Longevity of companion dog breeds: those at risk from early death (original) (raw)
Abstract
The companion dog is one of the most phenotypically diverse species. Variability between breeds extends not only to morphology and aspects of behaviour, but also to longevity. Despite this fact, little research has been devoted to assessing variation in life expectancy between breeds or evaluating the potential for phylogenetic characterisation of longevity. Using a dataset of 584,734 unique dogs located within the UK, including 284,734 deceased, we present variation in longevity estimates within the following: parental lineage (purebred = 1 breed, crossbred ≥ 2 breeds), breed (n = 155), body size (large, medium, small), sex (male, female) and cephalic index (brachycephalic, mesocephalic, dolichocephalic). Survival estimates were then partitioned amongst phylogenetic clades: providing evidence that canine evolutionary history (via domestication and associated artificial selection) is associated with breed lifespan. This information provides evidence to inform discussions regarding pedigree health, whilst helping current/prospective owners, breeders, policy makers, funding bodies and welfare organisations improve decision making regarding canine welfare.
Subject terms: Phylogenetics, Biological anthropology, Population dynamics
Introduction
The modern domesticated dog (Canis lupus familiaris) includes over 400 genetically distinct breeds, representing considerable variation in morphological, behavioural, and physiological phenotypes1,2. However, variability between breeds also extends to life expectancy3–5. In spite of this, little research has been devoted to assessing variation in longevity between canine sub-populations or evaluating the potential for phylogenetic characterisation of longevity. Providing these estimates would help veterinarians and researchers highlight health and welfare challenges within the field, whilst also managing stakeholder expectations (including current and prospective owners), with regards to future responsibilities and duration of dog-owner relationship.
Though it is widely accepted that dogs were domesticated from an ancient wolf ancestor6,7, findings diverge on the specific timing, location and number of domestication sites8–18. However, a date of around 16,000 cal BP (calibrated years before the present) is generally accepted as the timing of domestication, based on robust archaeological and genomic evidence6,15,17–19. Despite this ~ 16,000 year timeline, the spectacular phenotypic diversity observed amongst breeds is thought to have originated much more recently, largely through intense artificial selection. This has led to the development of closed breeding populations with limited genetic heterogeneity20. The loss of genetic diversity can be attributed to a significant population bottleneck event: the generation of pedigree breeds21–23. To ensure the persistence of pedigree diversity, strict breeding practices have been emplaced to restrict gene flow between breeds24. These practices include: the repeated use of popular sires, breeding to perpetuate desired physical or behavioural characteristics, promotion of the breed barrier rule (i.e., a dog can only become a registered member of a breed, if both parents are registered members), and population maintenance via inbreeding within closed familial lines25–28. Additionally, selection pressure towards phenotypic exaggeration to achieve breed standards and/or obtain a competitive advantage when judged at show standards, has been associated with hereditary pathology and conformation disorders29,30. Breeding practices focussed on physical appearance solely, often result in reduced attention to canine health, welfare, functionality, and behaviour31,32.
Longevity estimates for an average domestic dog varies between 10.0 and 13.7 years of age, with variation depending on populations analysed e.g., country and/or breed specific3–5,33–38. However, significant variation in longevity has been reported both within and between purebreds. For example, median estimates for West Highland White Terriers have been reported at 12.7 and 13.5 years of age, while estimates for Rottweilers have been substantially lower at 8.0 and 8.4 years of age3,4. Furthermore, variation in longevity has been reported between pure (parental lineage; PL = 1 breed) and crossbred (PL ≥ 2 breeds) dogs, with most research reporting longer life expectancies within crossbreeds34,36,39,40 (however see 41). Within a subset of UK and Japanese canine populations, a crossbred survival advantage of 1.2 and 1.3 years was reported in comparison with their purebred counterparts, respectively3,36. Consequently, it has been hypothesised that the longevity advantage presented in crossbred dogs may be due to the reduction in homozygous deleterious genes, along with non-genetic differences between and within pure and crossbred populations e.g., management styles31. The former would suggest the presence of ‘hybrid vigor’ within canine populations42. Hybrid vigor, or heterosis, is the increase in stature, biomass, and fertility that characterizes the progeny of crosses between diverse parents, such that the progeny is superior to the better of the two parents43. Inbreeding depression describes the converse effect, i.e., the decline in quantitative measures of fitness upon homozygosis of alleles (i.e., inbreeding)43. At present, there is little evidence to support the existence of either evolutionary process (i.e., hybrid vigor or inbreeding depression) within domestic dogs3,28,42. However, nearly 700 inherited disorders and traits have been described in the domestic dog44, including hip dysplasia, brachycephalic obstructive airway syndrome, cardiomyopathies, endocrine dysfunctions, and blood disorders11,29,45,46. As a result, the burden of disease within most purebred populations has become one of the most important issues in canine welfare. It is imperative that future discussions are open and multidisciplinary, as they have significant implications on the ethics of breeding practices, along with the quality of life and longevity of canine breeds.
In addition to breed status, body size, sex, and cephalic index (the ratio between the width and length of skull), have previously been associated with canine longevity33,47,48. Mammalian lifespan varies strongly with body size, such that large species tend to outlive smaller species49,50. However, within species, small body size is generally associated with greater longevity51. The domesticated dog aligns with this latter trend and body size is generally considered to be the greatest predictor of their lifespan28,35,47,48,52–59. Despite this fact, substantial survival differences are frequently reported between breeds of similar size56,60–62, with larger dogs reported to have the highest morbidity3,33,52,63. Female survival advantage is also well documented in mammalian species. A recent study compiling demographic data from 134 mammal populations (encompassing 101 species), reported an average 18.6% longer median lifespan in females, in comparison with conspecific males64. In spite of this, previous studies have suggested that sex bears little influence on canine longevity38,65. Popularity of brachycephalic (flat-faced) breeds has been increasing internationally, despite increasing scientific evidence highlighting the significant health and welfare challenges associated with this conformation, in comparison with mesocephalic (medium proportions) or dolichocephalic (long-faced) breeds66–68. By identifying and comparing differences in longevity between canine populations e.g., parental lineage (PL), breed, size, sex, and cephalic index, future studies can start to disentangle mechanisms linked to risk of early death. However, very little research attention has been dedicated to: (1) assessing variation in life expectancy between subsets of the canine populations, and (2) the phylogenetic characterisation of canine longevity. This knowledge gap is due, in part, to a lack of informative, comparable, and accessible datasets regarding canine mortality.
To address this shortcoming, we developed a research project involving 18 participants, including rehoming and welfare organisations, breed registries, pet insurance companies, veterinary corporations, and university based veterinary archives. This wealth of data allowed us to partition heterogeneity of survival estimates between populations, to compare estimated longevities, and identify those most at risk from early death. Improving our knowledge of canine survivorship provides an important welfare opportunity for the estimated 12 million companion dogs in the UK69. This information will assist current/prospective owners, breeders, policy makers, funding bodies and welfare organisations in decision-making to improve the welfare of companion dogs, while also contributing evidence to the canine pedigree health debate.
Results
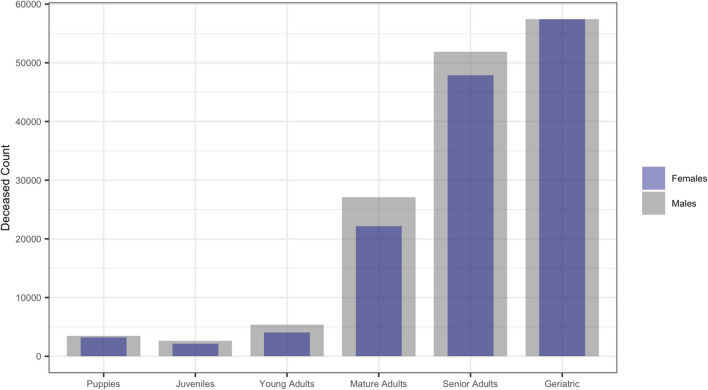
Merged, cleaned and deduplicated data included 584,734 individual dogs located within the United Kingdom (UK), including 284,734 deceased, from 18 sources (see ‘Methods: Data sources, cleaning and subsetting’ for further details). Individuals within data ranged from 0–24 years of age (x̃ = 12.2, μ = 8.56), consisting of the following demographics: 1.6% Puppies (n = 9298), 2.3% Juveniles (n = 13,226), 6.2% Young Adults (n = 36,269), 29.2% Mature Adults (n = 170,522), 34.0% Senior Adults (n = 198,963) and 26.7% Geriatric (n = 156,456). Distribution of deaths within age group, per sex, are shown in Fig. 1.
Figure 1.
Distribution of deaths from the raw data, within age group, per sex (superimposed). Note higher density of deaths within ‘Puppies’ in comparison with ‘Juveniles’, and male skewed ratios for all age groupings excluding ‘Geriatric’, where ratio is 1:1.
There was a 1.1:1 ratio of male (n♂ = 300,380) to female dogs (n♀= 284,354), and a 4.3:1 ratio of pure (nE = 473,423) to cross breeds (nX = 111,311). Within pure breeds, 20.9% were categorised as brachycephalic (n♂:n♀ 1.1:1), 63.4% were mesocephalic (n♂:n♀ 1.1:1) and 15.6% were dolichocephalic (n♂:n♀ 1.1:1). Over 50% of pure breeds were reported as one of the following twelve breeds: Labrador Retriever (9.14%), Staffordshire Bull Terrier (7.73%), English Cocker Spaniel (5.57%), German Shepherd Dog (4.21%), Jack Russel Terrier (4.06%), English Springer Spaniel (3.75%), Yorkshire Terrier (3.05%), Cavalier King Charles Spaniel (2.92%), Border Collie (2.75%), Shih Tzu (2.57%), West Highland White Terrier (2.52%) and French Bulldog (2.44%). Frequency of all purebreds in dataset are listed in Fig. S1.
