The trickster microbes that are shaking up the tree of life (original) (raw)
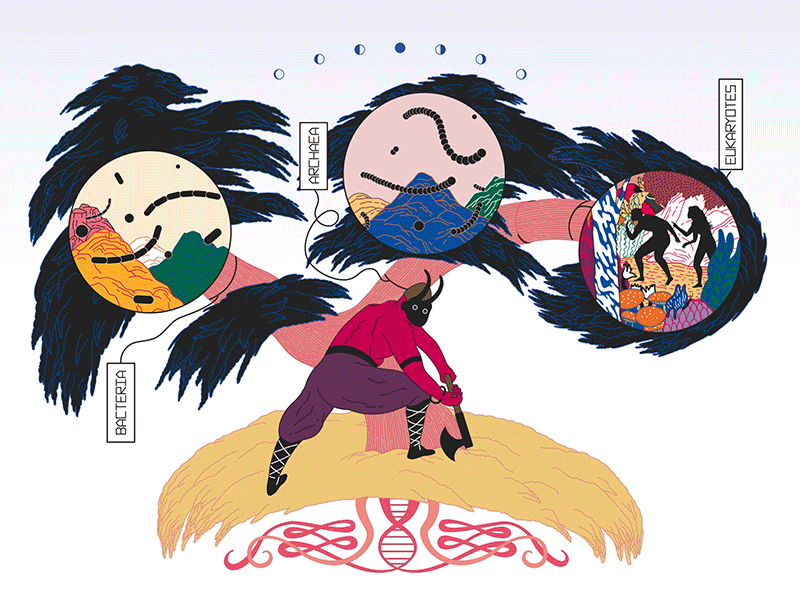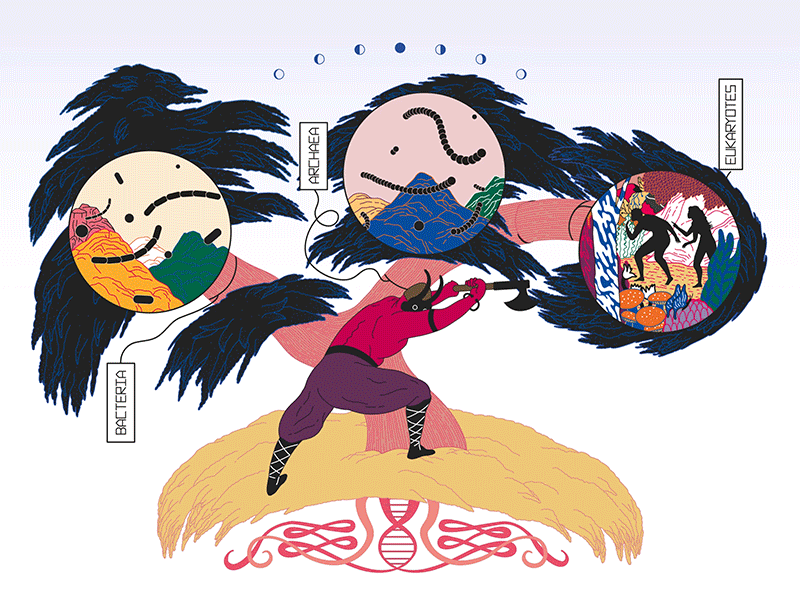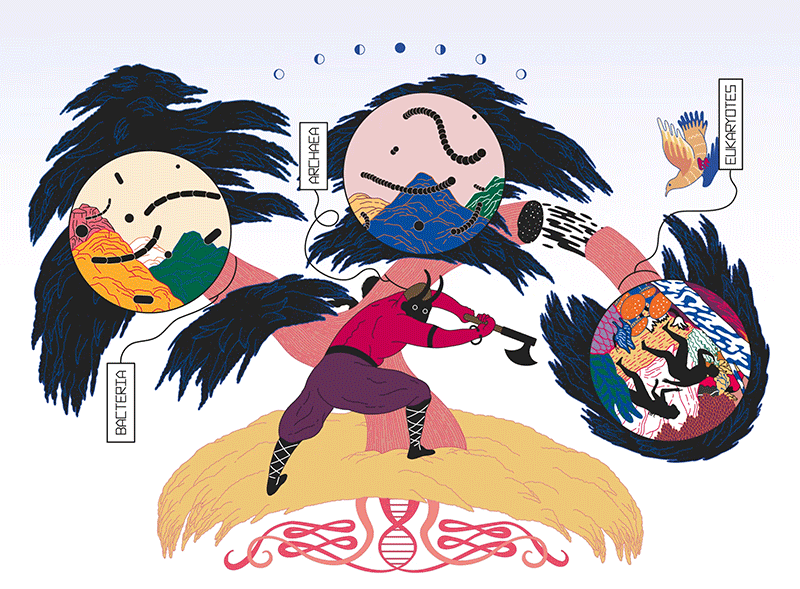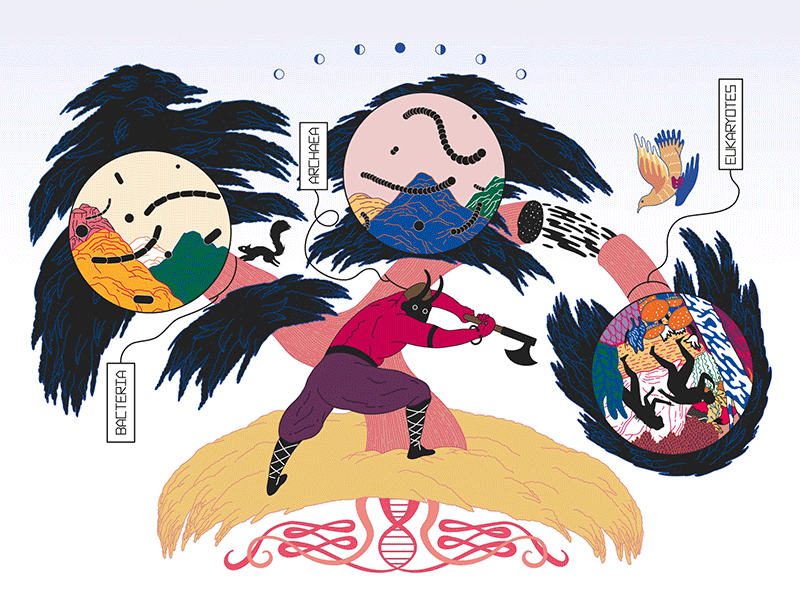
Illustration by Fabio Buonocore
Every mythology needs a good trickster, and there are few better than the Norse god Loki. He stirs trouble and insults other gods. He is elusive, anarchic and ambiguous. He is, in other words, the perfect namesake for a group of microbes — the Lokiarchaeota — that is rewriting a fundamental story about life’s early roots.
These unruly microbes belong to a category of single-celled organisms called archaea, which resemble bacteria under a microscope but are as distinct from them in some respects as humans are. The Lokis, as they are sometimes known, were discovered by sequencing DNA from sea-floor muck collected near Greenland1. Together with some related microbes, they are prodding biologists to reconsider one of the greatest events in the history of life on Earth — the appearance of the eukaryotes, the group of organisms that includes all plants, animals, fungi and more.
The discovery of archaea in the late 1970s led scientists to propose that the tree of life diverged long ago into three main trunks, or ‘domains’. One trunk gave rise to modern bacteria; one to archaea. And the third produced eukaryotes. But debates soon erupted over the structure of these trunks. A leading ‘three-domain’ model held that archaea and eukaryotes diverged from a common ancestor. But a two-domain scenario suggested that eukaryotes diverged directly from a subgroup of archaea.
