Identification of Critical Elements in the tRNA Acceptor Stem and TΨC Loop Necessary for Human Immunodeficiency Virus Type 1 Infectivity (original) (raw)
Abstract
A mutant human immunodeficiency virus type 1 (HIV-1) with a primer binding site (PBS) complementary to yeast tRNAPhe (psHIV-Phe), which relies on exogenous yeast tRNAPhe as reverse transcription primer, was used to investigate elements in the tRNA acceptor stem and TΨC stem-loop required for the tRNA primer selection and use in HIV-1 replication. tRNAPhe mutants with two- or four-nucleotide deletions in the 3′ end retained the capacity to complement replication of psHIV-Phe. tRNAPhe mutants with an extended 5′ end had reduced capacity for complementation, which could be restored by extension of the 3′ end of these tRNAPhe mutants with sequences complementary to the HIV-1 U5 region. Further analysis of mutations in the acceptor stem of tRNAPhe suggested that an intact acceptor stem RNA structure is important for complementation. Analysis of single-nucleotide changes in the TΨC stem-loop of tRNAPhe revealed an unexpected, essential role of this region for rescue of psHIV-Phe.
Human immunodeficiency virus (HIV) exclusively uses tRNA as a primer for reverse transcription. The tRNA
as a primer for reverse transcription. The tRNA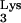 is selected from the cellular tRNA pools, encapsidated into HIV virions, and positioned at the primer binding site (PBS) to initiate reverse transcription (11). The tRNA-PBS interaction has been shown to be a major determinant for selection of the tRNA primer (4, 9, 10, 16). Substitution of the HIV type 1 (HIV-1) PBS with sequence complementary to alternative tRNAs resulted viruses which used these alternative tRNAs as primers for reverse transcription. However, these viruses reverted to use tRNA
is selected from the cellular tRNA pools, encapsidated into HIV virions, and positioned at the primer binding site (PBS) to initiate reverse transcription (11). The tRNA-PBS interaction has been shown to be a major determinant for selection of the tRNA primer (4, 9, 10, 16). Substitution of the HIV type 1 (HIV-1) PBS with sequence complementary to alternative tRNAs resulted viruses which used these alternative tRNAs as primers for reverse transcription. However, these viruses reverted to use tRNA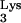 after a short term of in vitro culture (4, 9, 10, 16). The results of these studies pointed to a more complex interaction between the tRNA and viral genome. Additional interactions between HIV-1 RNA and tRNA
after a short term of in vitro culture (4, 9, 10, 16). The results of these studies pointed to a more complex interaction between the tRNA and viral genome. Additional interactions between HIV-1 RNA and tRNA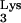 have also been suggested by in vitro chemical and enzymatic analysis of tRNA and U5-PBS complexes (6, 7).
have also been suggested by in vitro chemical and enzymatic analysis of tRNA and U5-PBS complexes (6, 7).
Elucidation of determinants within tRNA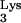 required for primer selection is essential to understand the mechanism of reverse transcription. Inherently, these experiments are difficult because of the inability to modulate the levels of tRNA
required for primer selection is essential to understand the mechanism of reverse transcription. Inherently, these experiments are difficult because of the inability to modulate the levels of tRNA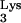 within the cell. To circumvent this problem, we have recently developed an in vivo complementation system to study the tRNA primer selection and use by HIV-1 (18). In this system, the infectivity of a defective HIV-1 (psHIV-Phe) depends on addition of an exogenous reverse transcription primer, yeast tRNAPhe. We have used this system to identify elements of tRNAPhe that are important for primer selection and use in HIV-1. In our previous study, the anticodon stem-loop and TψC stem-loop, but not the D stem-loop, of tRNAPhe were found to be important for psHIV-Phe infectivity (19). In this work, we further investigated the role of the tRNAPhe acceptor stem and TψC loop in the primer selection and use by HIV-1.
within the cell. To circumvent this problem, we have recently developed an in vivo complementation system to study the tRNA primer selection and use by HIV-1 (18). In this system, the infectivity of a defective HIV-1 (psHIV-Phe) depends on addition of an exogenous reverse transcription primer, yeast tRNAPhe. We have used this system to identify elements of tRNAPhe that are important for primer selection and use in HIV-1. In our previous study, the anticodon stem-loop and TψC stem-loop, but not the D stem-loop, of tRNAPhe were found to be important for psHIV-Phe infectivity (19). In this work, we further investigated the role of the tRNAPhe acceptor stem and TψC loop in the primer selection and use by HIV-1.