Small dogs contributed 53.1% of the purebred dataset (n♂:n♀ 1:1, nbreeds = 62), whilst medium and large sized dogs made up the remaining 17.6% (n♂:n♀ 1.1:1, nbreeds = 44) and 29.3% (n♂:n♀ 1.1:1, nbreeds = 49), respectively. Within small breeds, 27.4% were brachycephalic (n = 60,723, nbreeds = 11), 63.7% were mesocephalic (n = 141,024, nbreeds= 38) and 8.9% were dolichocephalic (n = 19,605, nbreeds = 13). Within medium breeds, 13.3% were brachycephalic (n = 9709, nbreeds = 1), 68.7% were mesocephalic (n = 50,030, nbreeds = 30) and 18.0% were dolichocephalic (n = 13,072, nbreeds = 13). Within large breeds, 14.4% were brachycephalic (n = 17,671, nbreeds = 7), 62.6% were mesocephalic (n = 76,699, nbreeds = 32) and 23.0% were dolichocephalic (n = 28,114, nbreeds = 10).
Survival analysis
Percentage of total population expiring within each age grouping, θXi, are listed within Table 1: highlighting that survival probability is greatest for Juveniles, similar amongst Puppies and Young Adults, and least for Geriatrics.
Table 1.
Percentage of total population expiring, θ, per age group Xi where i represents age grouping i.e.,XP,XJ,XY⋯ calculated as: θXi=NDXiN∗100. Includes the following statistics: Ni i.e., total sub-population size; NDi i.e., total number of deaths per sub-population; θXi i.e., % total population expiring per age group. ND (= 284,734) does not equate to N (= 584,734) as the dataset includes subjects alive at point of completion. Thus, θXG calculation incorporates remaining subjects.
| Age grouping | Ni (sub-population) | NDi (total no. of deaths, per sub-population) | θXi (% total population expiring per age group) |
|---|---|---|---|
| Puppies XP | 9285 | 6675 | 1.14 |
| Juveniles XJ | 13,316 | 4,770 | 0.82 |
| Young adults XY | 36,433 | 9,426 | 1.61 |
| Mature adults XM | 170,427 | 49,289 | 8.43 |
| Senior adults XS | 198,853 | 99,729 | 17.06 |
| Geriatric XG | 156,420 | 114,845 | 70.95 |
| Total | N = 584,734 | ND = 284,734 | – |
Averaging across all individuals, i.e., pure (E) and crossbred (X), median survival was estimated at 12.5 years (12.5–12.6, n = 584,734, events = 284,734). However, variation was evident between pure and crossbred individuals (p < 0.001). Median survival for pure breeds was 12.7 years (12.6–12.7, n = 473,681, events = 212,289), whilst median survival for crossbreds was 12.0 years (12.0–12.1, n = 111,053, events = 72,445, HR 1.1, 1.1–1.1, p < 0.001). Probability of purebred and crossbred survival per decimal year are presented in Supplementary Table 1. This table highlights 95% of purebred and crossbred individuals are deceased by 18.3 and 18.2 years, respectively.
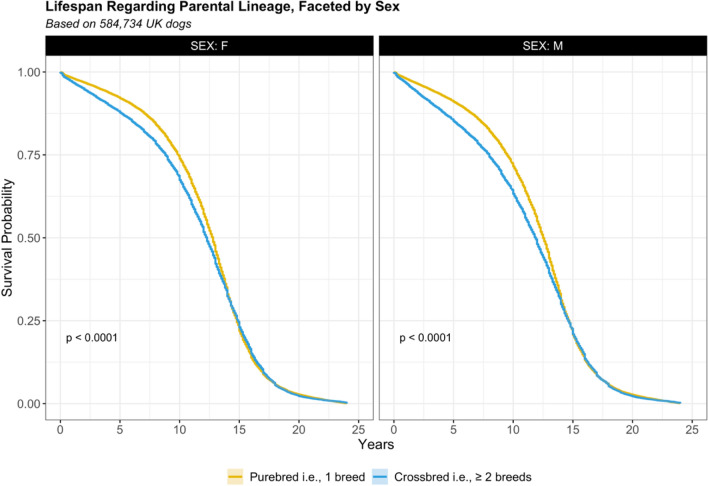
Median survival for any (i.e., E and X) female (♀) was 12.7 years (12.6–12.7, n = 283,984, events = 136,910), whilst median survival for any male (♂) was 12.4 years (12.4–12.4, n = 300,750, events = 147,824, HR 1.06, 1.06–1.07, p < 0.001). Probability of female and male survival per decimal year are presented in Supplementary Table 2, which highlight that 95% of both sexes are deceased by 18.3 years of age. Female survival advantage was apparent within both pure and cross bred individuals (x̃Ε = 12.8, 12.7–12.8, x̃Χ = 12.3, 12.2–12.3), in comparison with their male counterparts (x̃Ε = 12.5, 12.5–12.6, x̃Χ = 11.9, 11.8–12.0, p < 0.001; Fig. 2).
Figure 2.
Survival curves of pure (yellow) and cross (blue) bred individuals, faceted by sex. Survival functions based on Kaplan–Meier estimates by log rank test (p-value).
Longevity estimates for 155 recognised purebreds versus the crossbred group (x̃ = 12.0) are presented in in Supplementary Table 3. In comparison with the crossbred group, 47.1% (nbreeds = 73) of purebreds presented longer median estimates, 25.8% (nbreeds = 40) presented shorter median estimates, and the remaining 27.1% (nbreeds = 42) did not vary significantly from the crossbred group.
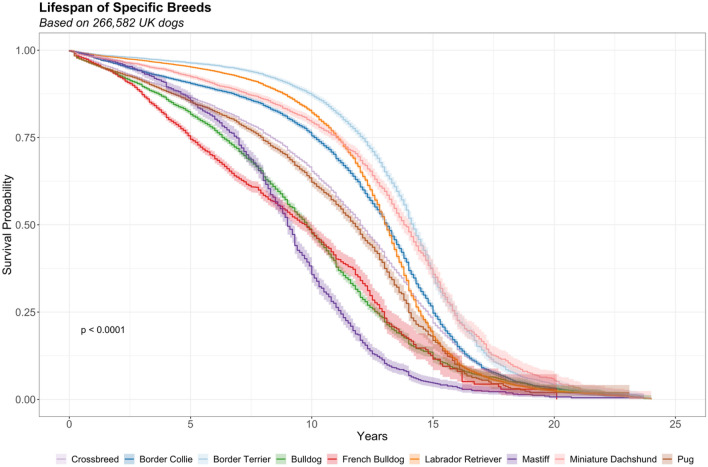
Longevity estimates for a subset of purebreds are visualised in Fig. 3. Significant variation in longevity was evident between breeds (p < 0.001): those most at risk from early death included Caucasian Shepherd Dog (x̃ = 5.4, HR 2.4, 1.6–3.7), Presa Canario (x̃ = 7.7, HR 3.0, 2.5–3.7), Cane Corso (x̃ = 8.1, HR 2.4, 2.0–3.0), Mastiff (x̃ = 9.0, HR 2.3, 2.2–2.4), St Bernard (x̃ = 9.3, HR 1.7, 1.5–1.8), Bloodhound (x̃ = 9.3, HR 1.7, 1.3–2.1), Affenpinscher (x̃ = 9.3, HR 1.7, 1.5–1.9), Neapolitan Mastiff (x̃ = 9.3, HR 1.8, 1.6–2.0), Bulldog (x̃ = 9.8, HR 1.8, 1.7–1.8) and French Bulldog (x̃ = 9.8, HR 2.2, 2.1–2.3). Those least at risk from early death included Lancashire Heeler (x̃ = 15.4, HR 0.5, 0.4–0.6), Tibetan Spaniel (x̃ = 15.2, HR 0.5, 0.4–0.5), Shiba Inu (x̃ = 14.6, HR 0.5, 0.4–0.6), Papillon (x̃ = 14.5, HR 0.6, 0.5–0.6), Lakeland Terrier (x̃ = 14.2, HR 0.7, 0.6–0.7), Schipperke (x̃ = 14.2, HR 0.6, 0.4–0.9), Border Terrier (x̃ = 14.2, HR 0.5, 0.5–0.6), Italian Greyhound (x̃ = 14.0, HR 0.7, 0.6–0.9) and Miniature Dachshund (x̃ = 14.0, HR 0.7, 0.6–0.7).
Figure 3.
Survival curves for 8 purebreds: Border Collie (dark blue, x̃ = 13.1), Border Terrier (light blue, x̃ = 14.2), Bulldog (green, x̃ = 9.8), French Bulldog (red, x̃ = 9.8), Labrador Retriever (orange, x̃ = 13.1), Mastiff (purple, x̃ = 9.0), Miniature Dachshund (pink, x̃ = 14.0) and Pug (brown, x̃ = 11.6). All purebreds vary significantly from crossbreds (light purple, x̃ = 12.0, p < 0.001). Survival functions based on Kaplan–Meier estimates by log rank test (p-value). Longevity estimates for 155 recognised purebreds versus the crossbred group are presented in in Supplementary Table 3.
