Two Human Orthologues of Eco1/Ctf7 Acetyltransferases Are Both Required for Proper Sister-Chromatid Cohesion (original) (raw)
Abstract
Genetic studies in yeast and Drosophila have uncovered a conserved acetyltransferase involved in sister-chromatid cohesion. Here, we described the two human orthologues, previously named EFO1/ESCO1 and EFO2/ESCO2. Similar to their yeast (Eco1/Ctf7 and Eso1) and fly (deco) counterparts, both proteins feature a conserved C-terminal domain consisting of a H2C2 zinc finger motif and an acetyltransferase domain that is able to catalyze autoacetylation reaction in vitro. However, no similarity can be detected outside of the conserved domain. RNA interference depletion experiment revealed that EFO1/ESCO1 and EFO2/ESCO2 were not redundant and that both were required for proper sister-chromatid cohesion. The difference between EFO1 and EFO2 also is reflected in their cell cycle regulation. In mitosis, EFO1 is phosphorylated, whereas EFO2 is degraded. Furthermore, both proteins associate with chromosomes, and the chromosome binding depends on the diverse N-terminal domains. We propose that EFO1 and EFO2 are targeted to different chromosome structures to help establish or maintain sister-chromatid cohesion.
INTRODUCTION
Sister-chromatid cohesion plays a fundamental role in chromosome partition during the cell cycle. Although the molecular components that physically hold sister chromatids together have been well characterized, the mechanism that leads to this linkage remains elusive. Genetic studies in budding yeast suggest that chromatid cohesion is established in two distinct steps. In the first step, the cohesin complexes, which directly participate in holding two sisters together, are loaded onto chromosomes before S phase. In vertebrates, this step depends on the formation of the prereplication complex and requires a conserved cohesin loading factor Scc2 (Furuya et al., 1998; Ciosk et al., 2000; Gillespie and Hirano, 2004; Rollins et al., 2004; Takahashi et al., 2004; Tonkin et al., 2004). On the other hand, the second step establishes the physical link between sisters during DNA replication. The distinction of the two steps was best demonstrated in budding yeast cells mutated for the ECO1/CTF7 gene (Skibbens et al., 1999; Toth et al., 1999). In this mutant, despite successful loading of the cohesin complexes, cohesion between the sisters was grossly defective.
ECO1/CTF7 encodes a poorly understood acetyltransferase (Ivanov et al., 2002). In budding yeast, Eco1/Ctf7 binds to chromosomes throughout the cell cycle. However, it is not required for the recruitment of the cohesin complexes before S phase, nor is it involved in the maintenance of cohesion after S phase. Instead, it has been implicated in the establishment of the physical links between the sisters during S phase, presumably at or immediately after the replication forks (Skibbens et al., 1999; Toth et al., 1999). The functional counterpart of Eco1/Ctf7 in fission yeast is called Eso1 (Tanaka et al., 2000). The Eso1 protein is much bigger than Eco1/Ctf7. Its N-terminal domain is similar to a repair DNA polymerase, and the C-terminal domain closely resembles Eco1/Ctf7. Remarkably, the polymerase domain is completely dispensable for chromatid cohesion, suggesting this particular organization bears little, if any, functional significance for the establishment of cohesion in fission yeast.
A recent genetic screen in Drosophila revealed a putative acetyltransferases, named deco, that also is required for proper sister-chromatid cohesion (Williams et al., 2003). The C-terminal domain of deco is highly homologous to the acetyltransferase domains of both Eco1/Ctf7 and Eso1, suggesting that deco is their homologue. The deco protein also contains a long N-terminal domain that exhibits no homology to any other known proteins. The finding of the Drosophila homologue suggests that a conserved mechanism is used in metazoan to establish chromatid cohesion.
