Purine Catabolism in Escherichia coli and Function of Xanthine Dehydrogenase in Purine Salvage (original) (raw)
Abstract
Escherichia coli is not known to utilize purines, other than adenine and adenosine, as nitrogen sources. We reinvestigated purine catabolism because a computer analysis suggested several potential ς54-dependent promoters within a 23-gene cluster whose products have homology to purine catabolic enzymes. Our results did not provide conclusive evidence that the ς54-dependent promoters are active. Nonetheless, our results suggest that some of the genes are metabolically significant. We found that even though several purines did not support growth as the sole nitrogen source, they did stimulate growth with aspartate as the nitrogen source. Cells produced 14CO2 from minimal medium containing [14C]adenine, which implies allantoin production. However, neither ammonia nor carbamoyl phosphate was produced, which implies that purine catabolism is incomplete and does not provide nitrogen during nitrogen-limited growth. We constructed strains with deletions of two genes whose products might catalyze the first reaction of purine catabolism. Deletion of one eliminated 14CO2 production from [14C]adenine, which implies that its product is necessary for xanthine dehydrogenase activity. We changed the name of this gene to xdhA. The xdhA mutant grew faster with aspartate as a nitrogen source. The mutant also exhibited sensitivity to adenine, which guanosine partially reversed. Adenine sensitivity has been previously associated with defective purine salvage resulting from impaired synthesis of guanine nucleotides from adenine. We propose that xanthine dehydrogenase contributes to this purine interconversion.
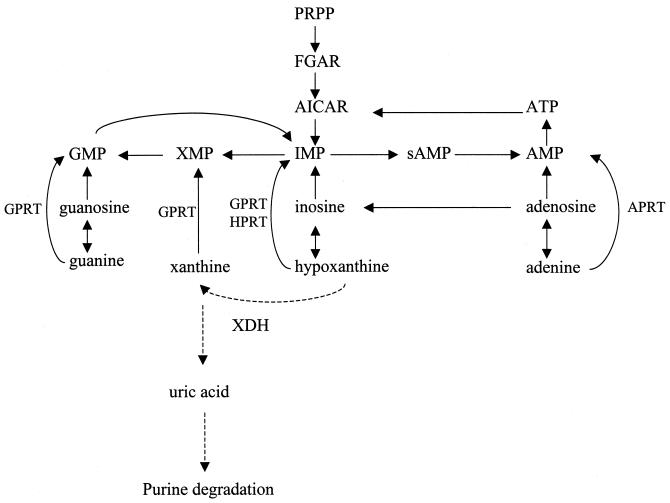
Escherichia coli converts exogenous purines (bases or nucleosides) to nucleotides via salvage pathways (Fig. 1) (22). Nucleosides are converted to nucleobases; for example, exogenous guanosine and inosine are degraded to guanine and hypoxanthine, respectively. Adenosine can be converted to two different nucleobases: adenine or hypoxanthine (via inosine). The purine nucleobases are then converted to the corresponding purine mononucleotides by adenine phosphoribosyltransferase (specific for adenine), hypoxanthine phosphoribosyltransferase (specific for hypoxanthine), and guanine phosphoribosyltransferase (which can salvage guanine, hypoxanthine, and xanthine).
FIG. 1.
Purine salvage pathways in E. coli. The solid lines show known pathways, while the dashed lines indicate reactions demonstrated in this paper. Abbreviations: PRPP, 5′-phospho-α-d-ribosyl-1-pyrophosphate; FGAR, 5′-phosphoribosyl-_N_-formylglycinamide; GPRT, guanine phosphoribosyltransferase; HPRT, hypoxanthine phosphoribosyltransferase; APRT, adenine phosphoribosyltransferase; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; and sAMP, adenylosuccinate.
Some microorganisms, e.g., Klebsiella pneumoniae, can utilize purines as carbon or nitrogen sources (19). Strains of Escherichia are not thought to degrade most purines, although there is considerable strain-to-strain variation (see references cited in reference 20). Several observations led us to reexamine the ability of E. coli to utilize purines as nitrogen sources. First, we identified five potential ς54-dependent promoters in a 23-gene cluster at min 65 of the E. coli chromosome. Such promoters often control genes whose products are involved in nitrogen assimilation, such as glutamine synthetase and enzymes of arginine catabolism (13, 17). BLAST analysis suggested that these genes might code for proteins that transport or catabolize purines. (The results of the BLAST searches are described in more detail below.) Therefore, we considered the possibility that nitrogen limitation induces enzymes of purine catabolism. Second, the failure to utilize a particular compound as a sole nitrogen source does not imply that this compound is not catabolized. For example, E. coli cannot degrade pyrimidines as the sole nitrogen source. Nonetheless, in the presence of certain amino acids, 14CO2 is produced from [14C]uracil or [14C]thymidine (2). A similar situation involves arginine catabolism. Aspartate is not absolutely required for arginine utilization, but aspartate greatly stimulates arginine utilization (17). Because the metabolism of a particular compound may depend on the presence of other compounds, we considered the possibility that this might be the case for purines.
In this paper, we present evidence for purine catabolism. Such catabolism requires aspartate and is not complete. We disrupted two different genes coding for proteins that might catalyze the first reaction in purine catabolism. One mutant eliminated purine catabolism. Despite the limited catabolism of purines, both mutants had an altered phenotype: they were both somewhat sensitive to adenine. We propose that xanthine dehydrogenase participates in purine salvage but not in aerobic purine catabolism.
MATERIALS AND METHODS
Strains and their construction.
The strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Strains, plasmids, and phage
| Strain, plasmid, or phage | Relevant features | Source or reference |
|---|---|---|
| Strains | ||
| HX1 | xdhA | This work |
| HX2 | W3110, but also Δb2881 | This work |
| HX3 | W3110, but also Δ_xdhA_ Δb2881 | This work |
| K4633 | recD1903::Tetr | D. Friedman, University of Michigan |
| KC1669 | Wild-type K. pneumoniae | R. Bender, University of Michigan |
| LR3-carAB | W3110, but also Δ_carAB_ with a chromosomal ′glnAp2-lacZ fusion | Lab strain |
| W3110 | Wild-type E. coli, lacI_q_lacL8 | Lab strain |
| Phage and plasmids | ||
| Kohara phage 465 | xdhA | 16 |
| pWM529 | Cloning vector, Ampr | 9 |
| pBlue-kan-FRT | _Sma_I-_Hin_dIII fragment containing Kanr-FRT cassette from pCP15 (4) inserted between _Eco_RI and _Hin_dIII sites of pBlueScript SK+; Ampr Kanr | A. Kiupakis, University of Texas at Dallas |
(i) Strain HX1 (Δ_xdhA_).