Longevity differed between purebreds of varying size (p < 0.001): median survival for small and medium sized breeds were 12.7 (12.7–12.8, n = 223,222, events = 112,063, HR 0.83, 0.82–0.84) and 12.5 years (12.4–12.6, n = 73,532, events = 35,787, HR 0.88, 0.87–0.89) respectively, with an accelerated time to death for large sized breeds at 11.9 years (11.9–11.9, n = 123,072, events = 61,976). Probability of survival per decimal year for small, medium, and large individuals are presented in Supplementary Table 4: highlighting that 95% of are deceased by 18.1, 18.3 and 17.9 respectively.
Lower survival estimates within large purebreds were apparent within both male and female individuals (x̃♂ = 11.8, 11.7–11.8; x̃♀ = 12.0, 12.0–12.1), in comparison with their small (x̃♂ = 12.7, 12.6–12.7; x̃♀ = 12.8, 12.7–12.8) and medium sized counterparts (x̃♂ = 12.3, 12.3–12.4; x̃♀ = 12.7, 12.6–12.8, p < 0.001). Large males and females presented a 1.28 (1.26–1.29) and 1.17 (1.15–1.18) fold faster time to death, respectively, than the longest living small sized females (p < 0.001). Variation in longevity between the three sizes are more apparent within males (Fig. S2).
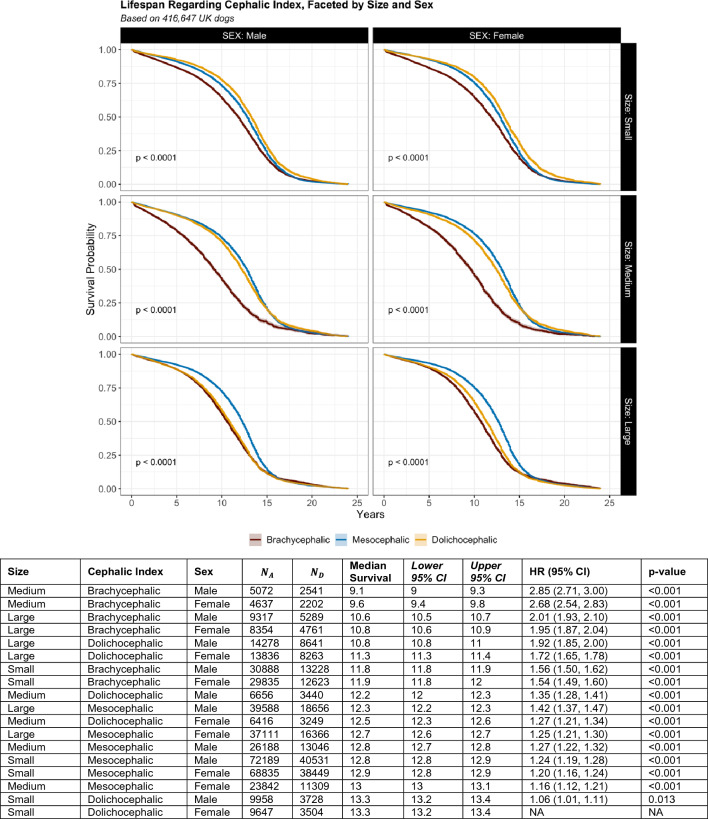
Median survival for mesocephalic purebreds was 12.8 years (12.7–12.8, n = 267,753, events = 138,357), with an accelerated time to death for brachycephalic and dolichocephalic purebreds at 11.2 years (11.2–11.3, n = 88,103, events = 40,644, HR 1.4, 1.4–1.4, p < 0.001) and 12.1 years (12.0–12.1, n = 66,369, events = 33,224, HR 1.1, 1.1–1.1, p < 0.001), respectively. An interaction was evident between cephalic index and size. In comparison with longest living small-dolichocephalic breeds (x̃ = 13.3, 13.2–13.4), brachycephalic-medium and brachycephalic-large sized breeds presented a 2.69 (2.59–2.79, x̃ = 9.4, 9.3–9.5, p < 0.001) and 1.92 (1.87–1.98, x̃ = 10.7, 10.6–10.8, p < 0.001) fold faster time to death, respectively (Fig. S3). However, a further interaction was noted, between cephalic index, size, and sex (Fig. 4). Similar to previous findings, in comparison with longest living small-dolichocephalic-female breeds (x̃ = 13.3, 13.2–13.4), medium-brachycephalic male and female breeds presented a 2.85 (2.71–2.3, x̃ = 9.1, 9.0–9.3, p < 0.001) and 2.69 (2.54–2.83, x̃ = 9.6, 9.4–9.8, p < 0.001) fold faster time to death, respectively. Furthermore, variation in longevity between the three cephalic indices were most apparent within medium sized dogs.
Figure 4.
Survival curves of brachycephalic (red), mesocephalic (blue) and dolichocephalic (yellow) purebred individuals, faceted by size and sex, along with associated table. Survival functions based on Kaplan–Meier estimates by log rank test (p-value). Table reports Kaplan–Meier survival estimates and cox proportional hazards regression model outputs for size, by sex. All groupings are compared with small-dolichocephalic-female individuals, as these represent the longest living group. Includes the following statistics: NA i.e., total number of individuals still alive; ND i.e., total number of deaths; Median Survival i.e., median age of death; Lower 95% Confidence Interval (CI) and Upper 95% CI; Hazards Ratio (Lower 95% CI and Upper 95% CI) and p-value.
Phylogenetic analyses
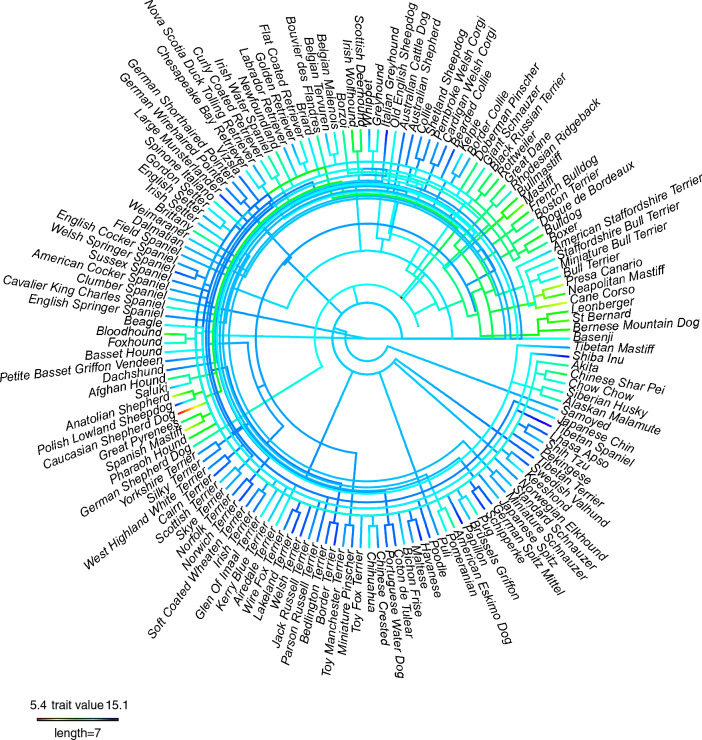
Pagel’s lambda value of 0.808 was obtained, suggesting that median lifespan exhibited a strong phylogenetic signal across the dog breeds included in these analyses. Distribution of median lifespan was strongly related to evolutionary history: with breeds within clades exhibiting closer lifespans than would be expected by chance (Fig. 5, see Fig. S4 for flat phylogeny).
Figure 5.
Circular phylogeny representing ancestor‐to‐descendant breed relationship, along with median lifespan (see Fig. S4 for flat phylogeny). Median lifespan for 148 purebreds, of which we had lifespan data, and could be assigned to tips of existing phylogenies113,114. Hotter colours represent lower median lifespans. The phylogenetic signal across the entire breed tree was very strong (Pagel’s Lambda = 0.808), suggesting that median lifespan was strongly affected by the evolutionary history of dog breeds.
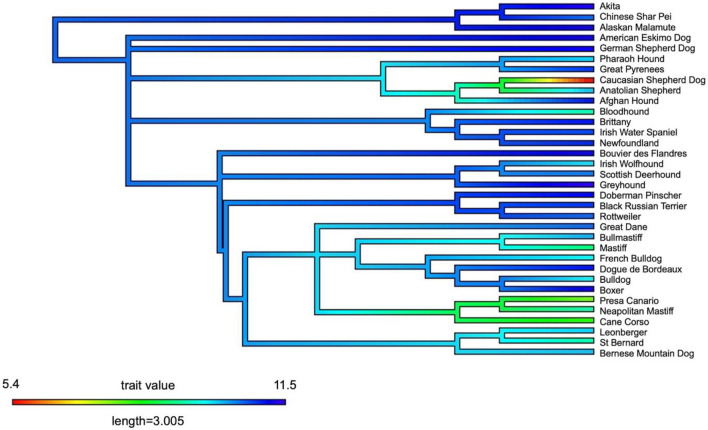
Figure 6 presents the median lifespans of the lowest quartile of breeds plotted onto the breed phylogeny. Clusters with lower median lifespans are localised within three clades: the first including breeds such as the Caucasian Shepherd Dog, the second containing the Mastiffs and Bulldogs, and the third including the Presa Canario, Neapolitan Mastiff and Cane Corso.
