93, 2870-2876.
N.Buzhynskyy, P.Sens, V.Prima, J.N.Sturgis, and S.Scheuring (2007). Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J,93, 2870-2876. 



 16962969
16962969  D.P.Kloer, C.Hagel, J.Heider, and G.E.Schulz (2006). Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure,14, 1377-1388.
D.P.Kloer, C.Hagel, J.Heider, and G.E.Schulz (2006). Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure,14, 1377-1388.  PDB code:
PDB code:  2ivf
2ivf 


 16830149
16830149  H.C.Raaijmakers, and M.J.Romão (2006). Formate-reduced E. coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem,11, 849-854.
H.C.Raaijmakers, and M.J.Romão (2006). Formate-reduced E. coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem,11, 849-854.  PDB code:
PDB code:  2iv2
2iv2 


 16885262
16885262  H.Ridley, C.A.Watts, D.J.Richardson, and C.S.Butler (2006). Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl Environ Microbiol,72, 5173-5180.
H.Ridley, C.A.Watts, D.J.Richardson, and C.S.Butler (2006). Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl Environ Microbiol,72, 5173-5180. 



 17024183
17024183  M.G.Madej, H.R.Nasiri, N.S.Hilgendorff, H.Schwalbe, and C.R.Lancaster (2006). Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J,25, 4963-4970.
M.G.Madej, H.R.Nasiri, N.S.Hilgendorff, H.Schwalbe, and C.R.Lancaster (2006). Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J,25, 4963-4970.  PDB code:
PDB code:  2bs2
2bs2 


 16802174
16802174  M.John, R.P.Schmitz, M.Westermann, W.Richter, and G.Diekert (2006). Growth substrate dependent localization of tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch Microbiol,186, 99.
M.John, R.P.Schmitz, M.Westermann, W.Richter, and G.Diekert (2006). Growth substrate dependent localization of tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch Microbiol,186, 99. 



 17139260
17139260  M.L.Rodrigues, T.F.Oliveira, I.A.Pereira, and M.Archer (2006). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J,25, 5951-5960.
M.L.Rodrigues, T.F.Oliveira, I.A.Pereira, and M.Archer (2006). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J,25, 5951-5960.  PDB code:
PDB code:  2j7a
2j7a 


 16597999
16597999  S.S.Ruebush, S.L.Brantley, and M.Tien (2006). Reduction of soluble and insoluble iron forms by membrane fractions of Shewanella oneidensis grown under aerobic and anaerobic conditions. Appl Environ Microbiol,72, 2925-2935.
S.S.Ruebush, S.L.Brantley, and M.Tien (2006). Reduction of soluble and insoluble iron forms by membrane fractions of Shewanella oneidensis grown under aerobic and anaerobic conditions. Appl Environ Microbiol,72, 2925-2935. 



 16433558
16433558  T.Teschner, L.Yatsunyk, V.Schünemann, H.Paulsen, H.Winkler, C.Hu, W.R.Scheidt, F.A.Walker, and A.X.Trautwein (2006). Models of the membrane-bound cytochromes: mössbauer spectra of crystalline low-spin ferriheme complexes having axial ligand plane dihedral angles ranging from 0 degree to 90 degrees. J Am Chem Soc,128, 1379-1389.
T.Teschner, L.Yatsunyk, V.Schünemann, H.Paulsen, H.Winkler, C.Hu, W.R.Scheidt, F.A.Walker, and A.X.Trautwein (2006). Models of the membrane-bound cytochromes: mössbauer spectra of crystalline low-spin ferriheme complexes having axial ligand plane dihedral angles ranging from 0 degree to 90 degrees. J Am Chem Soc,128, 1379-1389. 



 16172928
16172928  A.J.Watson, A.V.Hughes, P.K.Fyfe, M.C.Wakeham, K.Holden-Dye, P.Heathcote, and M.R.Jones (2005). On the role of basic residues in adapting the reaction centre-LH1 complex for growth at elevated temperatures in purple bacteria. Photosynth Res,86, 81.
A.J.Watson, A.V.Hughes, P.K.Fyfe, M.C.Wakeham, K.Holden-Dye, P.Heathcote, and M.R.Jones (2005). On the role of basic residues in adapting the reaction centre-LH1 complex for growth at elevated temperatures in purple bacteria. Photosynth Res,86, 81. 