BLAST analyses indicate that genes b2866, b2867 (ygeT), and b2868 (the b number is a GenBank identifier) code for proteins with homology to different domains of Drosophila melanogaster xanthine dehydrogenase (XDH). A strain with a deletion of b2866 has a phenotype consistent with a defect in XDH (described below). Therefore, we designated this gene xhdA. A 4.9-kb _Acc_65I-_Bgl_II fragment from Kohara phage 465 containing b2866 (GenBank accession number AE000370) was ligated between the _Acc_65I and _Bam_HI sites of the cloning vector pWM529. The 2.0-kb _Age_I-_Pvu_II region of the insert was replaced with the 1.6-kb _Eco_RI-_Hin_dIII fragment from plasmid pBlue-kan-FRT (Flp recombination target), which contains FRT sites flanking a Kanr cassette. The FRT sites permit subsequent excision of the region between them. The plasmid was linearized and transformed into E. coli strain K4633 (recD), and recombinants were selected on Luria-Bertani (LB) agar plates containing 50 μg of kanamycin per ml. The mutant allele was transduced from K4633 to W3110 by P1 transduction (10). Finally, the Kanr insert was excised from the FRT sites using plasmid pCP20, which is a temperature-sensitive plasmid that encodes a site-specific recombinase, as described previously (4). The net result should be an in-frame deletion of codons 70 to 728 of a potential 752-residue protein. The resulting strain was named HX1.
(ii) Strain HX2 (Δ_b2881_).
BLAST analysis indicates that the product of gene b2881 has homology to four of the five domains of D. melanogaster XDH. DNA containing this gene was obtained by PCR amplification of DNA from 300 bp upstream to 700 bp downstream, which resulted in an approximately 4-kb fragment. The PCR primers contained _Bam_HI and _Eco_RI sites, which allowed insertion of the fragment into the _Bam_HI and _Eco_RI sites of pWM529. A 2-kb region of the insert was removed between _Eco_RV and _Bsi_WI sites and replaced with a 1.6-kb _Sma_I-_Acc_65I fragment from pBlue-kan-FRT, that contained the Kanr-FRT cassette. The disrupted allele was introduced into W3110, and the Kanr-FRT cassette was removed in the same way as for HX1. This resulted in a deletion of codons 161 to 824 of a potential 956-residue protein.
(iii) Strain HX3 (Δ_xdhA_ Δb2881).
The b2881::Kanr-FRT allele (described for the construction of HX2) was transduced into HX1. The Kanr-FRT cassette was removed in the same way as for HX1. PCR was used to verify all of the deletions.
Cell growth.
The salts for the minimal medium have been described previously (15). In addition, minimal medium contained 0.4% glucose, 0.02% thiamine, and 0.1% of each nitrogen source unless otherwise noted. To measure the growth rates, cells were grown overnight in the medium to be tested, diluted into 10 ml of fresh minimal medium so that the initial turbidity was 10 Klett units (no. 42 filter), and incubated at 30°C at 220 rpm. Turbidity was measured at 1- to 3-h intervals. Generation times are presented as the means from triplicate cultures with the standard deviations.
Measurement of 14CO2 from [14C]adenine degradation.
[14C]adenine (0.5 μCi) (Amersham Pharmacia Biotech) was added to a 10-ml culture. The stock solution contained 287 mCi/mmol, which implies that the final concentration of adenine was 1.7 μM. 14CO2 was collected in a 25-ml Falcon tube which contained 0.8 ml of 2M NaOH and which was punctured at its midpoint. The top of the tube was sealed, and the whole tube was inserted into a rubber stopper, which sealed a 250-ml flask that contained the culture medium. After a 24- to 48-h incubation, the NaOH solution was mixed in a 4:1 ratio with Scintisafe Plus fluid (Fisher Scientific), and 14C was measured in a scintillation counter (Beckman LS6500). Cultures without cells were used to determine the background, which varied from 140 to 160 dpm per culture. This value was subtracted from experimental values, which were at least 15-fold over this background value. The experimental values were normalized to a culture _A_600 of 1.0. The final _A_600 of all of the cultures was slightly above 2.0. All determinations were done in duplicate, with the ranges of values indicated in the figures or the text.
Transcript analysis.
Total RNA from E. coli was extracted with hot phenol as described previously (1). The quality of the RNA was monitored by running 2 μl of the RNA preparation in a 0.7% agarose gel. The presence of two sharp bands corresponding to the 16S and 23S rRNAs suggested minimal RNA degradation.
For primer extension, the primer was labeled in a reaction mixture containing 70 mM Tris (pH 7.6), 10 mM MgCl2, 5 mM dithiothreitol, 30 μCi of [γ-32P]ATP, 0.1 μg of primer, and 3 U of T4 polynucleotide kinase (New England BioLabs) in a total volume of 10 μl. The primer for xdhA (b2866) was 5′-TATATCGTGCCCGCCCGGTGA-3′. Labeling reactions were done at room temperature for 1 h and then stopped by heat inactivation at 70°C for 10 min. One microliter of the resulting primer was mixed with 1 μg of RNA (determined from the _A_260) in a reaction mixture containing 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 6 mM MgCl2, and 4 mM dithiothreitol in 10 μl. This mixture was heated to 90°C for 5 min, cooled slowly to 42°C, and incubated for 90 min. Then, 5 μl of the same reaction mixture containing deoxynucleoside triphosphates (final concentrations were 0.2 mM for each nucleotide) and 1 U of avian myelobastosis virus reverse transcriptase (Gibco BRL) was added, and the reaction mixture was incubated for 90 min at 42°C. Reaction products were then denatured at 95°C for 7 min and resolved on a 6% polyacrylamide gel. The results were visualized using a PhosphorImager (Molecular Dynamics Storm 865).
RESULTS
Identification of possible genes of purine catabolism: computer analysis of ς54-dependent promoters.