The arguments, although heated at times, eventually stagnated, says microbiologist Phil Hugenholtz at the University of Queensland in Brisbane, Australia. Then the Lokis and their relatives blew in like “a breath of fresh air”, he says, and revived the case for a two-domain tree.
These newly discovered archaea have genes that are considered hallmarks of eukaryotes. And deep analysis of the organisms’ DNA suggests that modern eukaryotes belong to the same archaeal group. If that’s the case, essentially all complex life — everything from green algae to blue whales — originally came from archaea.
But many scientists remain unconvinced. Evolutionary tree building is messy, contentious work. And no one has yet published evidence to show that these organisms can be grown in the lab, which makes them difficult to study. The debate is still rancorous. Stalwarts on both sides are “very hostile to each other, and 100% believe there’s nothing correct in the other camp”, Hugenholtz says. Some decline to voice an opinion, for fear of offending senior colleagues.
What’s at stake is a deeper understanding of the biological leap that produced eukaryotes: “The biggest thing that happened since the origin of life,” according to evolutionary biologist Patrick Keeling at the University of British Columbia in Vancouver, Canada. Where they came from “is one of the most fundamental questions in understanding the nature of biological complexity”, he says. To answer that question, “we need to resolve who’s related to who”.
Two becomes three
For scientists half a century ago, life on Earth was split between two categories: eukaryotes, living things with cells that contain membrane-wrapped internal structures, such as a nucleus; and prokaryotes, single-celled organisms that generally lack internal membranes. Bacteria were the only prokaryotes that biologists knew about. Then, in 1977, evolutionary biologist Carl Woese and his colleagues described archaea as a third, distinct form of life — one that reached back billions of years2. Life, Woese said, should be divided into three bins rather than two.
He was not without his detractors. In the 1980s, evolutionary biologist James Lake at the University of California, Los Angeles, proposed that eukaryotes are sisters to archaea that he called eocytes, which means dawn cells3,4. The idea evolved into the two-domain scenario.
Lake and Woese fought bitterly over their competing models, culminating in a legendary shouting match in the mid-1980s. Afterwards, Woese “didn’t want to meet with Jim Lake”, says microbiologist Patrick Forterre at the Pasteur Institute in Paris. Lake does not dispute the acrimony. “That was really quite a debate, and there was an enormous amount of politics,” he says. Woese died in 2012.
Today, the argument over where eukaryotes came from has matured. Many on both sides agree that the origin of eukaryotes probably involved a step known as endosymbiosis. This theory, championed by the late biologist Lynn Margulis, holds that a simple host cell living eons ago somehow swallowed a bacterium, and the two struck up a mutually beneficial relationship. These captive bacteria eventually evolved into mitochondria — the cellular substructures that produce energy — and the hybrid cells became what are now known as eukaryotes.
The nature of the engulfing cell is where the two camps diverge. As the three-domain adherents tell it, the engulfer was an ancestral microbe, now extinct. According to Forterre, it was a “proto-eukaryote” — “neither a modern archaeon nor a modern eukaryote”. In this model, there were several major splits in early evolution. The first happened billions of years ago, when primeval organisms gave rise to both bacteria and an extinct group of microbes. This latter group diverged into archaea and the group that became eukaryotes.
In the two-domain world, however, a primeval organism gave rise to bacteria and archaea. And the organism that eventually swallowed the fateful bacterium was an archaeon. That would make all eukaryotes a sort of overachieving branch of the archaea — or, as some scientists call it, a ‘secondary domain’ (see ‘Domains in debate’).

Scrambled messages
Without a wayback machine for microbes, sorting through these hypo-theses is exceedingly difficult. The fossil record for the earliest eukaryotes is sparse, and examples can be inscrutable. Scientists must rely instead on the records that are written in the genomes of modern organisms, which themselves have been scrambled by the passage of time. “We’re trying to resolve something that happened probably a couple billion years ago, using modern sequence data,” says computational evolutionary biologist Tom Williams at the University of Bristol, UK. It is no easy task.