 15802249
15802249  B.C.Berks, T.Palmer, and F.Sargent (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol,8, 174-181.
B.C.Berks, T.Palmer, and F.Sargent (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol,8, 174-181. 



 16380425
16380425  C.R.Lancaster, U.S.Sauer, R.Gross, A.H.Haas, J.Graf, H.Schwalbe, W.Mäntele, J.Simon, and M.G.Madej (2005). Experimental support for the "E pathway hypothesis" of coupled transmembrane e- and H+ transfer in dihemic quinol:fumarate reductase. Proc Natl Acad Sci U S A,102, 18860-18865.
C.R.Lancaster, U.S.Sauer, R.Gross, A.H.Haas, J.Graf, H.Schwalbe, W.Mäntele, J.Simon, and M.G.Madej (2005). Experimental support for the "E pathway hypothesis" of coupled transmembrane e- and H+ transfer in dihemic quinol:fumarate reductase. Proc Natl Acad Sci U S A,102, 18860-18865.  PDB codes:
PDB codes:  2bs3 2bs4
2bs3 2bs4 


 15819632
15819632  M.L.Crouch, L.A.Becker, I.S.Bang, H.Tanabe, A.J.Ouellette, and F.C.Fang (2005). The alternative sigma factor sigma is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol Microbiol,56, 789-799.
M.L.Crouch, L.A.Becker, I.S.Bang, H.Tanabe, A.J.Ouellette, and F.C.Fang (2005). The alternative sigma factor sigma is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol Microbiol,56, 789-799. 



 15949761
15949761  P.K.Fyfe, A.V.Hughes, P.Heathcote, and M.R.Jones (2005). Proteins, chlorophylls and lipids: X-ray analysis of a three-way relationship. Trends Plant Sci,10, 275-282.
P.K.Fyfe, A.V.Hughes, P.Heathcote, and M.R.Jones (2005). Proteins, chlorophylls and lipids: X-ray analysis of a three-way relationship. Trends Plant Sci,10, 275-282. 



 15654871
15654871  R.A.Rothery, A.M.Seime, A.M.Spiers, E.Maklashina, I.Schröder, R.P.Gunsalus, G.Cecchini, and J.H.Weiner (2005). Defining the Q-site of Escherichia coli fumarate reductase by site-directed mutagenesis, fluorescence quench titrations and EPR spectroscopy. FEBS J,272, 313-326.
R.A.Rothery, A.M.Seime, A.M.Spiers, E.Maklashina, I.Schröder, R.P.Gunsalus, G.Cecchini, and J.H.Weiner (2005). Defining the Q-site of Escherichia coli fumarate reductase by site-directed mutagenesis, fluorescence quench titrations and EPR spectroscopy. FEBS J,272, 313-326. 



 14681400
14681400  A.Andreeva, D.Howorth, S.E.Brenner, T.J.Hubbard, C.Chothia, and A.G.Murzin (2004). SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res,32, D226-D229.
A.Andreeva, D.Howorth, S.E.Brenner, T.J.Hubbard, C.Chothia, and A.G.Murzin (2004). SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res,32, D226-D229. 



 15516557
15516557  A.C.Fisher, and M.P.DeLisa (2004). A little help from my friends: quality control of presecretory proteins in bacteria. J Bacteriol,186, 7467-7473.
A.C.Fisher, and M.P.DeLisa (2004). A little help from my friends: quality control of presecretory proteins in bacteria. J Bacteriol,186, 7467-7473. 



 15284442
15284442  A.Messerschmidt, H.Niessen, D.Abt, O.Einsle, B.Schink, and P.M.Kroneck (2004). Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc Natl Acad Sci U S A,101, 11571-11576.
A.Messerschmidt, H.Niessen, D.Abt, O.Einsle, B.Schink, and P.M.Kroneck (2004). Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc Natl Acad Sci U S A,101, 11571-11576.  PDB codes:
PDB codes:  1ti2 1ti4 1ti6 1vld 1vle 1vlf
1ti2 1ti4 1ti6 1vld 1vle 1vlf 


 15342602
15342602  C.Punginelli, B.Ize, N.R.Stanley, V.Stewart, G.Sawers, B.C.Berks, and T.Palmer (2004). mRNA secondary structure modulates translation of Tat-dependent formate dehydrogenase N. J Bacteriol,186, 6311-6315.
C.Punginelli, B.Ize, N.R.Stanley, V.Stewart, G.Sawers, B.C.Berks, and T.Palmer (2004). mRNA secondary structure modulates translation of Tat-dependent formate dehydrogenase N. J Bacteriol,186, 6311-6315. 