Genes induced by nitrogen limitation frequently possess ς54-dependent promoters. ς54 binds to promoters with a distinctive 17-bp sequence: TGGCACG(A/G)NNNNTTGC(A/T). We scanned the E. coli genome for potential ς54-dependent promoters using the SeqScan program (B. T. Nixon, Department of Biochemistry and Molecular Biology, Pennsylvania State University) (http://www.bmb.psu.edu/seqscan). The sequences are identified using a scoring matrix and are assigned a score ranging from 0 to 100. A score of 100 indicates a perfect one-to-one match, while a score of 0 indicates no matches. The program identifies sites with scores over 60. The complete results of this analysis will be the subject of a separate communication. Some highlights of this analysis are presented to indicate why we became interested in purine catabolism. The scores for the 12 known ς54-dependent promoters in E. coli range from 70.3 to 95.4, with a mean of 82.7. None of these promoters is within a gene. There are 217 possible ς54-dependent promoters outside of genes (or open reading frames) and only 48 (including known promoters) with a score greater than 70.
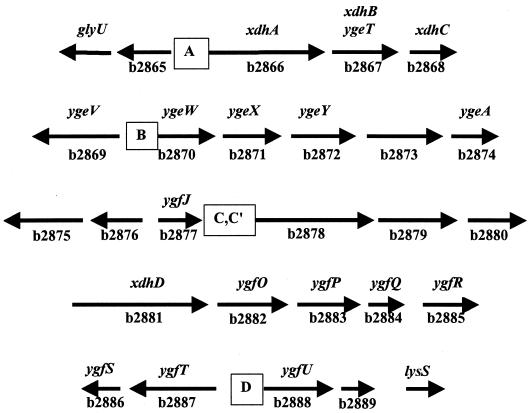
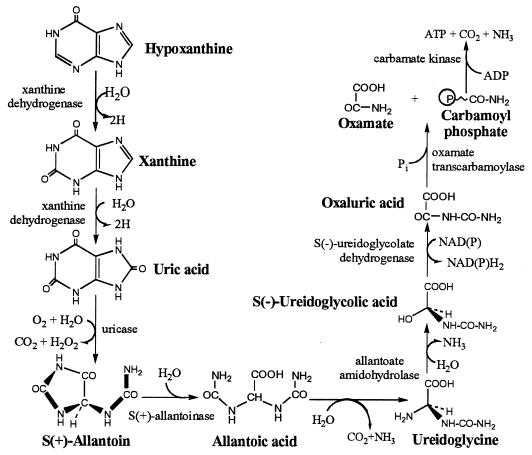
Five potential ς54-dependent promoters were found within an uncharacterized cluster of 23 genes at min 65 (Table 2). The chromosomal arrangement of these genes is graphically represented in Fig. 2. BLAST analysis of the putative gene products suggests that several could be involved in purine catabolism (Table 3). Genes b2866 to b2868 potentially encode a segmented XDH. (These genes are discussed below.) Gene b2881 potentially encodes another molybdenum-containing protein of the xanthine oxidase family. (This family is described in more detail below.) The products of genes b2873 and b2879 show homology to allantoinase, and the product of b2883 shows homology to allantoate amidohydrolase. These enzymes can be organized into a pathway similar to that found in other organisms (Fig. 3). The only enzymes of the proposed pathway not identified in the cluster were a potential uricase and a potential ureidoglycolate dehydrogenase. We will present evidence below that suggests that E. coli contains a uricase activity.
TABLE 2.
Possible ς54-dependent promoters at min 65 of the E. coli chromosome
| Promoter | Genea | Score | bp from structural geneb |
|---|---|---|---|
| A | xdhA (b2866) | 66 | 94 |
| B | ygeW (b2870) | 75.2 | 161 |
| C | (b2878) | 74.1 | 72 |
| C′ | (b2878) | 71.5 | 81 |
| D | ygfU (b2888) | 66.3 | −1 |
| 12 known promotersc | NAd | 70.3–95.4 | 30–82 |
FIG. 2.
Genes at min 65 of the E. coli genome. The diagram shows the arrangement of genes that BLAST analysis suggests are involved in purine catabolism. The locations of the possible ς54-dependent promoters are designated A, B, C, C′, and D.
TABLE 3.
Genes of putative purine catabolic operons
| Gene | b no. | Results of BLAST search | Possible function |
|---|---|---|---|
| b2866 | XDH (last part) | Subunit of XDH | |
| ygeT | b2867 | XDH (middle part) | Subunit of XDH |
| b2868 | XDH (first part) | Subunit of XDH | |
| ygeV | b2869 | ς54-dependent activator | Transcriptional activator |
| ygeW | b2870 | Ornithine transcarbamoylase | Oxamate transcarbamoylase |
| ygeX | b2871 | Threonine deaminase | ?a |
| ygeY | b2872 | _N_-Succinyl-l-diaminopimelic acid desuccinylase | ? |
| b2873 | Allantoinase | Allantoinase | |
| yqeA | b2874 | Carbamate kinase | Carbamate kinase |
| b2875 | Adenosyl-homocysteinase | Adenosine generation | |
| b2876 | Molybdenum utilization | ? | |
| ygfJ | b2877 | Molybdenum utilization | ? |
| b2878 | GOGAT-like protein | ? | |
| b2879 | Allantoinase | Allantoinase | |
| b2880 | Mo-containing hydrolyase | ? | |
| b2881 | XDH | XDH | |
| ygfO | b2882 | Xanthine, uracil permease | Transport |
| ygfP | b2883 | Allantoate amidohydrolase | Allantoate amidohydrolase |
| ygfQ | b2884 | Xanthine, uracil permease | Transport |
| ygfR | b2885 | Xanthine, uracil permease | Transport |
| ygfS | b2886 | GOGAT small subunit | ? |
| ygfT | b2887 | Formate-hydrogen lyase | ? |
| ygfU | b2888 | Xanthine, uracil permease | Transport |
| b2889 | ? | ? |
FIG. 3.
Possible purine catabolic pathway in E. coli.
Effect of purines on nitrogen-limited E. coli.