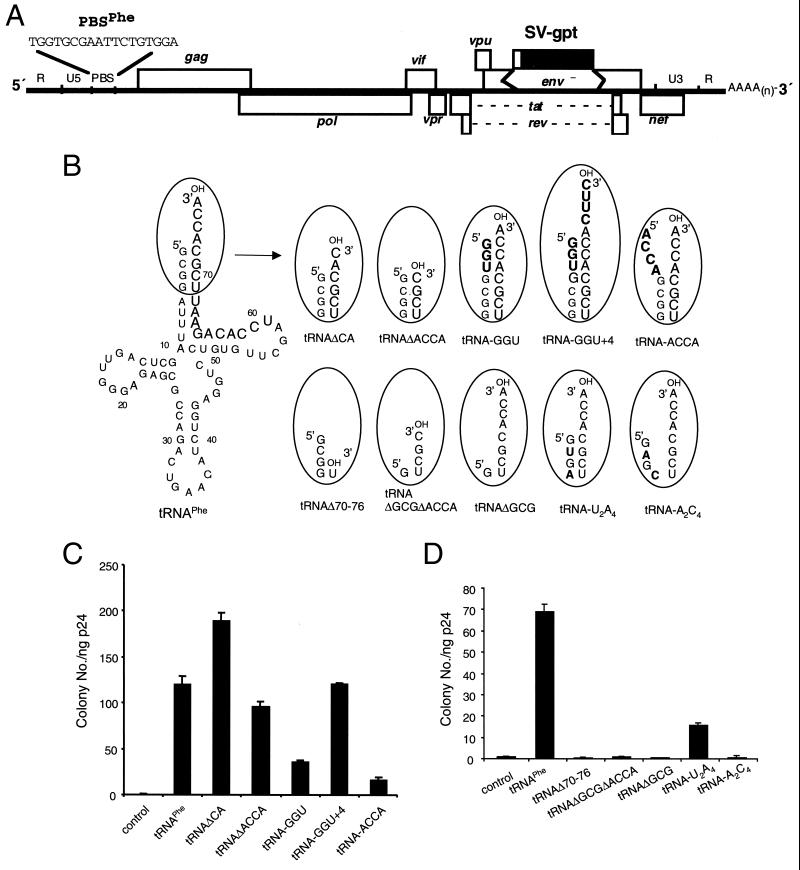
psHIV-Phe contains an HIV-1 proviral genome in which the env gene was deleted and replaced by a drug resistance gene, gpt (xanthine-guanosine phosphoribosyltransferase) (Fig. 1A) (18). The PBS region of psHIV-Phe was mutated to be complementary to the 3′-terminal 18 nucleotides of yeast tRNAPhe. Cotransfection of yeast tRNAPhe, psHIV-Phe provirus, and a plasmid (pLGRNL) encoding vesicular stomatitis virus G envelope protein (2) results in the production of a pseudovirus that can undergo a single round of infection. Successful infection leads to cells resistant to mycophenolic acid. The capacity of a tRNAPhe mutant to rescue psHIV-Phe can be determined from the number of drug-resistant colonies obtained following infection (18). To rescue psHIV-Phe, the tRNA mutant needs to be selected from the cellular tRNA pool, encapsidated into virions, positioned at the PBS region, and used in reverse transcription to generate full-length viral DNA.
FIG. 1.
Effect of tRNAPhe acceptor stem mutations on complementation of psHIV-Phe. (A) psHIV-Phe proviral genome. The defective psHIV-Phe proviral genome contains a PBS complementary to yeast tRNAPhe (PBSPhe). The env gene of HIV was substituted by the gpt gene under the control of simian virus 40 early promoter (SV-gpt). To generate pseudoviruses, this plasmid was cotransfected with the plasmid encoding vesicular stomatitis virus G protein either with or without tRNA. Successful infection of cells with the pseudoviruses confers resistance to mycophenolic acid. (B) Illustration of tRNAPhe mutants with nucleotide changes at the acceptor stem. The added or substituted nucleotides are depicted in bold. Only the terminal portion of each tRNAPhe mutant (circled) is illustrated. (C and D) Numbers of drug-resistant colonies derived from infection of the psHIV-Phe pseudoviruses complemented by indicated mutants. Values are means of data obtained from three independent experiments, with standard deviations depicted (error bars).