It is unclear how Eco1/Ctf7 family proteins participate in the cohesion establishment. In fission yeast, strong genetic interaction between Eso1 and Pds5 has been reported (Tanaka et al., 2001), suggesting that Eso1 functions through Pds5, a conserved nonstoichiometric component of the cohesin complex (Hartman et al., 2000; Panizza et al., 2000; Sumara et al., 2000; Tanaka et al., 2001). The exact role of Pds5 remains to be determined, and it remains controversial whether the function of Eco1/Ctf7 is dependent on its acetyltransferase activity (Ivanov et al., 2002; Brands and Skibbens, 2005). Furthermore, the acetyltransferase genetically and physically interacts with the DNA replication machinery (Skibbens et al., 1999; Madril et al., 2001; Edwards et al., 2003; Kenna and Skibbens, 2003). This has led to the suggestion that the acetyltransferase plays a direct role in the establishment of cohesion immediately after chromosome replication.
To investigate the mechanism of cohesion establishment in vertebrates, we have identified two human Eco1/Ctf7 orthologues. Both proteins have been described previously as the sequence homologues of Eco1/Ctf7 and named as EFO1 and EFO2 (Bellows et al., 2003). EFO2 was described as a partial sequence. In this report, we demonstrated that both EFO1 and EFO2 exhibit acetyltransferase activity in vitro. Small interfering RNA (siRNA) depletion experiments confirmed that EFO1 and EFO2 are required for the establishment of sister-chromatid cohesion, suggesting that they are orthologues of yeast Eco1/Ctf7. Surprisingly, EFO1 and EFO2 are not redundant because depletion of either protein led to a dramatic increase in defective cohesion. The uniqueness of these two proteins is likely due to their divers N-terminal domains, which are responsible for chromosome binding. Furthermore, EFO1 and EFO2 are regulated in mitosis by phosphorylation and protein degradation, respectively.
MATERIALS AND METHODS
Plasmid Construction, Recombinant Protein Expression and Purification, and Antibody Production
All mammalian cell expression plasmids are based on either pCS2 or pcDNA4. These constructs also were used to translate proteins in vitro in reticulocyte lysate or wheat germ extract (Promega, Madison, WI). Recombinant proteins were expressed in BL21(DE3) using pET28 vectors and, later, purified over TALON metal affinity resin (BD Bioscience Clontech, Palo Alto, CA). The eluded proteins were further polished over a HiTrap Q column. Rabbit polyclonal antibodies to EFO1 and EFO2 were raised at Genemed (South San Francisco, CA) using fragments of EFO1 (amino acid 1–203) and EFO2 (amino acid 216–317), respectively, and followed by affinity purification. Antibody to securin has been described previously (Zou et al., 1999). Antibodies to phospho-Histone H3, topoisomerase IIα, and β-tubulin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody to α-actin was purchased from Sigma (St. Louis, MO).
Tissue Culture, Cellular Fractionation, Immunofluorescence Microscopy, and Chromosome Spread
HeLa and 293T cells were cultured in DMEM, whereas HCT116 cells were cultured in McCoy's 5A, both supplemented with 10% fetal bovine serum. HeLa cells were synchronized at G1/S or premetaphase by double-thymidine block or thymidine-nocodazole arrest (Fang et al., 1998). For Western blot analysis, cells were lysed in the lysis buffer as described previously (Stemmann et al., 2001). For cellular fractionation, we followed the protocol described by Mendez and Stillman (2000). To prevent spontaneous polymerization of actin, which was used as the marker for the cytoplasm fractions, cytoclasin B was added to the final concentration of 10 μg/ml when the cells were lysed. For immunofluorescence microscopy, we followed the protocol used in the Mitchison laboratory (http://mitchison.med.harvard.edu/protocols/gen1.html). Methonal was used to fix the cells. When noted, 0.1% Triton X-100 extraction was performed to remove soluble proteins. In these cases, a different protocol (Waizenegger et al., 2000) was followed. For chromosome spread preparation, the protocol described by Henegariu et al. (2001) was followed exactly.