Figure 6.
Median lifespan of the lowest quartile of breeds plotted onto the breed phylogeny. Hotter colours represent lower median lifespans.
Discussion
Our study investigated variation in longevity estimates for dogs located within the UK, based on parental lineage, breed, body size, sex, cephalic index, and phylogeny. Our results partition heterogeneity of survival estimates across these populations: identifying groups most at risk from early death. More specifically, we provide longevity estimates for: (1) pure (PL = 1 breed) versus crossbred (PL ≥ 2 breeds) groupings, providing evidence to inform discussions regarding pedigree health; (2) 155 purebreds, representing a large range of morphological, behavioural, and physiological phenotypes; (3) varying body size, providing support for a negative correlation between size and longevity3,35,47,52,59; (4) sex, reinforcing documented female survival advantage within mammalian species64; and (5) cephalic index, expanding upon the brachycephalic health and welfare debate, regarding problems associated with conformation66,67. Furthermore, this is one of the first studies to map longevity estimates against evolutionary history (via domestication and associated artificial selection), providing evidence that canine ancestral lineage is associated with breed lifespan (however, see 41,59,70).
Breed specific estimates of survival are not only informative to veterinarians and researchers, but also to current and prospective owners, looking to fully understand their future responsibilities, and potential duration of dog-owner relationship. Whilst this study provides evidence to inform discussions around canine pedigree health, it is important to note that unbiased lifespan data necessarily requires the inclusion of live dogs whose cause of death is yet unknown, and furthermore, for those deceased—reason for death is not included within our dataset. Death may be due to euthanasia (based on physiological or behavioural concerns), trauma, disease, or natural causes. Thus, we cannot identify direct risk factors for early death. Instead, by comparing survivorship across parental lineage, breed, body size, sex, cephalic index, and phylogeny, we can identify groupings and/or lineages that demand further consideration.
Previous studies have reported significantly lower life expectancies for purebreds, in comparison with their crossbred counterparts34,36,40,71, leading to widespread belief that crossbred dogs are substantially healthier than purebreds72,73. We found that 47.1% of purebreds present a greater median survival estimate than crossbreds, only 25.8% present a shorter life expectancy than crossbreds, and 27.1% did not vary significantly. Consequently, our results are not in agreement with previous findings. However, due to methodological limitations, we do not reject the existence of hybrid vigour within the population. Within our data, purebreds and crossbreds are evaluated as a binary variable. This may be problematic as crossbreds are heterogenous: presenting differing degrees of between-breed genetic diversity. For example, the crossbred group ranges from individuals produced from a complex line of unknown hybrids to purebred hybrids, often labelled as ‘designer crossbreeds’ e.g., ‘Labradoodle’, Labrador Retriever x Poodle or ‘Jack-Chi’, Jack Russell Terrier x Chihuahua. Within purebred hybrids, while the first filial (F1) generation may benefit from hybrid vigor43 and display fewer inbreeding depression effects than their ancestral purebreds63,74,75, subsequent generations i.e., F2, F3…Fx, are likely to return a higher inbreeding co-efficient. Thus, they lose the health benefits from the initial purebred hybrid cross42. As we do not have access to detailed information regarding individual lineage, we cannot assess the presence of hybrid vigor, or inbreeding depression, within a quantitative context. However, given the growing popularity of specific 'designer cross’ breeding programs76,77, along with the proposed strategy to abandon or ‘outcross’ purebred populations significantly affected by inherited disorders and/or extreme characteristics20,28,78,79, it is important that future studies partition heterogeneity of survival estimates amongst mixed (PL > 2 breeds or unknown), known crossbred (PL = 2 breeds e.g., 'designer cross’) and ancestral purebred lineages. Furthermore, as mixed breeds have been reported to live 1.2 years longer than size-matched purebred dogs, and individual breeding level has been negatively associated with juvenile survival and adult lifespan28, future studies should consider incorporating individual-level phenotype i.e., lifespan, body size and PL, from genotyped dogs, to disentangle the effect of breed and body size upon longevity. Such research would help establishing the true extent of any suggested hybrid health risks and/or benefits by improving power for detecting subtler effects of inbreeding, as well as specific genetic variants that may affect lifespan.
This study evaluated 584,734 pure and crossbred dogs (including 284,734 deceased), from 18 organisations, including rehoming and welfare organisations, breed registries, pet insurance companies, veterinary corporations, and university based veterinary archives. The greater effective sample size of the aggregated data, has the potential to improve model estimates80, by providing a more comprehensive and representative picture of the study population. However, it is not without its shortcomings. Specific sources may introduce representation bias, providing incomplete information which impacts estimates, or reporting termination/death related statistics that differ from the general canine population. Pet insurance data have previously been used for assessing longevity estimates for pet populations37,81,82. Despite this fact, their value has been affected by the following biases: older animals are often uninsured; some health conditions are excluded (especially where repeated); presence of financial thresholds on claims; and some policies providing age-limited life cover. Additionally, not all pets will be represented in these data since not all owners are able, or choose, to insure their pet. This can lead to some breeds being under-represented within insurance data: especially those of high disease risk, such as brachycephalic breeds, due to the elevated cost of insurance83. Previous studies have also reported owners of purebreds are more likely to financially invest in their dogs, showing a greater willingness to pursue more extensive veterinary treatment at referral hospitals, than owners of mixed breed dogs84–86. This would result in a greater representation of purebred dogs within veterinary datasets (as seen in the present study). Furthermore, the growing popularity of certain breeds may result in an over overrepresentation of younger individuals within the data, which increases the risk of underestimating their lifespan87. Finally, due to the nature of breed registries, it is likely that these data are biased towards pedigree or purebred dogs—despite crossbreds being recorded within ‘separate’/alternate registries88. With the aim of mitigating some aforementioned biases, we incorporated rehoming and welfare charity datasets, where crossbred dogs have been reported to represent 44–87.8% of the UK rehoming population68,89. However, we recommend future researchers address biases by checking for breed survival differences between sources (not available here due to restrictions stipulated within data sharing agreements with project participants), and addressing missing data via imputation or other statistical techniques90,91.
Despite previous research reporting a survival advantage for neutered animals3,65,92,93 due to data limitations, we had to omit neuter status from this analysis. Neuter status is a time-dependent variable likely to have a non-linear relationship with age, both with regards to probability of the procedure having been carried out, and the probability of attaining the updated status within multiple data sources94,95. As such, the available data was deemed unreliable and excluded from further analysis.
Whilst these findings provide valuable insight into breed longevity, it is important to acknowledge that purebred dog populations and, thus, their population management are influenced by environment and location. Consequently, these results are representative of the UK canine population only, and should not be used to make assessments within other regions/countries. For example, within certain countries, breed registries have adopted independent policies regarding the importation, breeding, and registration of foreign dogs. Some registries effectively disallow breeding of imported dogs and prevent recognition of the resulting puppies—whilst others allow for the controlled importation and breeding of ‘foreign’ animals. The latter policy introduces genetic diversity: potentially impacting the overall longevity of their canine population96. Furthermore, variation in public attitude towards dogs (owned or free roaming) and responsible dog ownership practices, will alter population management approaches97. Consequently, further research is needed to explore the impact of regional dog population management practices and breed registry policies, upon the lifespan of local purebred dogs.
Variations in popularity of dog breeds are often evident as large and impulsive fluctuations, that are usually considered the hallmark of fashions and fads98,99. An acute increase in breed demand and impulse buying100,101 have resulted in puppies becoming lucrative commodities in an industry driven by profitability, often at the expense of canine welfare102,103. Recent studies have reported an increase in rates of dogs suffering from physiological and psychological issues caused by inappropriate breeding practices, pre-purchase handling, early life environment and husbandry102,104–107. Much of this supply may originate from large-scale, industrial-style operations, in which breeders may attempt to lower overheads through poor husbandry and deficient hygiene standards: operations that are colloquially dubbed ‘puppy farms’102,104. However, puppy welfare issues extend to some regulated and small-scale breeders due to limited regulation and/or the practice of inbreeding to ‘fix’ characteristics associated with breed standards. This can lead to increased risk of hereditary pathology and exaggerated physical characteristics20 including orthopaedic disorders29,108, skin disease29,109, aural disease29,46,110, ocular disease111,112, and breathing difficulties105,106. Gaps in regulation and the inability to reliably quantify supply sources have significant implications on the ‘dog market’, including the ethics of breeding practices. It is therefore imperative that future discussions regarding optimal canine breeding practices are open and multidisciplinary. As such, we hope these findings empower stakeholders, providing evidence required to vocalise opinions and collectively consider the impact of (1) biology e.g., genotype113, phenotype28, conformation66,114, or physiology115, and (2) non-biological factors e.g., owner demographics116,117, management styles31,118, or breed function5, upon risk of early death within the UK canine population.
Methods
Data sources, cleaning and subsetting
Dogs Trust data were combined with datasets sourced from 17 external project participants. Data sources included breed registries (45.0%), veterinary corporations (26.5%), pet insurance companies (17.1%), animal welfare charities (5.9%), and academic institutions (5.5%). Project participants who provided data to this project include: Battersea Dogs and Cats Home; Blue Cross; SSPCA; Raystede; Wood Green, The Animals Charity; Edinburgh Dog and Cat Home; PDSA; Mayhew; The Insurance Emporium (The Equine and Livestock Insurance Company Limited); NCI Insurance; Cardif Pinnacle; Agria Pet Insurance Ltd; Direct Line; Medivet; Vets4Pets; Savsnet (Small Animal Veterinary Surveillance Network, University of Liverpool); and The Kennel Club (UK).