The identification of possible ς54-dependent genes that might code for enzymes of a purine catabolic pathway led us to reinvestigate purine catabolism in E. coli during nitrogen-limited growth. E. coli could use adenine or adenosine, but not hypoxanthine, xanthosine, inosine, or allantoin, as the sole nitrogen source (data not shown). This does not necessarily imply that these compounds are not catabolized. It is possible that there is a low level of purine catabolism that is insufficent to support growth. Therefore, we examined whether purines stimulated growth with 0.1% aspartate as the sole nitrogen source, which supports a generation time of 7.23 ± 0.97 h. The addition of 0.07% hypoxanthine, 0.05% guanosine, 0.1% inosine, and 0.1% xanthosine reduced the generation times (in hours) to 4.80 ± 0.12, 4.15 ± 0.25, 4.33 ± 0.01, and 4.59 ± 0.17, respectively. In contrast, 0.1% allantoin did not stimulate growth (data not shown). This observation does not prove that purines are catabolized. It is possible that purine salvage reduces aspartate consumption for de novo purine synthesis, which results in elevated intracellular aspartate and more rapid aspartate catabolism. The fact that all of the purines stimulated growth to the same extent is consistent with this possibility.
Extent of purine catabolism in E. coli.
To determine whether the stimulatory effect of purines results from catabolism or simply purine salvage, we tested for the presence of products of purine catabolism. We first examined whether growth with [U-14C]adenine produced 14CO2, which would be generated from the first two reactions of purine catabolism but not from a salvage pathway. We also examined purine catabolism by K. pneumoniae, which can use purines and allantoin as the sole carbon or nitrogen source (21). For cells grown with 0.1% aspartate as the nitrogen source and 1.7 μM [U-14C]adenine, duplicate 10-ml cultures of E. coli generated 1,310 dpm of 14CO2, whereas similar cultures of K. pneumoniae produced 22,300 dpm. (The range of values was 3% or less of the mean.) These results establish that xanthine is catabolized to allantoin in E. coli. The 17-fold-less-efficient purine degradation in E. coli compared to that in K. pneumoniae may account for the former's inability to utilize most purines as nitrogen sources.
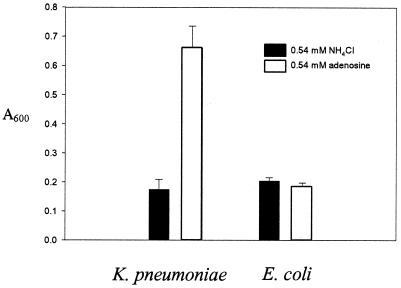
We next examined whether purines are catabolized past allantoin. If purines are degraded to ureidoglycine, then assimilatable ammonia would be produced. Therefore, we examined the final cell density of cells grown with adenine as the sole nitrogen source. One molecule of ammonia is generated by adenosine deaminase during the conversion of adenine or adenosine to hypoxanthine (Fig. 1). A second molecule of ammonia would be generated only if ureidoglycine is generated. If so, then E. coli grown with a limiting concentration of adenine will grow to a higher cell density than that grown with an equivalent concentration of NH4Cl. However, no significant difference in cell density was observed for cells grown with an equivalent concentration of adenine or NH4Cl (Fig. 4). We conclude that in E. coli adenine is degraded to allantoin and possibly to allantoic acid, but no further. In contrast, the cell density of K. pneumoniae grown with adenosine was four times that of a culture with NH4Cl, which suggests that four of the five nitrogens in adenine can be assimilated.
FIG. 4.
Final cell densities with NH4Cl or adenosine as the nitrogen source. Wild-type K. pneumoniae and E. coli strains were grown in minimal medium with either 0.54 mM NH4Cl or 0.54 mM adenosine at 37°C for 48 h. The experiment was done three times. The error bars indicate the standard deviations.
To further confirm that the proposed pathway of purine catabolism is not complete in E. coli, we tested whether purine catabolism produces carbamoyl phosphate. A carAB mutant lacks carbamoyl phosphate synthetase and therefore requires arginine and uridine. We grew a carAB mutant in minimal medium with 0.2% NH4Cl (the primary nitrogen source), 0.02% arginine, and 0.0005% uridine. The uridine is limiting: a culture with 0.0005% uridine gives an _A_600 of 0.194, which is about 7% of the growth with saturating uridine. When 0.03% adenosine was added to this medium, the final _A_600 of the culture was 0.200. We conclude that adenosine catabolism does not produce carbamoyl phosphate. E. coli requires 312 μmol of carbamoyl phosphate per g (dry weight) for pyrimidine synthesis (calculated from information provided in reference 12). The experiment to determine the cell density in which adenine was the sole nitrogen source (described in the preceding paragraph) requires the liberation and assimilation of ammonia. E. coli requires about 10,283 g-atoms of nitrogen per g (dry weight) (also calculated from reference 12). Therefore, the test for carbamoyl phosphate generation is 33 times more sensitive than that for ammonia generation. In summary, the final enzymes of the proposed purine catabolic pathway do not appear to be functional under the growth conditions of the experiment.
Putative genes for XDH in E. coli.
Despite the absence of extensive purine catabolism, we wanted to know if the purine-dependent growth stimulation was caused by purine catabolism or purine salvage. To distinguish between these possibilities, we focused a genetic analysis on genes that potentially specify XDH, because the XDH reaction initiates purine catabolism and is required for CO2 formation from purines. Homology searches have a good chance to identify such enzymes because of several unique structural and sequence determinants. The xanthine oxidase family is the largest and most diverse group of molybdenum cofactor (Mo-co)-containing enzymes, which usually transfer an oxygen atom to or from a substrate in a two-electron transfer reaction (7). This family includes xanthine oxidase/dehydrogenase, aldehyde oxidase, 4-hydroxybenzoyl coenzyme A reductase, quinoline oxidoreductase, and CO dehydrogenases.