Acetyltransferase Activity Assay
The acetyltransferase activity assay was carried out according to the protocol described previously (Gu and Roeder, 1997). Approximately 0.5 μg of recombinant EFO1 and EFO2 was used in each reaction.
siRNA Depletion
Two rounds of siRNA oligonucleotide transfection were performed to reduce the expression level of the targeted genes. HeLa cells at 80% confluence were transfected with siRNA oligonucleotides in six-well plate following a calcium phosphate-based protocol (Pear et al., 1993), except that DNA was replaced with siRNA in the transfection. Specifically, in day 1, 30 nM (final concentration) siRNA oligonucleotide in 2 ml of DMEM was used in each transfection, and the transfection complex was removed after 10 h. The transfection was repeated on the third day. Finally, the cells were harvested on the fifth day for metaphase spread and fluorescence-activated cell sorting (FACS) analysis. The sense sequences of the oligonucleotides for EFO1 and EFO2 are GGUCAUCAAAGGCAGUAUUTT and CCUGCAUUGCUCUCAAUAATT, respectively.
RESULTS
The Similarity of EFO1 and EFO2 with Eco1/Ctf7 Is Limited at Their C Termini
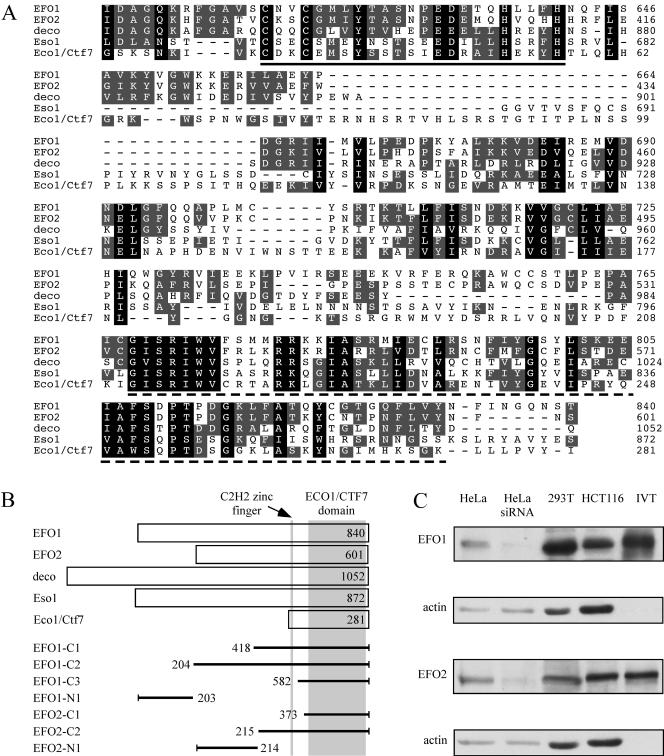
A BLAST search identified two human proteins (GI: 38566492 and GI: 51466732) whose C-terminal domains (∼200 residues) shared 21–33% identity and 38–54% similarity to that of Eco1/Ctf7, Eso1, or deco (Figure 1A). Within the conserved region, a C2H2 zinc finger and a putative acetyltransferase domain are organized in the same manner as Eco1/Ctf7, Eso1, and deco (Figure 1B). The larger protein is identical to a previously published protein named EFO1 and the smaller protein contains the partial sequences named EFO2 (Bellows et al., 2003). Interestingly, the smaller protein also is encoded by a recently identified disease gene named ESCO2 (Vega et al., 2005). Mutations in ESCO2 are responsible for a devastating childhood disease known as Roberts syndrome. In this article, we use EFO1 to describe the bigger homologue and EFO2 to describe the smaller homologue.
Figure 1.
Identification of human homologues of Eco1/Ctf7. (A) Sequence alignment of the conserved C-terminal region. The residues conserved among all five sequences are shaded with black, and the residues conserved among any three of the five are shaded in gray. The C2H2 zinc finger and the acetyltransferase domain is highlighted a solid and a dash line, respectively. (B) Organization of the five Eco1/Ctf7 family proteins. Conserved zinc finger and acetyltransferase domain are indicated in gray. The fragments of EFO1 and EFO2 used in this study also are presented in the diagram. (C) Western blot of endogenous EFO1 and EFO2 in HeLa, 293T, and HCT116 cells. Their migrations on SDS-PAGE were compared with the EFO1 and EFO2 protein transcribed and translated in wheat germ extract (IVT) using their cDNA. As negative controls, lysates of HeLa cells transfected with EFO1- or EFO2-siRNA oligonucleotides also were blotted. Neither EFO1 nor EFO2 was detected in the wheat germ extract in the absence of the plasmids (our unpublished data).