Data requested included: breed (free text), crossbred (Y/N/unknown), sex (M/F/unknown), date of birth (DOB; MM/YYYY), postcode area (i.e., first one or two characters), first three characters of dog name (common, not pedigree name), last six characters of microchip number, last six characters of additional microchip number (if more than one known), status (alive/dead) and termination date (death or end of policy due to death; DD/MM/YY). Not all canine-centric variables were sent by project participants and no data were collected regarding owner unique identifiers. Longevity (age of dog in decimal years) reflects the period between DOB and termination date (if deceased), or date data were received (if assumed alive).
Data cleaning included the removal of non-canids, classifying all individuals into (1) The UK Kennel Club (KC)119 and/or Fédération Cynologique Internationale (FCI)120 recognised ‘purebred’ breeds (PL = 1 breed), or (2) ‘crossbred’ breeds (PL ≥ 2 breeds). Breeds within the dataset may be present in one or both KC and FCI lists. Those that are not included in either but appear in the data were used as outgroups for phylogenetic analyses e.g., American Hairless Terrier, Apennine Wolf, Pastore della Lessinia e del Lagorai, Pastor della Sila and Wolf & Golden Jackal (Supplementary Table S5). Forty-four KC and/or FCI breeds were reclassified to match nodes, i.e., breeds, within existing canine phylogenies121,122. For example, ‘American Akita’ and ‘Japanese Akita Inu’ were collapsed into ‘Akita’, which is present in both KC119 and FCI120 breed lists. These changes (listed in Supplementary Table S5; under ‘Phylogeny Reclassification’), were necessary due to breed inconsistency across the data sources and were instituted via majority agreement by canine behaviour and research experts at Dogs Trust.
Body size classifications: small, medium, and large, were obtained from KC119 and FCI120 grey literature. Breed average ratio between the width and length of skull i.e., cephalic index: brachycephalic (flat-faced), mesocephalic (medium proportions) or dolichocephalic (long-faced), were sourced from O’Neill et al., 2020123. Breeds present within our data, but omitted from the above lists, were assigned body size and cephalic index classification via majority agreement by a group of canine behaviour and research experts at Dogs Trust (Supplementary Table S5). Body size and cephalic index data are only available for purebred individuals, due to phenotypic variation within crossbreds.
Month and year of birth were routinely recorded by rehoming and welfare centres, breed registries, veterinary corporations, and pet insurance companies. Unfortunately, the specific date was less commonly reported. Consequently, all age estimates were based on ‘MM/YYYY’ data. Individuals were grouped into one of six age categories: ‘Puppies’ XP aged 0 to < 6 months, ‘Juveniles’ XJ aged 6 to < 12 months, ‘Young Adults’ XY aged 12 to < 24 months, ‘Mature Adults’ XM aged 2 to < 7 years, ‘Senior’ XS aged 7 to < 12 years and ‘Geriatric’ XG aged ≥ 12. These categories were developed to capture age-related developmental trajectories for most dog breeds124.
As data were obtained from multiple sources, duplication of individuals was probable. Deduplication consisted of a four-phase process outlined in Supplementary Note S1. For the purposes of this study, deduplicated data were then subset to rows where the following variables were complete: crossbred (Y/N); status (alive/dead), breed (n = 155), sex (M/F) and age (decimal years). To remove improbably aged dogs, DOB was restricted to ≤ 24.0 years, based on previously reported maximum canine longevity3. Each group (e.g., Affenpinscher or Yorkshire Terrier) within an independent variable (e.g., breed), were only included in analysis, if an adequate number of individuals (and events) were available i.e., > 20 alive and > 20 deceased individuals (Clark et al. 2003). Resulting alive NA, deceased ND and total N sample sizes for each independent variable are listed in Table 2, and associated samples sizes per group within each independent variable, are listed in Supplementary Tables S6–S11.
Table 2.
Samples sizes regarding alive NA, deceased ND and total N individuals, per independent variable (post censoring).
| Independent variable | Sample sizes | ||
|---|---|---|---|
| NA | ND | N | |
| Breed Parental lineage Sex Age group | 300,000 | 284,734 | 584,734 |
| Body size | 210,000 | 209,826 | 419,826 |
| Cephalic index | 210,000 | 212,225 | 422,225 |
Statistical analyses
All analyses were conducted using the statistical programming software R version 4.0.4 (2021-02-15)125. As the dataset included a large number of subjects alive at point of completion i.e., not all subjects died during the study, this produces censored values. Having a large number of censored values decreases the equivalent number of subjects exposed (at risk) at later times, making survival estimates less reliable than they would be for the same number of subjects with less censoring126. Ignoring censored values, or simply equating their observed survival time with the unobserved total survival time (i.e., assuming all censored survival times occur immediately after their censoring times), would bias the results127. Fortunately, censoring is common within these data, such that specific statistical methods have been developed for appropriate analysis. Maximum Likelihood Estimation is the most notable method for analysing censored data. This method adjusts for whether the subjects observed were censored or not. It also utilizes all the information available. The statistical approaches that fall within the likelihood method include those used within this study128.
Kaplan–Meier survival curves129,130 were estimated per group, within each independent variable i.e., parental lineage, breed, size, sex, and cephalic index, using the surviminer package131. Median survival estimates are presented in place of mean values, as extreme values from non-normally distributed longevity distributions provide unreliable estimates87. Upper and lower 95% confidence intervals immediately follow median survival estimates or hazard ratios, unless otherwise stated. To test for treatment differences between survival curves, pairwise log-rank tests were conducted. Differences in proportional risks of mortality between categories were tested using a Cox proportional hazards model.
Percentage of total population expiring (Table 1), θ, per age group Xi where i represents age grouping i.e.,XP,XJ,XY⋯ were calculated as:
As dataset includes subjects alive at point of completion (NA), total number of deaths (ND) does not equate with total population (N). This produces ‘censored values’ (discussed above)126. Thus, θXG calculation incorporates remaining subjects, as ND would eventually equate to N, as measurements continued:
θXG=N-∑NDXP,XJ,XY,XM,XS∗100
Pagel’s Lambda132 was calculated using the phylosig function from the phytools133 package, with the aim of determining whether the median lifespan of the purebreds included in the analysis held a phylogenetic signal. The presence of a phylogenetic signal for a given trait suggests clustering of that trait on the phylogeny. Lambda is a metric that quantifies the level to which the similarity of a continuous trait is due to evolutionary history and can take any value within the range 0 to 1. A signal strength of 0 means that there is no phylogenetic signal, i.e., correlations between values of the trait do not reflect the relatedness of the tips of the phylogenetic tree. A signal strength of 1 suggests that the values of the tips are highly dependent upon evolutionary history. The phylogeny of domestic dogs was based on that of Parker et al., (2017)121 and supplemented with extra breeds from Talenti et al., (2018)122. In total 148 purebreds were included, for which we had lifespan data. All branch lengths were set to equal 1 before analysis, as the original tree(s) did not include branch length information. To visualise the variation in median lifespan across the phylogeny of purebreds, specifically those clades in which reduction in median lifespan clustered, we used the contMap function within phytools133. For visualisation we restricted this to purebreds within the lower quartile of median lifespans, i.e., the 34 purebreds with the lowest median lifespan.
Ethics approval and consent to participate
Ethical approval for this study was granted by Dogs Trust Ethical Review Board (Reference Number: ERB038). All methods were performed in accordance with the relevant guidelines and regulations.
Supplementary Information
Acknowledgements
We are grateful to the following project participants, who kindly provided data to this project: Battersea Dogs and Cats Home; Blue Cross; SSPCA; Raystede; Wood Green, The Animals Charity; Edinburgh Dog and Cat Home; PDSA; Mayhew; The Insurance Emporium (The Equine and Livestock Insurance Company Limited); NCI Insurance; Cardif Pinnacle; Agria Pet Insurance Ltd; Direct Line; Medivet; Vets4Pets; Savsnet (Small Animal Veterinary Surveillance Network, University of Liverpool); and The Kennel Club (UK). We are grateful to Dr. David Wong and Dr. Zhibin Zhao at the University of Manchester, for developing the deduplication pipeline; and Dr. Dan O’Neill at the Royal Veterinary College for facilitating communication with veterinary corporations. Internal to Dogs Trust, we are grateful to Kieran Huggan (Operations) for collating Dogs Trust data.
Author contributions
K.M.M. collected the data; K.M.M., C.L.W. & J.B. designed methodology; K.M.M. & J.B. analysed the data; K.M.M. led the writing of the manuscript. K.M.M., C.L.W., J.B., M.M.U., R.A.C. and R.M.C. conceived the ideas, contributed critically to the drafts, and gave final approval for publication.
Funding
This research received no external funding. All data collection and analyses were supported by Dogs Trust.
Data availability
Datasets generated and analysed during the current study are not publicly available due to restrictions stipulated within data sharing agreements with project participants. However, the corresponding author will consider facilitating collaborative discussions with all parties involved, on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in the legend of Figure 3. Full information regarding the correction made can be found in the correction for this Article.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/16/2024
A Correction to this paper has been published: 10.1038/s41598-024-59331-w
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-50458-w.