 15028701
15028701  F.Peters, M.Rother, and M.Boll (2004). Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J Bacteriol,186, 2156-2163.
F.Peters, M.Rother, and M.Boll (2004). Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J Bacteriol,186, 2156-2163. 



 15311335
15311335  J.J.Moura, C.D.Brondino, J.Trincão, and M.J.Romão (2004). Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J Biol Inorg Chem,9, 791-799.
J.J.Moura, C.D.Brondino, J.Trincão, and M.J.Romão (2004). Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J Biol Inorg Chem,9, 791-799. 



 15551861
15551861  O.Einsle, and P.M.Kroneck (2004). Structural basis of denitrification. Biol Chem,385, 875-883.
O.Einsle, and P.M.Kroneck (2004). Structural basis of denitrification. Biol Chem,385, 875-883. 



 15321667
15321667  R.P.Schmitz, and G.Diekert (2004). The fdh operon of Sulfurospirillum multivorans. FEMS Microbiol Lett,237, 235-242.
R.P.Schmitz, and G.Diekert (2004). The fdh operon of Sulfurospirillum multivorans. FEMS Microbiol Lett,237, 235-242. 



 15498942
15498942  T.J.Stevens, K.Mizuguchi, and I.T.Arkin (2004). Distinct protein interfaces in transmembrane domains suggest an in vivo folding model. Protein Sci,13, 3028-3037.
T.J.Stevens, K.Mizuguchi, and I.T.Arkin (2004). Distinct protein interfaces in transmembrane domains suggest an in vivo folding model. Protein Sci,13, 3028-3037. 



 15486691
15486691  T.Tomiki, and N.Saitou (2004). Phylogenetic analysis of proteins associated in the four major energy metabolism systems: photosynthesis, aerobic respiration, denitrification, and sulfur respiration. J Mol Evol,59, 158-176.
T.Tomiki, and N.Saitou (2004). Phylogenetic analysis of proteins associated in the four major energy metabolism systems: photosynthesis, aerobic respiration, denitrification, and sulfur respiration. J Mol Evol,59, 158-176. 



 12594934
12594934  F.Baymann, E.Lebrun, M.Brugna, B.Schoepp-Cothenet, M.T.Giudici-Orticoni, and W.Nitschke (2003). The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci,358, 267-274.
F.Baymann, E.Lebrun, M.Brugna, B.Schoepp-Cothenet, M.T.Giudici-Orticoni, and W.Nitschke (2003). The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci,358, 267-274. 



 12823811
12823811  J.Simon, M.Sänger, S.C.Schuster, and R.Gross (2003). Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol Microbiol,49, 69-79.
J.Simon, M.Sänger, S.C.Schuster, and R.Gross (2003). Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol Microbiol,49, 69-79. 



 12940994
12940994  K.Hatzixanthis, T.Palmer, and F.Sargent (2003). A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol,49, 1377-1390.
K.Hatzixanthis, T.Palmer, and F.Sargent (2003). A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol,49, 1377-1390. 



 14511372
14511372  M.G.Almeida, S.Macieira, L.L.Gonçalves, R.Huber, C.A.Cunha, M.J.Romão, C.Costa, J.Lampreia, J.J.Moura, and I.Moura (2003). The isolation and characterization of cytochrome c nitrite reductase subunits (NrfA and NrfH) from Desulfovibrio desulfuricans ATCC 27774. Re-evaluation of the spectroscopic data and redox properties. Eur J Biochem,270, 3904-3915.
M.G.Almeida, S.Macieira, L.L.Gonçalves, R.Huber, C.A.Cunha, M.J.Romão, C.Costa, J.Lampreia, J.J.Moura, and I.Moura (2003). The isolation and characterization of cytochrome c nitrite reductase subunits (NrfA and NrfH) from Desulfovibrio desulfuricans ATCC 27774. Re-evaluation of the spectroscopic data and redox properties. Eur J Biochem,270, 3904-3915. 