All of the tRNAPhe mutants in this study were generated by in vitro transcription using cDNA templates. Each cDNA template contained a T7 promoter followed by the coding sequence of a tRNAPhe mutant. After gel purification and quantitation, the in vitro-transcribed tRNAPhe mutants were cotransfected with psHIV-Phe provirus and pLGRNL to generate pseudoviruses. The amount of the pseudoviruses generated was measured by a p24 enzyme-linked immunosorbent assay and normalized for each infection. The intracellular stability of each mutant was analyzed by using biotin-labeled or 35S-labeled tRNA mutants for transfection (19). All tRNA mutants had stability profiles similar to that of tRNAPhe up to 36 h after transfection (the time the pseudoviruses were harvested) under our experimental conditions (data not shown).
Importance of nucleotides in the tRNAPhe acceptor stem for rescue of psHIV-Phe.
The 3′-end 18 nucleotides of the tRNA primer that bind with the viral PBS region are included in the tRNA acceptor stem and TΨC stem-loop (Fig. 1B). The last four nucleotides (ACCA) of the tRNAPhe 3′ end are single stranded, as with most other eukaryotic tRNAs. To test whether the 3′ single-stranded nucleotides of the tRNAPhe are essential for psHIV-Phe complementation, we designed two deletion mutants in which two (tRNAΔCA) or four (tRNAΔACCA) nucleotides were removed from the tRNAPhe 3′ end (Fig. 1B). The mutant tRNAΔCA rescued psHIV-Phe as effectively as the wild-type tRNAPhe. The mutant tRNAΔACCA also rescued psHIV-Phe, although with slightly lower efficiency than the wild-type tRNAPhe (Fig. 1C). These results suggest that the ACCA sequence on the tRNAPhe 3′ end was not absolutely required for the rescue of psHIV-Phe. This conclusion is based on the assumption that the deleted ACCA sequence was not added back by cellular enzymes following cotransfection. We could not rule out the possibility of the deleted CA sequence in tRNAΔCA being added back by the cellular ATP(CTP):tRNA nucleotidyltransferases (CCA-adding enzymes) after cotransfection (17). However, mutant tRNAΔACCA is an unlikely substrate for CCA-adding enzymes due to the deletion of 3′ CCA along with the discriminator nucleotide A73, an important identity determinant of the tRNA (14, 15). The 3′-end CCA sequence is universal among all mature eukaryotic tRNAs and is important for their natural function in protein synthesis. The lack of requirement for this sequence in the tRNA primer, coupled with our previous finding that the D stem-loop of tRNAPhe was not required for complementation (19), may indicate that HIV-1 can use tRNAs that are not functional in protein synthesis. If this is the case, HIV-1 might have evolved to use such defective tRNAs so as to avoid competition with the cellular protein synthesis machinery. Further experiments will be needed to address this possibility.
To further explore the effects of the acceptor stem alternation on the capacity of tRNAPhe to rescue virus, we generated three tRNAPhe mutants and examined their capacity to rescue psHIV-Phe (Fig. 1B). Mutant tRNA-GGU contained three additional nucleotides (GGU) predicted to base pair with the tRNAPhe 3′-end single-stranded region. This mutant showed decreased capacity for complementation compared to the wild-type tRNAPhe (Fig. 1C). Four nucleotides were added to the 3′ end of tRNA-GGU to generate a mutant (tRNA-GGU+4) with a single-stranded 3′ region (Fig. 1B). These 3′ additional nucleotides (CUUC) in tRNA-GGU+4 are complementary to the PBS-upstream sequence of psHIV-Phe. The four-nucleotide extension (CUUC) of the tRNA would be predicted to result in a four-nucleotide deletion in U5 at the 3′ end of the provirus. Interestingly, tRNA-GGU+4 restored psHIV-Phe infectivity with an efficiency similar to that of wild-type tRNAPhe (Fig. 1C). In contrast, tRNA-ACCA with an ACCA sequence at the 5′ end, which was predicted to allow the tRNA 3′ ACCA sequence to remain unpaired (Fig. 1B), had even lower virus rescue capacity than tRNA-GGU (Fig. 1C).