Using a human fetal thymus marathon cDNA library (BD Biosciences Clonetech), we performed 5′ rapid amplification of cDNA ends (RACE) to determine the N-terminal ends of these proteins. We found that EFO2 is 92-residue shorter than the sequence in GenBank (GI: 51466732), which was predicted based on genomic sequence. The RACE result of EFO2 was confirmed by the following three observations. First, a PCR reaction, using a primer targeting the 5′ end of the predicted sequence (GI: 51466732) and a primer annealing to a sequence ∼600 base pairs downstream, failed to produce any signal from the same cDNA library mentioned above (our unpublished data). The downstream primer worked in many different PCR reactions. Second, the EFO2 homologues in mouse and rat from the current databases have a similar length with the open reading frame (ORF) determined by our RACE experiment. Third, the endogenous EFO2 protein detected by Western blot migrated at the same rate on SDS-PAGE as the in vitro-translated EFO2 encoded by the shorter ORF (Figure 1C). Therefore, we concluded that we have correctly determined the 5′ end of EFO2. On the other hand, our 5′ RACE result of EFO1 was in agreement with the GenBank sequence (Figure 1C).
Like Eso1 and deco, both EFO1 and EFO2 are significantly larger than Eco1/Ctf7. However, between the two human homologues, the homology is remarkably restricted in the zinc finger–acetyltransferase domain, which is identified as the Eco1/Ctf7 domain in the rest of the article. The similarity between EFO1 and EFO2 reaches 77% (59% identity) within the Eco1/Ctf7 domain, whereas no significant homology can be detected outside of this region. The diversity of their N-terminal domains suggests that these two human homologues may perform distinct functions in sister-chromatid cohesion. The N-terminal domain of EFO1 reportedly shares some homology to a linker histone (Bellows et al., 2003). On the other hand, the N-terminal domain of EFO2 does not resemble any other proteins in the current databases.
EFO1 and EFO2 Exhibited Acetyltransferase Activity
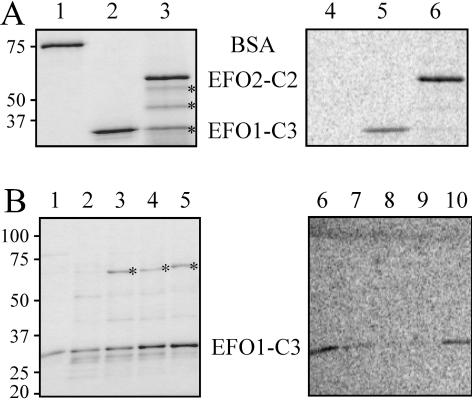
To test whether both EFO1 and EFO2 are indeed acetyltransferase, we purified the recombinant EFO1 and EFO2 from Escherichia coli. Because both proteins are fairly large and form insoluble inclusion bodies in E. coli (our unpublished data), we expressed only the His6-tagged C-terminal fragments that contain the putative acetyltransferase domain (Figures 1B and 2). The recombinant proteins were further purified over a HiTrap Q column and quantified by Coomassie Blue staining (Figure 2A, left). Without any known substrates, their activity was measured by autoacetylation, a reaction also documented for Eco1/Ctf7, EFO1, and other acetyltransferases (Ivanov et al., 2002; Bellows et al., 2003). In this assay, both proteins (∼0.5 μg) were incubated with [14C]acetyl-CoA, and, in both cases, 14C labeling was detected (Figure 2A, right). As control, the same amount of bovine serum albumin (BSA) also was included in the same reaction, and no labeling was detected. Therefore, the 14C labeling was unlikely due to spontaneous acetylation. Furthermore, point mutations at the conserved residues in the acetyltransferase domain led to significant decreases in autoacetylation of EFO1 (Figure 2B, lanes 7–9). Therefore, we concluded that the EFO1 and EFO2 proteins possess acetyltransferase activity.
Figure 2.