References
- 1.Neff MW, Rine J. A fetching model organism. Cell. 2006;124:229–231. doi: 10.1016/j.cell.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Zeder MA. Documenting Domestication: New Genetic and Archaeological Paradigms. University of California Press; 2006. [Google Scholar]
- 3.O’Neill DG, Church DB, McGreevy PD, Thomson PC, Brodbelt DC. Longevity and mortality of owned dogs in England. Vet. J. 2013;198:638–643. doi: 10.1016/j.tvjl.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Lewis TW, Wiles BM, Llewellyn-Zaidi AM, Evans KM, O’Neill DG. Longevity and mortality in Kennel Club registered dog breeds in the UK in 2014. Can. Genet. Epidemiol. 2018;5:10. doi: 10.1186/s40575-018-0066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teng KT, Brodbelt DC, Pegram C, Church DB, O’Neill DG. Life tables of annual life expectancy and mortality for companion dogs in the United Kingdom. Sci. Rep. 2022;12:6415. doi: 10.1038/s41598-022-10341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freedman AH, et al. Genome sequencing highlights the dynamic early history of dogs. PLOS Genetics. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vilà C, et al. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 8.Germonpré M, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: Osteometry, ancient DNA and stable isotopes. J. Archaeol. Sci. 2009;36:473–490. doi: 10.1016/j.jas.2008.09.033. [DOI] [Google Scholar]
- 9.Germonpré M, et al. Palaeolithic dogs and the early domestication of the wolf: A reply to the comments of Crockford and Kuzmin (2012) J. Archaeol. Sci. 2013;40:786–792. doi: 10.1016/j.jas.2012.06.016. [DOI] [Google Scholar]
- 10.vonHoldt BM, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson G, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc. Natl. Acad. Sci. 2012;109:8878–8883. doi: 10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thalmann O, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013;342:871–874. doi: 10.1126/science.1243650. [DOI] [PubMed] [Google Scholar]
- 13.Larson G, Bradley DG. How much is that in dog years? The advent of canine population genomics. PLoS Genet. 2014;10:e1004093. doi: 10.1371/journal.pgen.1004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake AG, Coquerelle M, Colombeau G. 3D morphometric analysis of fossil canid skulls contradicts the suggested domestication of dogs during the late Paleolithic. Sci. Rep. 2015;5:8299. doi: 10.1038/srep08299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morey DF, Jeger R. Paleolithic dogs: Why sustained domestication then? J. Archaeol. Sci. Rep. 2015;3:420–428. [Google Scholar]
- 16.Morey DF, Jeger R. From wolf to dog: Late Pleistocene ecological dynamics, altered trophic strategies, and shifting human perceptions. Histor. Biol. 2017;29:895–903. doi: 10.1080/08912963.2016.1262854. [DOI] [Google Scholar]
- 17.Frantz LAF, et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science. 2016;352:1228–1231. doi: 10.1126/science.aaf3161. [DOI] [PubMed] [Google Scholar]
- 18.Perri A. A wolf in dog’s clothing: Initial dog domestication and Pleistocene wolf variation. J. Archaeol. Sci. 2016;68:1–4. doi: 10.1016/j.jas.2016.02.003. [DOI] [Google Scholar]
- 19.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 20.Farrell LL, Schoenebeck JJ, Wiener P, Clements DN, Summers KM. The challenges of pedigree dog health: Approaches to combating inherited disease. Can. Genet. Epidemiol. 2015;2:3. doi: 10.1186/s40575-015-0014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruz F, Vilà C, Webster MT. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol Biol Evol. 2008;25:2331–2336. doi: 10.1093/molbev/msn177. [DOI] [PubMed] [Google Scholar]
- 22.Leroy G. Genetic diversity, inbreeding and breeding practices in dogs: Results from pedigree analyses. Vet. J. 2011;189:177–182. doi: 10.1016/j.tvjl.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Marsden CD, et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. 2016;113:152–157. doi: 10.1073/pnas.1512501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akey JM, et al. Tracking footprints of artificial selection in the dog genome. Proc. Natl. Acad. Sci. 2010;107:1160–1165. doi: 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1706–1716. doi: 10.1101/gr.3736605. [DOI] [PubMed] [Google Scholar]
- 26.Calboli FCF, Sampson J, Fretwell N, Balding DJ. Population structure and inbreeding from pedigree analysis of purebred dogs. Genetics. 2008;179:593–601. doi: 10.1534/genetics.107.084954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellanby RJ, et al. Population structure and genetic heterogeneity in popular dog breeds in the UK. Vet. J. 2013;196:92–97. doi: 10.1016/j.tvjl.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Yordy J, et al. Body size, inbreeding, and lifespan in domestic dogs. Conserv Genet. 2020;21:137–148. doi: 10.1007/s10592-019-01240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher L, Diesel G, Summers JF, McGreevy PD, Collins LM. Inherited defects in pedigree dogs. Part 1: disorders related to breed standards. Vet J. 2009;182:402–411. doi: 10.1016/j.tvjl.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Lewis TW, Woolliams JA, Blott SC. Optimisation of breeding strategies to reduce the prevalence of inherited disease in pedigree dogs. Anim. Welf. 2010;19:93–98. doi: 10.1017/S0962728600002281. [DOI] [Google Scholar]
- 31.McGreevy PD, Nicholas FW. Some practical solutions to welfare problems in dog breeding. Anim. Welf. 1999;8:329–341. doi: 10.1017/S0962728600021965. [DOI] [Google Scholar]
- 32.Trees L, et al. Strengthening legislation around dog breeding. Vet. Rec. 2023;193:116–117. doi: 10.1002/vetr.3337. [DOI] [PubMed] [Google Scholar]
- 33.Michell AR. Longevity of British breeds of dog and its relationships with sex, size, cardiovascular variables and disease. Vet. Rec. 1999;145:625–629. doi: 10.1136/vr.145.22.625. [DOI] [PubMed] [Google Scholar]
- 34.Proschowsky HF, Rugbjerg H, Ersbøll AK. Mortality of purebred and mixed-breed dogs in Denmark. Prevent. Vet. Med. 2003;58:63–74. doi: 10.1016/S0167-5877(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 35.Adams VJ, Evans KM, Sampson J, Wood JLN. Methods and mortality results of a health survey of purebred dogs in the UK. J. Small Anim. Pract. 2010;51:512–524. doi: 10.1111/j.1748-5827.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 36.Inoue M, Kwan NCL, Sugiura K. Estimating the life expectancy of companion dogs in Japan using pet cemetery data. J. Vet. Med. Sci. 2018;80:1153–1158. doi: 10.1292/jvms.17-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue M, Hasegawa A, Hosoi Y, Sugiura K. A current life table and causes of death for insured dogs in Japan. Prevent. Vet. Med. 2015;120:210–218. doi: 10.1016/j.prevetmed.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Montoya, M. et al. Life expectancy tables for dogs and cats derived from clinical data. Front. Vet. Sci. 10 (2023). [DOI] [PMC free article] [PubMed]
- 39.Patronek GJ, Waters DJ, Glickman LT. Comparative longevity of pet dogs and humans: Implications for gerontology research. J. Gerontol. Ser. A. 1997;52A:B171–B178. doi: 10.1093/gerona/52A.3.B171. [DOI] [PubMed] [Google Scholar]
- 40.Wallis, L. J., Szabó, D., Erdélyi-Belle, B. & Kubinyi, E. Demographic change across the lifespan of pet dogs and their impact on health status. Front. Vet. Sci. 5 (2018). [DOI] [PMC free article] [PubMed]
- 41.Urfer SR, Kaeberlein M, Promislow DEL, Creevy KE. Lifespan of companion dogs seen in three independent primary care veterinary clinics in the United States. Can. Med. Genet. 2020;7:7. doi: 10.1186/s40575-020-00086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholas FW, Arnott ER, McGreevy PD. Hybrid vigour in dogs? Vet. J. 2016;214:77–83. doi: 10.1016/j.tvjl.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Birchler JA, Yao H, Chudalayandi S. Unraveling the genetic basis of hybrid vigor. Proc. Nat. Acad. Sci. 2006;103:12957–12958. doi: 10.1073/pnas.0605627103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faculty of Veterinary Science, University of Sydney. OMIA—Online Mendelian Inheritance in Animals. https://www.omia.org/ (2021).
- 45.Ackerman, L. J. The Genetic Connection: A Guide to Health Problems in Purebred Dogs. (American Animal Hosp Assoc, 2011).