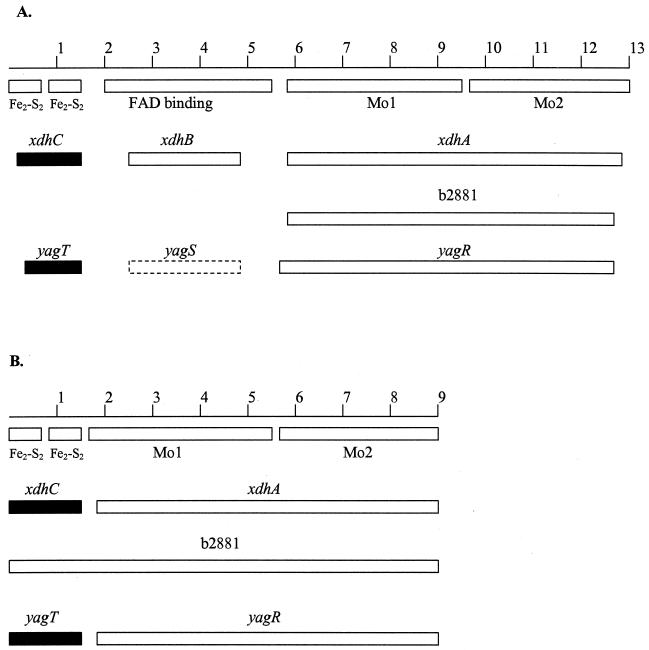
We searched the E. coli genome for genes coding for members of the xanthine oxidase family. We have graphically aligned all E. coli proteins with homology to D. melanogaster XDH (Fig. 5A) and Desulfovibrio gigas aldehyde oxidoreductase (Fig. 5B). We chose these two proteins for comparison because structure-function relations have been defined for these proteins. D. gigas aldehyde oxidoreductase has four domains (Fig. 5B) (14). The first two domains contain a total of 150 amino acid residues, and each domain binds an [Fe2-S2] cluster. The second iron-binding domain assumes a structure that has not been previously found in other iron-binding proteins. The third domain, called Mo1, binds to Mo-co and contains 386 residues. The fourth domain, called Mo2, contains 326 residues, binds to Mo-co, and also interacts with the two iron-binding domains. The XDH from D. melanogaster has regions homologous to these four domains. It also contains an extra domain that binds FAD, which is located between the second iron-binding domain and Mo1 of the D. gigas enzyme (Fig. 5A) (14).
FIG. 5.
Xanthine oxidase family genes in E. coli. The reference genes for comparison are the xanthine dehydrogenase (rosy locus) gene of D. melanogaster (A) and the aldehyde oxidoreductase of D. gigas (B). The numbers refer to hundreds of amino acid residues. The boxes immediately below the first line of each section indicate the extents of the domains. The Fe2-S2 domains bind the iron-sulfur clusters, the FAD domain binds FAD, and Mo1 and Mo2 are two separate Mo-co-binding domains. The solid boxes with gene designations above them indicate that there were 36 to 38% identity and 50 to 57% similarity to the reference genes. The open boxes indicate 23 to 27% identity and 37 to 46% similarity. The dashed box for yagS signifies that it is not homologus to the D. melanogaster XDH gene. Instead, it is 24% identical and 37% similar to xdhB.
Three gene products have homology to the two Mo-co-binding domains of the reference proteins: those of xdhA, b2881, and yagR. (We have renamed b2866, b2867, and b2868 xdhA, xdhB, and xdhC, respectively. These designations are based on results of the homology searches, precedents with other bacterial XDHs, and genetic results presented elsewhere in this paper. To prevent any confusion that could result from renaming the genes after a complete discussion of these issues, we adopted the new designations at the point when these genes are considered in some detail.) The xdhA and b2881 genes are in the 23-gene cluster identified earlier, while yagR (GenBank accession number AE000136) is unlinked. xdhC (which is contiguous with xdhB and xdhA) and yagT (contiguous with yagS and yagT) specify proteins that have homology to the iron-binding regions of the two reference proteins. Only one potential protein, the product of xdhB, has homology to the FAD-binding region of the D. melanogaster enzyme. The yagS product is not homologous to the FAD-containing domain of D. melanogaster XDH, but it is homologous to XdhB (Fig. 5A).
The product of b2881 is homologous to the entire D. gigas enzyme without any gaps, but it lacks the FAD-binding domain of D. melanogaster XDH. The absence of this domain precludes dehydrogenase activity, i.e., electron transfer to NAD (or NADP), but does not preclude xanthine oxidase activity, i.e., electron transfer to O2. This is consistent with the observation that the D. gigas enzyme does not catalyze electron transfer from xanthine to an artificial electron acceptor but can nonetheless oxidize xanthine (3).
It is not uncommon that separate genes specify different domains of XDH (7). Therefore, the products of xdhA and the two genes downstream from xdhA may form a heterotrimeric XDH. The products of yagR, yagS, and yagT may have a similar potential. There are two precedents for heterotrimeric XDHs in bacteria. Heterotrimeric XDHs are found in Veillonella atypica and Eubacterium barkeri, and their subunit sizes are virtually identical to those for the proposed E. coli enzymes (6, 18).
Phenotypes of HX1 (Δ_xdhA_), HX2 (Δb2881), and HX3 (Δ_xdhA_ Δb2881). (i) CO2 generation.
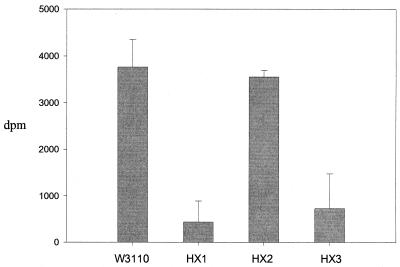
We examined the effects of disruptions of genes for two potential xanthine-oxidizing enzymes: xdhA, the first gene of a possible operon xdhABC operon, and b2881. We did not disrupt the yag genes. We were unable to directly verify the loss of xanthine dehydrogenase or xanthine oxidase activity, since we could not assay either activity from W3110 or strains with xdhABC or b2881 expressed from a high-copy-number plasmid. Instead, we examined 14CO2 production from [U-14C]adenine by strains HX1, HX2, and HX3 grown in minimal medium supplemented with aspartate and histidine. We added histidine in the hope that it would optimize purine catabolism and result in a more sensitive assay to detect purine catabolism. Histidine inhibits the formation of guanine nucleotides from adenine via ATP to 5-aminoimidazole-4-carboxamide ribonucleotide, which involves the first part of the histidine synthetic pathway. 5-Aminoimidazole-4-carboxamide ribonucleotide is subsequently converted to IMP and finally to GMP (8). If this pathway is inhibited, the only mechanism of GMP formation from adenine is via hypoxanthine. Increased hypoxanthine formation may stimulate purine catabolism. This complex reasoning is apparently correct, since histidine increased 14CO2 production from adenine by W3110 from 1,300 dpm per culture to 4,000 dpm per culture (the results are normalized to a constant cell density). Therefore, changes in 14CO2 production caused by the deletion of xdhA or b2881 can be more easily observed.