Since the tRNAPhe mutant without the 3′ ACCA sequence could rescue psHIV-Phe, we next asked how many nucleotides could be removed from the tRNA 3′ region without compromising virus rescue capability. tRNAΔ70-76 contains an additional three-nucleotide deletion at the 3′ end compared to mutant tRNAΔACCA. Unlike tRNAΔACCA, tRNAΔ70-76 failed to rescue psHIV-Phe (Fig. 1D). Inadequate complementarity between tRNAΔ70-76 and the viral PBS, or the reduced base-pairing potential within the acceptor stem of tRNAΔ70-76, might have precluded this mutant from selection and use as the tRNA primer. To address these possibilities, a mutant (tRNAΔGCGΔACCA) was constructed such that three 5′-end nucleotides (G1C2G3) were deleted from the previously described mutant, tRNAΔACCA (Fig. 1B). Although tRNAΔGCGΔACCA would have the same complementarity with the PBS as in tRNAΔACCA, this mutant would contain a reduced number of base pairs in the acceptor stem as does tRNAΔ70-76; this mutant could not rescue psHIV-Phe (Fig. 1D). Furthermore, an additional mutant, tRNAΔGCG, which contained a wild-type 3′ end and the 5′ G1C2G3 deletion, also failed to rescue the virus (Fig. 1B and D). These results pointed to the possibility that the nucleotide length of the base-paired acceptor stem was important for psHIV-Phe rescue. To further explore this possibility, mutants tRNA-A2C4 and tRNA-U2A4 were designed (Fig. 1B). With nucleotide substitutions at the same positions, tRNA-A2C4 was predicted to have reduced base pairing in the acceptor stem, whereas tRNA-U2A4 was predicted to maintain all base pairs in the acceptor stem. Remarkably, the tRNA-A2C4 mutant lost the capacity to rescue psHIV-Phe (Fig. 1D). The tRNA-U2A4 did rescue virus, although not as effectively as wild-type tRNAPhe. Collectively, these results indicate that the base pairing in the tRNA acceptor stem, facilitated by three GC base pairs, was important for the tRNA to rescue psHIV-Phe. Furthermore, the 5′ end of the tRNA is important for the rescue of psHIV-Phe, since additions or deletions of nucleotides at the tRNA 5′ end impaired the virus rescue capacity of tRNAPhe. The role of the tRNA 5′ end in primer selection and use remains to be clarified. A cross-linking study suggested that the 5′ end of tRNA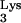 interacts with the C-terminal portion of HIV-1 RT (13). In a structure model of the HIV-1 RNA-tRNA
interacts with the C-terminal portion of HIV-1 RT (13). In a structure model of the HIV-1 RNA-tRNA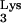 initiation complex, the 5′ end of the tRNA was proposed to form a helix with the 5′ strand of the TψC stem (6, 7).
initiation complex, the 5′ end of the tRNA was proposed to form a helix with the 5′ strand of the TψC stem (6, 7).
Elements in the TΨC stem-loop are essential for psHIV-Phe complementation.
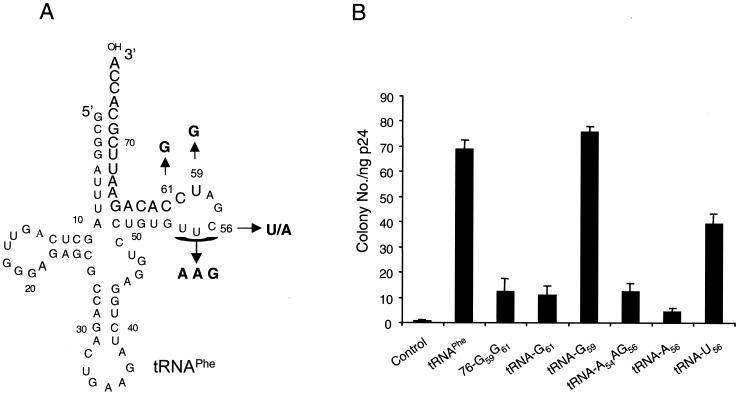
Mutations were introduced to the tRNA TψC stem-loop to explore the importance of the tRNA nucleotides that are complementary to the 3′ end of the PBS (Fig. 2A). Surprisingly, mutant tRNA-G59G61 failed to rescue psHIV-Phe, despite the remaining of 15-nucleotide complementarity between this mutant and the PBS (Fig. 2B). Further dissection of the G59G61 mutation indicated that the G61 mutation resulted in a tRNA unable to rescue psHIV-Phe (Fig. 2). To further explore this result, the highly conserved TΨC sequence was replaced with AAG to generate mutant tRNA-A54AG (Fig. 2A). Analysis of this tRNA mutant revealed an impaired capacity to rescue psHIV-Phe (Fig. 2B).