- 46.Summers JF, Diesel G, Asher L, McGreevy PD, Collins LM. Inherited defects in pedigree dogs Part 2: Disorders that are not related to breed standards. Vet. J. 2010;183:39–45. doi: 10.1016/j.tvjl.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Galis F, Van Der Sluijs I, Van Dooren TJM, Metz JAJ, Nussbaumer M. Do large dogs die young? J. Exp. Zool. B Mol. Dev. Evol. 2007;308B:119–126. doi: 10.1002/jez.b.21116. [DOI] [PubMed] [Google Scholar]
- 48.Fleming JM, Creevy KE, Promislow DEL. Mortality in North American dogs from 1984 to 2004: an investigation into age-, size-, and breed-related causes of death. J. Vet. Int. Med. 2011;25:187–198. doi: 10.1111/j.1939-1676.2011.0695.x. [DOI] [PubMed] [Google Scholar]
- 49.de Magalhães JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J. Gerontol. Ser. A. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ricklefs RE. Life-history connections to rates of aging in terrestrial vertebrates. Proc. Nat. Acad. Sci. 2010;107:10314–10319. doi: 10.1073/pnas.1005862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blagosklonny MV. Big mice die young but large animals live longer. Aging (Albany NY) 2013;5:227–233. doi: 10.18632/aging.100551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greer KA, Canterberry SC, Murphy KE. Statistical analysis regarding the effects of height and weight on life span of the domestic dog. Res. Vet. Sci. 2007;82:208–214. doi: 10.1016/j.rvsc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Kraus C, Pavard S, Promislow DEL. The size-life span trade-off decomposed: Why large dogs die young. Am. Nat. 2013;181:492–505. doi: 10.1086/669665. [DOI] [PubMed] [Google Scholar]
- 54.Salvin HE, McGreevy PD, Sachdev PS, Valenzuela MJ. The effect of breed on age-related changes in behavior and disease prevalence in cognitively normal older community dogs, Canis lupus familiaris. J. Vet. Behavior. 2012;7:61–69. doi: 10.1016/j.jveb.2011.06.002. [DOI] [Google Scholar]
- 55.Teng KT, McGreevy PD, Toribio J-ALML, Dhand NK. Trends in popularity of some morphological traits of purebred dogs in Australia. Can. Genet. Epidemiol. 2016;3:2. doi: 10.1186/s40575-016-0032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urfer SR, Greer K, Wolf NS. Age-related cataract in dogs: A biomarker for life span and its relation to body size. AGE. 2011;33:451–460. doi: 10.1007/s11357-010-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Deeb B, Pendergrass W, Wolf N. Cellular proliferative capacity and life span in small and large dogs. J. Gerontol. Ser. A. 1996;51A:B403–B408. doi: 10.1093/gerona/51A.6.B403. [DOI] [PubMed] [Google Scholar]
- 58.Nam, Y. et al. Dog size and patterns of disease history across the canine age spectrum: Results from the dog aging project. 10.1101/2022.05.03.490110 (2022). [DOI] [PMC free article] [PubMed]
- 59.da Silva J, Cross BJ. Dog life spans and the evolution of aging. Am. Nat. 2023;201:E140–E152. doi: 10.1086/724384. [DOI] [PubMed] [Google Scholar]
- 60.Urfer SR, Wang M, Yang M, Lund EM, Lefebvre SL. Risk factors associated with lifespan in pet dogs evaluated in primary care veterinary hospitals. J. Am. Anim. Hosp. Assoc. 2019;55:130–137. doi: 10.5326/JAAHA-MS-6763. [DOI] [PubMed] [Google Scholar]
- 61.Bonnett BN, Egenvall A, Hedhammar A, Olson P. Mortality in over 350,000 insured Swedish dogs from 1995–2000: I. Breed-, gender-, age- and cause-specific rates. Acta Vet. Scand. 2005;46:105–120. doi: 10.1186/1751-0147-46-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kraus C, Snyder-Mackler N, Promislow DEL. How size and genetic diversity shape lifespan across breeds of purebred dogs. GeroScience. 2023;45:627–643. doi: 10.1007/s11357-022-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bannasch D, et al. The effect of inbreeding, body size and morphology on health in dog breeds. Can. Med. Genetics. 2021;8:12. doi: 10.1186/s40575-021-00111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemaître J-F, et al. Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proc. Nat. Acad. Sci. 2020;117:8546–8553. doi: 10.1073/pnas.1911999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffman JM, O’Neill DG, Creevy KE, Austad SN. Do female dogs age differently than male dogs? J. Gerontol. Ser. A. 2018;73:150–156. doi: 10.1093/gerona/glx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Packer RMA, O’Neill DG, Fletcher F, Farnworth MJ. Great expectations, inconvenient truths, and the paradoxes of the dog-owner relationship for owners of brachycephalic dogs. PLOS ONE. 2019;14:e0219918. doi: 10.1371/journal.pone.0219918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Packer R, O’Neill D. Health and Welfare of Brachycephalic (Flat-faced) Companion Animals: A Complete Guide for Veterinary and Animal Professionals. CRC Press; 2021. [Google Scholar]
- 68.Carter AJ, Martin JH. Demographic changes in UK rescue centre dog population between 2014 and 2018. J. Appl. Anim. Welf. Sci. 2021;24:347–356. doi: 10.1080/10888705.2020.1839755. [DOI] [PubMed] [Google Scholar]
- 69.UK Pet Food. UK Pet Population 2023. https://www.ukpetfood.org/information-centre/statistics/uk-pet-population.html (2023).
- 70.Bellumori TP, Famula TR, Bannasch DL, Belanger JM, Oberbauer AM. Prevalence of inherited disorders among mixed-breed and purebred dogs: 27,254 cases (1995–2010) J. Am. Vet. Med. Assoc. 2013;242:1549–1555. doi: 10.2460/javma.242.11.1549. [DOI] [PubMed] [Google Scholar]
- 71.Egenvall A, Hedhammar A, Bonnett BN, Olson P. Gender, age, breed and distribution of morbidity and mortality in insured dogs in Sweden during 1995 and 1996. Vet. Rec. 2000;146:519–525. doi: 10.1136/vr.146.18.519. [DOI] [PubMed] [Google Scholar]
- 72.Starkey MP, Scase TJ, Mellersh CS, Murphy S. Dogs really are man’s best friend—Canine genomics has applications in veterinary and human medicine! Brief. Funct. Genomics. 2005;4:112–128. doi: 10.1093/bfgp/4.2.112. [DOI] [PubMed] [Google Scholar]
- 73.Farrow T, Keown A, Farnworth M. An exploration of attitudes towards pedigree dogs and their disorders as expressed by a sample of companion animal veterinarians in New Zealand. N. Z. Vet. J. 2014;62:267–273. doi: 10.1080/00480169.2014.902340. [DOI] [PubMed] [Google Scholar]
- 74.Liberg O, et al. Severe inbreeding depression in a wild wolf (Canis lupus) population. Biol. Lett. 2005;1:17–20. doi: 10.1098/rsbl.2004.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ali MB, et al. Genetic analysis of the modern Australian labradoodle dog breed reveals an excess of the poodle genome. PLOS Genetics. 2020;16:e1008956. doi: 10.1371/journal.pgen.1008956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turcsán B, Miklósi Á, Kubinyi E. Owner perceived differences between mixed-breed and purebred dogs. PLOS ONE. 2017;12:e0172720. doi: 10.1371/journal.pone.0172720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burnett E, et al. How much is that doodle in the window? Exploring motivations and behaviours of UK owners acquiring designer crossbreed dogs (2019–2020) Can. Med. Genet. 2022;9:8. doi: 10.1186/s40575-022-00120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeppsson S. Purebred dogs and canine wellbeing. J. Agric. Environ. Ethics. 2014;27:417–430. doi: 10.1007/s10806-013-9470-y. [DOI] [Google Scholar]
- 79.Donner J, et al. Frequency and distribution of 152 genetic disease variants in over 100,000 mixed breed and purebred dogs. PLOS Genetics. 2018;14:e1007361. doi: 10.1371/journal.pgen.1007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goodman D. Methods for joint inference from multiple data sources for improved estimates of population size and survival rates. Mar. Mammal Sci. 2004;20:401–423. doi: 10.1111/j.1748-7692.2004.tb01169.x. [DOI] [Google Scholar]
- 81.Egenvall A, et al. Mortality of life-insured swedish cats during 1999–2006: Age, breed, sex, and diagnosis. J. Vet. Internal Med. 2009;23:1175–1183. doi: 10.1111/j.1939-1676.2009.0396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonnett BN, Egenvall A. Age patterns of disease and death in insured swedish dogs, cats and horses. J. Comp. Pathol. 2010;142:S33–S38. doi: 10.1016/j.jcpa.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Chaters, G., Trees, A. J. & Laing, G. Higher insurance premiums revealed for popular brachycephalic BREEDs—The Veterinary Policy Research Foundation. https://vetpolicy.uk/2020/07/09/higher-insurance-premiums-revealed-for-popular-brachycephalic-breeds/ (2020).
- 84.Brown CM. The future of the North American Veterinary Teaching Hospital. J. Vet. Med. Educ. 2003;30:197–202. doi: 10.3138/jvme.30.3.197. [DOI] [PubMed] [Google Scholar]
- 85.Dotson MJ, Hyatt EM. Understanding dog–human companionship. J. Bus. Res. 2008;61:457–466. doi: 10.1016/j.jbusres.2007.07.019. [DOI] [Google Scholar]
- 86.Rooney, N., Pead, M. & Sargan, D. Pedigree dog breeding in the UK: A major welfare concern? 1–78 http://www.terrierman.com/PDE-RSPCA-FULL.pdf (2009).
- 87.Clark TG, Bradburn MJ, Love SB, Altman DG. Survival analysis Part I: Basic concepts and first analyses. Br. J. Cancer. 2003;89:232–238. doi: 10.1038/sj.bjc.6601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.The Kennel Club. Registration Rules and Regulations (B Regulations). https://www.thekennelclub.org.uk/about-us/about-the-kennel-club/the-kennel-club-codes/registration-rules-and-regulations/#:~:text=Dogs%20registered%20on%20the%20Activity,rally%20competitions%20and%20canicross%20races. (2023).