HX1 and HX3 both showed reduced 14CO2 production, while 14CO2 from HX2 was the same as that from the wild type (Fig. 6). These results indicate that XdhA is required for CO2 production and for XDH activity. HX2 had normal CO2 generation, which implies that the b2881 product does not have XDH activity with the low concentration of adenine (1.7 μM) present in the medium.
FIG. 6.
14CO2 production from [14C]adenine. E. coli strains W3110, HX1 (Δ_xdhA_), HX2 (Δb2881), and HX3 (Δ_xdhA_ Δb2881) were grown in minimal medium containing 0.1% aspartate, 0.03% histidine, and 1.7 μM [U-14C]adenine. Methods and Materials describes all other procedures, including data processing. Each value is the mean of two determinations, and the range of values (error bars) is shown.
(ii) Adenine toxicity.
During these experiments, we noticed that the mutants grew slower than the wild-type strain when adenine was present. This was not due to a contaminant in the radioactive adenine, since unlabeled adenine gave a similar effect. Adenine at high concentrations (>0.1%) inhibits the growth of wild-type E. coli (reference 8 and references cited therein). Certain mutants defective in purine salvage are much more sensitive to adenine. An hpt gpt double mutant, which is deficient in both hypoxanthine and guanine phosphoribosyltransferases, fails to grow with 0.002% adenine (8). There are two components to this toxicity in the double mutant. First, adenine (actually adenine nucleotides, which are synthesized from adenine by adenine phosphoribosyltransferase) inhibits de novo purine synthesis. Second, there is inefficient (or mutationally blocked) formation of guanine nucleotides from adenine, since the hpt gpt mutant cannot phosphoribosylate either hypoxanthine or xanthine (8). An important piece of evidence that supports the hypothesis that adenine toxicity results from failure to synthesize GMP is the observation that guanosine reverses the inhibition (8).
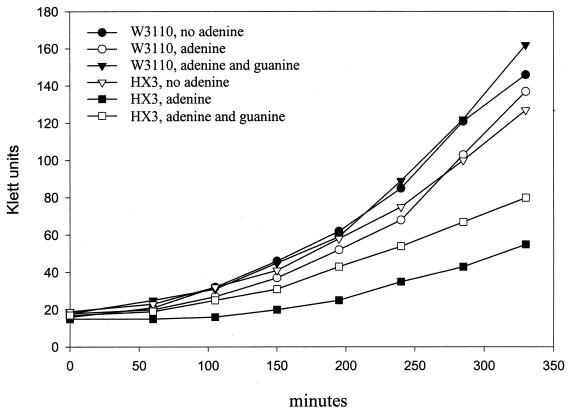
To investigate their enhanced adenine toxicity, we grew HX1, HX2, and HX3 with various concentrations of adenine as the sole nitrogen source. A 0.03% concentration of adenine completely inhibited the growth of HX1 (Δ_xdhA_) and HX3 (Δ_xdhA_ Δb2881); 0.1% adenine completely inhibited the growth of HX2 (Δb2881), whereas W3110 still grew with 0.1% adenine (Table 4). The growth of HX3 was also monitored in minimal medium with 0.1% NH4Cl plus 0.03% adenine as the nitrogen source. The expectation is that adenine will be toxic until it is metabolized, and then the cells will grow at a normal rate. Levine and Taylor studied the toxicity by adding adenine to such a culture and observing the immediate effects on growth (8). We performed the experiments and plotted the results in the same way to facilitate comparisons. HX3 showed a prolonged lag phase (Fig. 7) but did not appear to be as sensitive to adenine as the hpt gpt double mutant, which could not grow with 0.002% adenine (Fig. 7) (8). Guanosine can suppress the adenine toxicity of an hpt gpt double mutant (8). Similarly, 0.06% guanosine partially suppressed the adenine toxicity in HX3 (Fig. 7). In summary, the adenine toxicity and its suppression by guanosine imply that XdhA and possibly the product of b2881 contribute to the conversion of adenine to guanine nucleotides during purine salvage.
TABLE 4.
Growth with adenine as the nitrogen sourcea
| Strain | Growthb with the following adenine conc (%): | |||
|---|---|---|---|---|
| 0.01 | 0.03 | 0.06 | 0.10 | |
| W3110 | + | + | + | + |
| HX1 (Δ_xdhA_) | + | − | − | − |
| HX2 (Δb2881) | + | + | + | − |
| HX3 (Δ_xdhA_ Δb2881) | + | − | − | − |
FIG. 7.
Adenine sensitivity of HX3 (Δ_xdhA_ Δb2881) and reversal by guanosine. W3110 (wild type) and HX3 were grown exponentially in cultures containing 0.1% NH4Cl as the nitrogen source. These cells were inoculated into fresh medium containing 0.1% NH4Cl with no addition, with 0.03% adenine, or with 0.03% adenine plus 0.06% guanosine. Growth was monitored at 30°C. The experiment was done twice, and the same pattern was observed.
(iii) Growth rates.
Surprisingly, HX1 (Δ_xdhA_) grew faster than wild-type E. coli in nitrogen-limited minimal medium with 0.1% aspartate as the sole nitrogen source: the doubling times were 5.70 ± 0.25 and 7.36 ± 0.31 h, respectively. However, the mutant grew normally in nitrogen-rich ammonia-containing minimal medium (1.92 ± 0.2 and 1.75 ± 0.16 h, respectively).
HX1 grew as well as wild-type E. coli when grown with 0.1% aspartate plus 0.07% hypoxanthine as the nitrogen sources (data not shown). Therefore, hypoxanthine stimulates growth in the absence of the catabolic pathway, which suggests that purine salvage is responsible for the growth stimulation.
Transcript analysis of xdhA.