FIG. 2.
Mutations in the tRNAPhe TΨC stem-loop affect complementation of psHIV-Phe. (A) Illustrations of the positions of the nucleotides mutated in designated tRNAPhe mutants. (B) The psHIV-Phe rescue capacity of the tRNAPhe mutants as measured by drug-resistant colony numbers. Values are means of data obtained from three independent experiments, with standard deviations depicted (error bars).
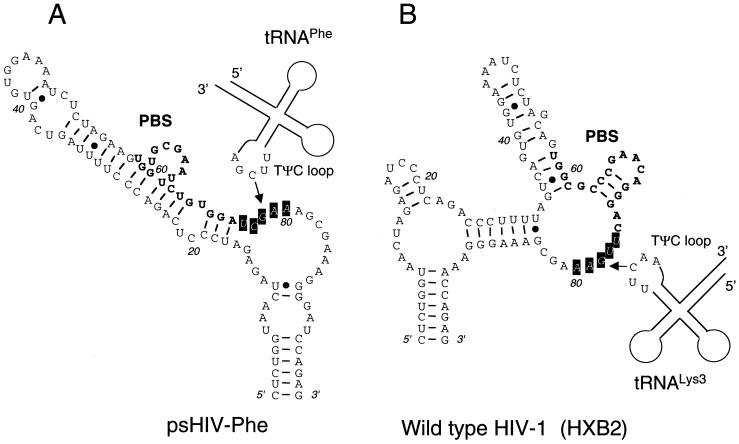
Examination of the psHIV-Phe RNA sequence immediately downstream of the PBS (5′-201UCGAA205-3′) revealed complementarity with the tRNAPhe TψC loop sequence (5′-54TΨCGA58-3′). RNA structure modeling of this region using the M-fold program predicted a non-base-paired loop structure 3′ to the PBS (Fig. 3A) (8, 12, 20). Therefore, the inability of tRNA-A54AG to rescue the virus might be due to disruption of potential interaction between the TψC loop and the PBS-downstream loop sequence of HIV-1. To address this possibility, we introduced in the TΨC loop a single mutation (C56→A56) which would not favor the proposed interaction between TΨC loop and the PBS-downstream sequence (Fig. 2A). Strikingly, the resultant mutant tRNA-A56 almost completely lost the capacity to rescue psHIV-Phe. Furthermore, a C56→U56 substitution at the same position resulted in a mutant (tRNA-U56) with considerably enhanced virus rescue capacity compared to tRNA-A56 (Fig. 2B). Unlike A56, U56 could base pair with G203 in the PBS-downstream sequence to restore the complementarity between TΨC loop and the PBS-downstream sequence, supporting the possibility of a functional interaction between TΨC loop of tRNAPhe and the PBS-downstream sequence of psHIV-Phe. The initial step in the tRNA-PBS binding might be through an RNA loop-loop interaction at the PBS 3′ region rather than at the PBS 5′ end located in a stem, which would necessitate unwinding of the PBS prior to the initial interaction (Fig. 3). HIV-1 nucleocapsid protein, which promotes tRNA annealing to the viral RNA (1), does not promote the unwinding of tRNA in the absence of the viral RNA genome (3, 5). Thus, the complementarity between the tRNA TΨC loop and viral PBS-downstream sequence might facilitate an initial interaction to trigger the nucleocapsid protein to unwind the tRNA and promote annealing with the PBS. The fact that tRNAPhe mutants with 3′-end nucleotide deletions would still complement psHIV-Phe also supports this idea. Further studies using the complementation system and the tRNAPhe mutants described in this study should help to delineate the mechanism of tRNA selection and use in HIV-1 replication.
in the absence of the viral RNA genome (3, 5). Thus, the complementarity between the tRNA TΨC loop and viral PBS-downstream sequence might facilitate an initial interaction to trigger the nucleocapsid protein to unwind the tRNA and promote annealing with the PBS. The fact that tRNAPhe mutants with 3′-end nucleotide deletions would still complement psHIV-Phe also supports this idea. Further studies using the complementation system and the tRNAPhe mutants described in this study should help to delineate the mechanism of tRNA selection and use in HIV-1 replication.