 12910261
12910261  M.G.Bertero, R.A.Rothery, M.Palak, C.Hou, D.Lim, F.Blasco, J.H.Weiner, and N.C.Strynadka (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol,10, 681-687.
M.G.Bertero, R.A.Rothery, M.Palak, C.Hou, D.Lim, F.Blasco, J.H.Weiner, and N.C.Strynadka (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol,10, 681-687.  PDB code:
PDB code:  1q16
1q16 


 12948771
12948771  M.Jormakka, B.Byrne, and S.Iwata (2003). Formate dehydrogenase--a versatile enzyme in changing environments. Curr Opin Struct Biol,13, 418-423.
M.Jormakka, B.Byrne, and S.Iwata (2003). Formate dehydrogenase--a versatile enzyme in changing environments. Curr Opin Struct Biol,13, 418-423. 



 12815340
12815340  M.Machius (2003). Structural biology: a high-tech tool for biomedical research. Curr Opin Nephrol Hypertens,12, 431-438.
M.Machius (2003). Structural biology: a high-tech tool for biomedical research. Curr Opin Nephrol Hypertens,12, 431-438. 



 12948779
12948779  P.D.Barker (2003). Designing redox metalloproteins from bottom-up and top-down perspectives. Curr Opin Struct Biol,13, 490-499.
P.D.Barker (2003). Designing redox metalloproteins from bottom-up and top-down perspectives. Curr Opin Struct Biol,13, 490-499. 



 14640690
14640690  Z.Zhao, R.A.Rothery, and J.H.Weiner (2003). Effects of site-directed mutations on heme reduction in Escherichia coli nitrate reductase A by menaquinol: a stopped-flow study. Biochemistry,42, 14225-14233.
Z.Zhao, R.A.Rothery, and J.H.Weiner (2003). Effects of site-directed mutations on heme reduction in Escherichia coli nitrate reductase A by menaquinol: a stopped-flow study. Biochemistry,42, 14225-14233. 



 11959503
11959503  B.Byrne, and S.Iwata (2002). Membrane protein complexes. Curr Opin Struct Biol,12, 239-243.
B.Byrne, and S.Iwata (2002). Membrane protein complexes. Curr Opin Struct Biol,12, 239-243. 



 12220497
12220497  H.Raaijmakers, S.Macieira, J.M.Dias, S.Teixeira, S.Bursakov, R.Huber, J.J.Moura, I.Moura, and M.J.Romão (2002). Gene sequence and the 1.8 A crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure,10, 1261-1272.
H.Raaijmakers, S.Macieira, J.M.Dias, S.Teixeira, S.Bursakov, R.Huber, J.J.Moura, I.Moura, and M.J.Romão (2002). Gene sequence and the 1.8 A crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure,10, 1261-1272.  PDB code:
PDB code:  1h0h
1h0h 


 12165429
12165429  J.Simon (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev,26, 285-309.
J.Simon (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev,26, 285-309. 



 11952800
11952800  S.Biel, J.Simon, R.Gross, T.Ruiz, M.Ruitenberg, and A.Kröger (2002). Reconstitution of coupled fumarate respiration in liposomes by incorporating the electron transport enzymes isolated from Wolinella succinogenes. Eur J Biochem,269, 1974-1983.
S.Biel, J.Simon, R.Gross, T.Ruiz, M.Ruitenberg, and A.Kröger (2002). Reconstitution of coupled fumarate respiration in liposomes by incorporating the electron transport enzymes isolated from Wolinella succinogenes. Eur J Biochem,269, 1974-1983. 

 The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right.
The most recent references are shown first. Citation data come partly from CiteXplore and partly from an automated harvesting procedure. Note that this is likely to be only a partial list as not all journals are covered by either method. However, we are continually building up the citation data so more and more references will be included with time. Where a reference describes a PDB structure, the PDB codes are shown on the right. 