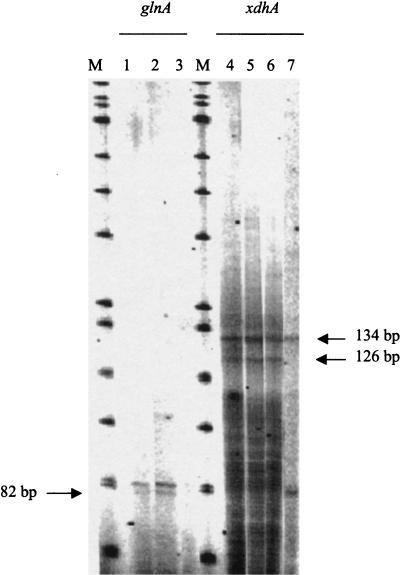
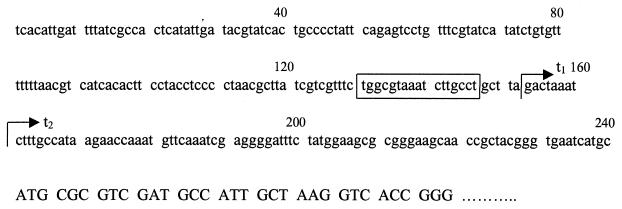
We initially studied the 23-gene cluster at min 65 because it contained five possible ς54-dependent promoters. Such promoters are often associated with expression during nitrogen limitation. However, we detected 14CO2 from [U-14C]adenine from E. coli grown not only in nitrogen-limited minimal medium but also in nitrogen-rich (ammonia-containing) minimal medium and amino acid-rich LB broth (data not shown). These results show that XdhA (the primary enzyme responsible for CO2 generation) is not solely under nitrogen regulation. To verify this regulation and to locate the transcriptional start site, we mapped the 5′ end of the xdhA transcript by primer extension analysis. Transcript mapping was done for xdhA, which appears to be the first gene in its operon. (We did not examine transcription from b2881 because it is not clear where the operon starts.) The results of primer extension analysis of xdhA are shown in Fig. 8. As a positive control, primer extension was also done for the glnA gene, which is actively transcribed under nitrogen-limiting conditions. The expected results for the glnA transcript were observed: there was significantly less transcript in nitrogen-rich ammonia-containing medium. The DNAs made from primer extension of xdhA transcripts were 126 and 134 bases, which correspond to start sites 80 and 88 bases, respectively, upstream from the presumed initiation codon (Fig. 9). (Adjacent sequencing reactions were used to determine the exact sizes of the transcripts, and these results are not shown.) Both transcripts were present for cells grown in nitrogen-limited medium, without or with hypoxanthine (Fig. 8, lanes 4 and 5, respectively), and for cells grown in nitrogen-rich medium (lane 6). The larger transcript, but not the smaller transcript, was also observed from an rpoN (encoding ς54) mutant grown in LB broth (lane 7, Fig. 8). The smaller transcript might be ς54 dependent. Its first nucleotide is 14 bases from the potential ς54 recognition site (promoter A [Table 2]), which is not unusual. However, if this is actually the 5′ end of the mRNA, and not a degradation product, then transcription begins in a pyrimidine-rich region, which seems unlikely. The upstream transcript begins in a purine-rich region, which is reasonable. Furthermore, the putative, −10 region (TAAATCTT) is AT rich, which is consistent with an authentic binding site for RNA polymerase. In the absence of results from transcription with purified components or mutational alteration of the putative binding site for ς54, the results must be considered preliminary. Nonetheless, we suspect that the smaller transcript is a degradation product of the larger and that the potential ς54-dependent promoter is not functional. In any case, there is clearly transcription independent of ς54, which is consistent with the conditions in which CO2 is generated from adenine.
FIG. 8.
xdhA start site of transcription. Primer extensions were done using a primer annealing to glnA (lanes 1 to 3) or xdhA (lanes 4 to 7). RNA was extracted from either W3110 grown in various minimal media (lanes 1 to 6) or an isogenic rpoN mutant grown in LB medium (lane 7). The minimal media contained the following nitrogen sources: 0.1% aspartate (lanes 1 and 4), 0.1% aspartate and 0.08% hypoxanthine (lanes 2 and 5), or 0.1% NH4Cl (lanes 3 and 6). Lanes M, size markers.
FIG. 9.
Nucleotide sequence upstream of the xdhA structural gene. The first three lines show 240 bases upstream from the xdhA structural gene. The fourth line shows the sequence at the amino-terminal coding region of xdhA. The t1 transcript is the most upstream of two transcripts, while t2 is the downstream transcript. The boxed region shows the possible ς54-binding region.
DISCUSSION
Purine catabolism in E. coli.
We have presented evidence that E. coli catabolizes purines, although not sufficiently to support aerobic growth as sources of nitrogen (with the exception of adenine and adenosine). The best evidence for such catabolism is the generation of 14CO2 from [14C]adenine, which implies functional XDH and uricase activities, and the production of allantoin. It has recently been shown that allantoin can be used as the sole nitrogen source in anaerobically grown E. coli (5). Cusa et al. (5) showed that E. coli contains allantoinase, allantoate amidohydrolase, and ureidoglycolate dehydrogenase, which collectively catalyze the conversion of allantoin to oxaluric acid. Such a metabolism implies that E. coli possesses most of the enzymes of purine catabolism (Fig. 3). The inability to use purines aerobically might be due to inadequate transport, weak promoters, or the absence of appropriate regulation.
XDH activity and purine salvage in E. coli.
XDH catalyzes two reactions: the conversion of hypoxanthine to xanthine and the conversion of xanthine to uric acid. The second reaction is the first committed step in purine catabolism. It diverts purines away from the salvage pathways. 14CO2 production from [U-14C]adenine results from the activity of uricase on uric acid and implies XDH activity, which is required for uric acid formation. Deletion of xdhA eliminated 14CO2 production, which implies that XdhA is required for this activity. All known XDHs have domains that bind Fe-S clusters and FAD, and separate genes can specify these domains. The two genes just downstream of xdhA code for proteins with homology to the FAD-binding domain and the iron-binding domain of D. melanogaster XDH, respectively. Therefore, we suggest that b2867 (ygeT) and b2868 should be renamed xdhB and xdhC, respectively. We propose that xdhA, xdhB, and xdhC code for components of a heterotrimeric XDH.
An unexpected finding was that deletion of xdhA resulted in sensitivity to exogenous adenine. Studies by Levine and Taylor showed that the adenine toxicity in mutants with defective purine salvage is caused by failure to synthesize guanine nucleotides from adenine (8). To account for the phenotype of the xdhA mutant, we propose that GMP can be more efficiently replenished from xanthine (via XMP) than from hypoxanthine (via IMP and XMP) and that deletion of xdhA impairs the former pathway. The partial suppression of adenine toxicity by exogenous guanosine supports this explanation. Such a proposal also accounts for the unusual kinetic parameters for the purine salvage enzymes, which we propose are physiologically relevant. The _K_m of guanosine phosphoribosyltransferase for xanthine (40 μM) is four times lower than that for hypoxanthine (170 μM) and three times lower than the _K_m of hypoxanthine phosphoribosyltransferase for hypoxanthine (120 μM) (22).