- 89.Diesel G, Brodbelt D, Pfeiffer DU. Characteristics of relinquished dogs and their owners at 14 rehoming centers in the United Kingdom. J. Appl. Animal Welf. Sci. 2010;13:15–30. doi: 10.1080/10888700903369255. [DOI] [PubMed] [Google Scholar]
- 90.Herring AH, Ibrahim JG. Likelihood-based methods for missing covariates in the cox proportional hazards model. J. Am. Stat. Assoc. 2001;96:292–302. doi: 10.1198/016214501750332866. [DOI] [Google Scholar]
- 91.Chen H, Little R. A test of missing completely at random for generalised estimating equations with missing data. Biometrika. 1999;86:1–13. doi: 10.1093/biomet/86.1.1. [DOI] [Google Scholar]
- 92.Joonè CJ, Konovalov DA. The effect of neuter status on longevity in the Rottweiler dog. Sci. Rep. 2023;13:17845. doi: 10.1038/s41598-023-45128-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Urfer SR, Kaeberlein M. Desexing dogs: A review of the current literature. Animals. 2019;9:1086. doi: 10.3390/ani9121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Hagen MAE, Ducro BJ, van den Broek J, Knol BW. Life expectancy in a birth cohort of Boxers followed up from weaning to 10 years of age. Am. J. Vet. Res. 2005;66:1646–1650. doi: 10.2460/ajvr.2005.66.1646. [DOI] [PubMed] [Google Scholar]
- 95.Pegram C, et al. Associations between neutering and early-onset urinary incontinence in UK bitches under primary veterinary care. J. Small Animal Pract. 2019;60:723–733. doi: 10.1111/jsap.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wade CM, Nuttall R, Liu S. Comprehensive analysis of geographic and breed-purpose influences on genetic diversity and inherited disease risk in the Doberman dog breed. Can. Med. Genetics. 2023;10:7. doi: 10.1186/s40575-023-00130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith LM, et al. Attitudes towards free-roaming dogs and dog ownership practices in Bulgaria, Italy, and Ukraine. PLOS ONE. 2022;17:e0252368. doi: 10.1371/journal.pone.0252368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Herzog H. Forty-two thousand and one dalmatians: Fads, social contagion, and dog breed popularity. Soc. Animals. 2006;14:383–397. doi: 10.1163/156853006778882448. [DOI] [Google Scholar]
- 99.Herzog HA, Bentley RA, Hahn MW. Random drift and large shifts in popularity of dog breeds. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004;271:S353–S356. doi: 10.1098/rsbl.2004.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morgan L, et al. Human–dog relationships during the COVID-19 pandemic: Booming dog adoption during social isolation. Humanit. Soc. Sci. Commun. 2020;7:1–11. doi: 10.1057/s41599-020-00649-x. [DOI] [Google Scholar]
- 101.Ho, J., Hussain, S. & Sparagano, O. Did the COVID-19 pandemic spark a public interest in pet adoption? Front. Vet. Sci.8 (2021). [DOI] [PMC free article] [PubMed]
- 102.Maher J, Wyatt T. European illegal puppy trade and organised crime. Trends Organ Crim. 2021;24:506–525. doi: 10.1007/s12117-021-09429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maher, J. & Wyatt, T. Rural-urban dynamics in the UK illegal puppy trade: Trafficking and trade in ‘man’s best friend’. Int. J. Rural Law Policy9 (2019).
- 104.RSPCA. Sold a pup? Exposing the breeding, trade and sale of puppies. Royal Society for the Prevention of Cruelty to Animals: London, UK, 2016; pp. 26–31. https://view.pagetiger.com/RSPCAPuppyTradeReport (2016).
- 105.Dogs Trust. The puppy smuggling scandal: an investigation into the illegal entry of dogs into Great Britain under the Pets Travel Scheme. https://www.dogstrust.org.uk/downloads/2014%20Puppy%20smuggling%20report.pdf (2014).
- 106.Dogs Trust. Puppy Smuggling: Puppies still paying as Government delays. https://www.dogstrust.org.uk/downloads/2020%20Puppy%20smuggling%20report.pdf (2020).
- 107.Norman C, Stavisky J, Westgarth C. Importing rescue dogs into the UK: Reasons, methods and welfare considerations. Vet. Rec. 2020;186:248–248. doi: 10.1136/vr.105380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.LaFond E, Breur GJ, Austin CC. Breed susceptibility for developmental orthopedic diseases in dogs. J. Am. Animal Hosp. Assoc. 2002;38:467–477. doi: 10.5326/0380467. [DOI] [PubMed] [Google Scholar]
- 109.Hodgman SFJ. Abnormalities and defects in pedigree dogs–I. An investigation into the existence of abnormalities in pedigree dogs in the British Isles. J. Small Animal Pract. 1963;4:447–456. doi: 10.1111/j.1748-5827.1963.tb01301.x. [DOI] [Google Scholar]
- 110.Hayes GM, Friend EJ, Jeffery ND. Relationship between pharyngeal conformation and otitis media with effusion in Cavalier King Charles spaniels. Vet. Rec. 2010;167:55–58. doi: 10.1136/vr.b4886. [DOI] [PubMed] [Google Scholar]
- 111.Packer RMA, Hendricks A, Burn CC. Impact of facial conformation on canine health: Corneal ulceration. PLOS ONE. 2015;10:e0123827. doi: 10.1371/journal.pone.0123827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hecht J, Horowitz A. Seeing dogs: Human preferences for dog physical attributes. Anthrozoös. 2015;28:153–163. doi: 10.2752/089279315X14129350722217. [DOI] [Google Scholar]
- 113.Sándor, S. & Kubinyi, E. Genetic pathways of aging and their relevance in the dog as a natural model of human aging. Front. Genet.10 (2019). [DOI] [PMC free article] [PubMed]
- 114.Edmunds GL, et al. Dog breeds and body conformations with predisposition to osteosarcoma in the UK: A case-control study. Can. Med. Genet. 2021;8:2. doi: 10.1186/s40575-021-00100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jimenez AG. The physiological conundrum that is the domestic dog. Integr. Comp. Biol. 2021;61:140–153. doi: 10.1093/icb/icab005. [DOI] [PubMed] [Google Scholar]
- 116.Shih HY, Paterson MBA, Phillips CJC. Breed group effects on complaints about canine welfare made to the royal society for the prevention of cruelty to animals (RSPCA) Queensland, Australia. Animals. 2019;9:390. doi: 10.3390/ani9070390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCoy BM, et al. Social determinants of health and disease in companion dogs: A cohort study from the Dog Aging Project. Evolut. Med. Public Health. 2023;11:187–201. doi: 10.1093/emph/eoad011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McKenzie BA, Chen F, LaCroix-Fralish ML. The phenotype of aging in the dog: How aging impacts the health and well-being of dogs and their caregivers. J. Am. Vet. Med. Assoc. 2022;260:963–970. doi: 10.2460/javma.22.02.0088. [DOI] [PubMed] [Google Scholar]
- 119.The Kennel Club. Breeds A to Z. https://www.thekennelclub.org.uk/search/breeds-a-to-z/ (2021).
- 120.Fédération Cynologique Internationale. FCI: the largest canine organisation of the world. https://www.fci.be/en/FCI-the-largest-canine-organisation-of-the-world-90.html (2021).
- 121.Parker HG, et al. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 2017;19:697–708. doi: 10.1016/j.celrep.2017.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Talenti A, et al. Studies of modern Italian dog populations reveal multiple patterns for domestic breed evolution. Ecol. Evolut. 2018;8:2911–2925. doi: 10.1002/ece3.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O’Neill DG, et al. Unravelling the health status of brachycephalic dogs in the UK using multivariable analysis. Sci. Rep. 2020;10:17251. doi: 10.1038/s41598-020-73088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Harvey, N. D. How old is my dog? Identification of rational age groupings in pet dogs based upon normative age-linked processes. Front. Vet. Sci.8 (2021). [DOI] [PMC free article] [PubMed]
- 125.Team, R. C. R: A language and environment for statistical computing. Vienna: R foundation for statistical computing. (No Title) (2021).
- 126.Moore DF. Applied Survival Analysis Using R. Berlin: Springer; 2016. [Google Scholar]
- 127.Schober P, Vetter TR. Survival analysis and interpretation of time-to-event data: The tortoise and the hare. Anesth. Analg. 2018;127:792–798. doi: 10.1213/ANE.0000000000003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Turkson AJ, Ayiah-Mensah F, Nimoh V. Handling censoring and censored data in survival analysis: A standalone systematic literature review. Int. J. Math. Math. Sci. 2021;2021:e9307475. doi: 10.1155/2021/9307475. [DOI] [Google Scholar]
- 129.Cox DR, Oakes D. Analysis of survival data. Berlin: CRC Press; 1984. [Google Scholar]
- 130.Pollock KH, Winterstein SR, Bunck CM, Curtis PD. Survival analysis in telemetry studies: The staggered entry design. J. Wildl. Manag. 1989;53:7–15. doi: 10.2307/3801296. [DOI] [Google Scholar]
- 131.Kassambara A, Kosinski M, Biecek P, & Fabian S. Survminer: Drawing survival curves using ggplot2.
- 132.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 133.Revell LJ. phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol. Evolut. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets generated and analysed during the current study are not publicly available due to restrictions stipulated within data sharing agreements with project participants. However, the corresponding author will consider facilitating collaborative discussions with all parties involved, on reasonable request.