FIG. 3.
RNA secondary structures of the PBS and surrounding regions in psHIV-Phe (A) and in wild-type HIV-1 (HXB2 strain) (B) predicted by the M-fold structure modeling program. The PBS regions are in bold. The boxed nucleotides in a loop region represent the PBS-downstream sequence complementary to the TψC loop sequence of tRNAs. Potential loop-loop interactions between the tRNA TΨC loop and the PBS-downstream sequence of the viral RNA genome are depicted. The drawings are for illustration only and are not to scale.
Acknowledgments
We thank Steve Harvey and Stephen Hajduk for helpful comments and Dee Martin for preparation of the manuscript.
C.D.M. acknowledges the continued support from MAR. The pseudovirus culture was carried out in the UAB AIDS Center virus core facility (supported by grant AI-27767). This work was supported by grants AI34749 and GM56544 to C.D.M.
REFERENCES
- 1.Barat C, Lullien V, Schatz O, Keith G, Nugeyre M T, Gruninger-Leitch F, Barre-Sinoussi F, LeGrice S F, Darlix J L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan B, Weidemaier K, Yip W T, Barbara P F, Musier-Forsyth K. Intra-tRNA distance measurements for nucleocapsid protein-dependent tRNA unwinding during priming of HIV reverse transcription. Proc Natl Acad Sci USA. 1999;96:459–464. doi: 10.1073/pnas.96.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das A T, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA
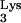 J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar] - 5.Gregoire C J, Gautheret D, Loret E P. No tRNA
 unwinding in a complex with HIV NCp7. J Biol Chem. 1997;272:25143–25148. doi: 10.1074/jbc.272.40.25143. [DOI] [PubMed] [Google Scholar]
unwinding in a complex with HIV NCp7. J Biol Chem. 1997;272:25143–25148. doi: 10.1074/jbc.272.40.25143. [DOI] [PubMed] [Google Scholar] - 6.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA
 (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
(template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar] - 7.Isel C, Westhof E, Massire C, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18:1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 9.Kang S M, Wakefield J K, Morrow C D. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology. 1996;222:401–414. doi: 10.1006/viro.1996.0437. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Mak J, Arts E J, Gu Z, Kleiman L, Wainberg M A, Parnaik M A. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J Virol. 1994;68:6198–6206. doi: 10.1128/jvi.68.10.6198-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 13.Mishima Y, Steitz J A. Site-specific crosslinking of 4-thiouridine-modified human tRNA
 to reverse transcriptase from human immunodeficiency virus type I. EMBO J. 1995;14:2679–2687. doi: 10.1002/j.1460-2075.1995.tb07266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
to reverse transcriptase from human immunodeficiency virus type I. EMBO J. 1995;14:2679–2687. doi: 10.1002/j.1460-2075.1995.tb07266.x. [DOI] [PMC free article] [PubMed] [Google Scholar] - 14.Reichert A, Rothbauer U, Morl M. Processing and editing of overlapping tRNAs in human mitochondria. J Biol Chem. 1998;273:31977–31984. doi: 10.1074/jbc.273.48.31977. [DOI] [PubMed] [Google Scholar]
- 15.Reichert A S, Morl M. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res. 2000;28:2043–2048. doi: 10.1093/nar/28.10.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakefield J K, Wolf A G, Morrow C D. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA
 J Virol. 1995;69:6021–6029. doi: 10.1128/jvi.69.10.6021-6029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
J Virol. 1995;69:6021–6029. doi: 10.1128/jvi.69.10.6021-6029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar] - 17.Wolin S L, Matera A G. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Yu Q, Morrow C D. Complementarity between 3′ terminal nucleotides of tRNA and primer binding site is a major determinant for selection of the tRNA primer used for initiation of HIV-1 reverse transcription. Virology. 1999;254:160–168. doi: 10.1006/viro.1998.9542. [DOI] [PubMed] [Google Scholar]
- 19.Yu Q, Morrow C D. Essential regions of the tRNA primer required for HIV-1 infectivity. Nucleic Acids Res. 2000;28:4783–4789. doi: 10.1093/nar/28.23.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]