Current gene-sequencing technologies have pushed the debate forward. Until recently, scientists who sought to identify the bacteria or archaea in a particular habitat had to grow the organisms in the lab. Now, researchers can assess microbial diversity in a sample of water or soil by fishing out the DNA and analysing it using mathematical tools, a technique called meta-genomics. In 2002, scientists knew of two categories (or phyla) of archaea. Today, thanks to metagenomics, the number of groupings has exploded.
Evolutionary scientists have been quick to take advantage of the growing bounty. Using the latest powerful modelling techniques, they have created a forest of evolutionary trees detailing the familial relationships among archaea. The results, in many cases, place eukaryotes within the archaeal ranks.
“The weight of evidence, in our view, really did shift toward the two-domain, eocyte tree,” says Williams. But for some, the debate was still short on data.
Then, in 2015, a group led by Thijs Ettema, an evolutionary microbiologist at Uppsala University in Sweden at the time, published DNA sequences for Lokiarchaeota, found in sediments dredged up five years earlier1. Within two years, Ettema’s team and other researchers had announced the discovery of three new archaeal phyla related to the Lokis5,6. The entire grouping of new phyla was named Asgard after the realm of the Norse gods.
The Asgard archaea are tiny in size, but they have proved to be mighty. They have reinvigorated debate about the true number of life’s domains. And they are providing tantalizing hints about the nature of the cells that gave rise to the first eukaryotes — at least to two-domain proponents.
Like their namesake, Lokiarchaeota and their kin evade easy description. They are unquestionably archaea, but their genomes include a smorgasbord of genes that are similar to some found in eukaryotes. Loki DNA, for example, contains genetic instructions for actins, proteins that form a skeleton-like framework in eukaryotic cells. The genes seemed so out of place that the researcher who spotted them initially worried that contamination was to blame. “I said, ‘Hmm, how is that possible? Can it be that this is really an archaeal genome?’” recalls evolutionary microbiologist Anja Spang at the Royal Netherlands Institute for Sea Research in Texel.
Evolutionary modelling reinforced the tight linkage between the Asgard archaea and eukaryotes. The trees built by Ettema’s team place all eukaryotes in the Asgard grouping5,7.
Now, many researchers are using data from these archaea to formulate a better picture of the eukaryotic precursor. It might already have had some features typical of eukaryotes before it took in the mitochondrial predecessor. “It probably had some very primitive membrane-biology processes going on,” Ettema says.
According to an analysis published this year7, the ancestor of the Asgard archaea probably fed on carbon-based molecules, such as fatty acids and butane. This diet would have generated byproducts that could nourish partner bacteria. Such food-sharing agreements — common among microbes — could have evolved into a more intimate relationship. An archaeon might have snuggled up next to its bacterial partner to ease nutrient exchanges, leading eventually to the ultimate embrace.
Such scenarios still provoke doubts, however. Chief among the unconvinced is Forterre. After combing through the Asgard paper, he and his colleagues published an exhaustive rebuttal8 of the work.
Misleading markers?
In a charge that infuriates Ettema, Forterre and his group suggested that some eukaryotic-like sequences found in the Lokis were a result of contamination. A Loki protein called elongation factor 2, for example, was “likely contaminated by eukaryotic sequences”, Forterre’s team wrote in its critique. Forterre now says he’s uncertain about this point.
But he and his colleagues still stand by their criticism of the Asgard evolutionary trees. Even those who are master tree-builders concede that it is tricky to untangle how organisms living two billion years ago were related to each other. Biologists reconstruct these relationships by modelling how a particular ‘marker’ — usually a protein or a gene — has changed over time in the organisms of interest.
Forterre’s group says that Ettema’s team unintentionally chose misleading markers to build its tree. Forterre and his group did their own tree analysis using two large proteins as markers because, by virtue of their size, big proteins are more likely to record the desired information. The result was a three-domain tree.