Our results suggest a possible, but minor, function for the product of b2881. A mutant lacking this protein produces a normal amount of CO2 from 1.7 μM adenine, which implies that this protein does not have XDH activity. In contrast, the toxicity with 0.1% adenine (7 mM) suggests that this protein might oxidize hypoxanthine, which we propose results in xanthine formation and more efficient GMP production. This is only partially consistent with the results of the homology analysis. There are four families of Mo-co-containing proteins, and they are not homologous to each other (7). Therefore, the homology analysis clearly shows that the b2881 product is a member of the xanthine oxidase family. The deduced protein contains four of the five domains of D. melanogaster XDH but lacks the FAD-binding domain, which suggests that it may not have dehydrogenase activity, although it may have oxidase activity. These seemingly inconsistent results can be reconciled if the b2881 product oxidizes high intracellular levels of hypoxanthine and perhaps xanthine. This would not be surprising, since members of the xanthine oxidase family can have broad substrate specificies (3, 11). In any case, our results do not conclusively define a function for this protein.
In summary, our results suggest that the products of xdhA and probably those of xdhB and xdhC are components of an XDH isozyme and that XDH participates in purine salvage. This is a new function for XDH with an interesting implication. XDH is the second enzyme (adenosine deaminase is the first) that imparts a bias to GMP synthesis and away from AMP synthesis during purine salvage.
XDH and nitrogen limitation.
We initiated this study because a computer analysis of ς54-dependent genes suggested that nitrogen limitation might induce a purine catabolic pathway. However, our results do not provide evidence that purine catabolism produces nitrogen for cell growth. Nonetheless, there is a link between nitrogen limitation and purine metabolism. An xdhA mutant grew faster than wild-type E. coli with aspartate as the sole nitrogen source. A possible explanation for such an effect is that blocking the conversion of hypoxanthine to xanthine results in a higher intracellular level of aspartate, which can now be more efficiently utilized. However, there is no simple explanation that could account for the differences in the aspartate concentration, especially since the extracellular concentration is high (aspartate is the nitrogen source in the medium), which should imply that the intracellular concentration is also high. An alternate explanation is that accumulation of an intermediate in purine salvage stimulates aspartate catabolism. At this time, it is not clear why HX1 grows faster with aspartate as the nitrogen source.
In summary, our study of possible purine catabolism showed that purines stimulate growth of nitrogen-limited cells, E. coli has XDH activity, XDH contributes to purine salvage, and mutants deficient in XDH activity have altered amino acid catabolism during nitrogen-limited growth.
ACKNOWLEDGMENTS
Grants MCB-9723003 from the National Science Foundation and GM47965 from the National Institute of General Medical Sciences supported this work.
We gratefully acknowledge R. Bender and D. Friedman, both from the University of Michigan, for strains and Alexandros Kiupakis for composing Fig. 2 and 3.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Ban J, Vitale L, Kos E. Thymine and uracil catabolism in Escherichia coli. J Gen Microbiol. 1972;73:267–272. doi: 10.1099/00221287-73-2-267. [DOI] [PubMed] [Google Scholar]
- 3.Barata B A, LeGall J, Moura J J. Aldehyde oxidoreductase activity in Desulfovibrio gigas: in vitro reconstitution of an electron-transfer chain from aldehydes to the production of molecular hydrogen. Biochemistry. 1993;32:11559–11568. doi: 10.1021/bi00094a012. [DOI] [PubMed] [Google Scholar]
- 4.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 5.Cusa E, Obradors N, Baldoma L, Badia J, Aguilar J. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J Bacteriol. 1999;181:7479–7484. doi: 10.1128/jb.181.24.7479-7484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gremer L, Meyer O. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur J Biochem. 1996;238:862–866. doi: 10.1111/j.1432-1033.1996.0862w.x. [DOI] [PubMed] [Google Scholar]
- 7.Kisker C, Schindelin H, Baas D, Retey J, Meckenstock R U, Kroneck P M. A structural comparison of molybdenum cofactor-containing enzymes. FEMS Microbiol Rev. 1998;22:503–521. doi: 10.1111/j.1574-6976.1998.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 8.Levine R A, Taylor M W. Mechanism of adenine toxicity in Escherichia coli. J Bacteriol. 1982;149:923–930. doi: 10.1128/jb.149.3.923-930.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandecki W, Hayden M A, Shallcross M A, Stotland E. A totally synthetic plasmid for general cloning, gene expression and mutagenesis in Escherichia coli. Gene. 1990;94:103–107. doi: 10.1016/0378-1119(90)90474-6. [DOI] [PubMed] [Google Scholar]
- 10.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 11.Morpeth F F, Bray R C. Inhibition of xanthine oxidase by various aldehydes. Biochemistry. 1984;23:1332–1338. doi: 10.1021/bi00301a047. [DOI] [PubMed] [Google Scholar]
- 12.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 13.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 14.Romao M J, Archer M, Moura I, Moura J J, LeGall J, Engh R, Schneider M, Hof P, Huber R. Crystal structure of the xanthine oxidase-related aldehyde oxido-reductase from D. gigas. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- 15.Rothstein D M, Pahel G, Tyler B, Magasanik B. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc Natl Acad Sci USA. 1980;77:7372–7376. doi: 10.1073/pnas.77.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider B L, Kiupakis A K, Reitzer L J. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol. 1998;180:4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrader T, Rienhofer A, Andreesen J R. Selenium-containing xanthine dehydrogenase from Eubacterium barkeri. Eur J Biochem. 1999;264:862–871. doi: 10.1046/j.1432-1327.1999.00678.x. [DOI] [PubMed] [Google Scholar]
- 19.Tyler B. Regulation of the assimilation of nitrogen compounds. Annu Rev Biochem. 1978;47:1127–1162. doi: 10.1146/annurev.bi.47.070178.005403. [DOI] [PubMed] [Google Scholar]
- 20.Vogels G D, Van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev. 1976;40:403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong S H. The use of purine compounds as sole sources of carbon and nitrogen by Klebsiella species. Microbios. 1988;56:57–62. [PubMed] [Google Scholar]
- 22.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 561–579. [Google Scholar]