EFO1 and EFO2 catalyzed autoacetylation. (A) In vitro autoacetylation of EFO1 and EFO2. Purified recombinant EFO1-C3 (lanes 2 and 5) and EFO2-C2 (lanes 3 and 6) fragments were tested for their autoacetylation activity. BSA was used as the negative control (lanes 1 and 3). For each reaction, 0.5 μg of the recombinant protein was added. After reaction, the mixtures were analyzed on SDS-PAGE, and proteins were visualized by Coomassie Blue staining (left). The molecular weight markers (kilodaltons) are indicated on the left side of the gel. The asterisks indicate the degradation fragments of EFO2-C2. The same gel was dried and exposed to a phosphorimage screen to detect 14C labeling (right). (B) Mutations in the conserved acetyltransferase domain of EFO1 reduced the activity of autoacetylation. Various point mutations were introduced into the EFO1-C3 fragments within the conserved putative acetyltransferase domain. Recombinant proteins were then purified from E. coli, and their acetyltransferase activity was determined as described in A. The amount of protein in the assay was measured by Coomassie Blue staining (left). The same gel was exposed to phosphorimage screen (right). The activity of the G768D (lanes 2 and 7), R779/780G (lanes 3 and 8), and K782E/I783V (lanes 4 and 9) mutants was significantly reduced compared with that of the wild-type control (lanes 1 and 5). The E889G (lanes 5 and 10) mutation slightly reduced the amount of 14C labeling. The bands indicated by asterisks were likely a copurified heat-shock protein. The mutant proteins were less stable as their wild-type counterpart, indicated by the presence of the degradation fragments. The molecular weight (kilodaltons) of the size markers is indicated on the left.
Depletion of EFO1 and/or EFO2 by siRNA Caused Defective Sister-Chromatid Cohesion
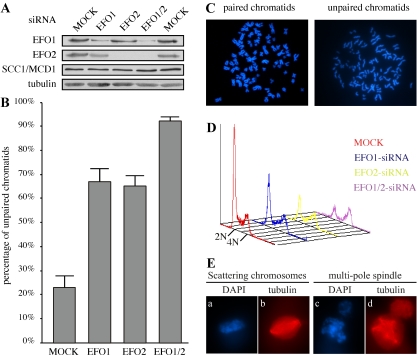
To demonstrate whether EFO1 and EFO2 are involved in chromatid cohesion, we depleted their expression with specific siRNA oligonucleotides. As shown in Figure 3A, the expression levels of EFO1 and EFO2 were reduced up to 85% after two rounds of transfection with the respective siRNA oligonucleotides. The level of EFO2 was lower in the EFO1-depleted cells. This was most likely due to the fact that the EFO1-depleted cells were enriched in mitosis when EFO2 became unstable (see below). Furthermore, double depletion was achieved by transfecting both siRNA oligonucleotides simultaneously. The expression levels of cohesinSCC1/MCD1 were not altered by the depletion (Figure 3A).
Figure 3.
Defective chromatid cohesion in HeLa cells depleted of EFO1 and/or EFO2. (A) The expression levels of SCC1/MCD1, EFO1, and EFO2 in HeLa cells depleted of EFO1 and/or EFO2. (B) Quantification of the percentage of the unpaired chromosomes in the metaphase spreads prepared from the aforementioned siRNA cells. The standard derivations were calculated based on at least four independent double-blinded experiments and plotted as the error bars. (C) Examples of the metaphase chromosome spreads. The spread of paired sister-chromatids were prepared from mock-transfected HeLa cells, and the spread of unpaired sister-chromatids were prepared from EFO1-siRNA cells. (D) FACS analysis of the siRNA cells described in Figure 2A. (E) Stacks of confocal images of mitotic cells with scattering chromosomes (a and b) and multipole spindles (c and d). The images were taken from double-depleted cells. Chromosomes (blue) were stained with DAPI, and spindles (red) were stained with a monoclonal antibody to α-tubulin.
To examine whether sister chromatid cohesion was affected by the depletions, chromosome spreads were prepared and examined for unpaired chromosomes. To avoid potential artifacts caused by the treatment of microtubule poisons nocodazole, we prepared chromosome spreads from cells with or without 2-h nocodazole treatment. The results were nearly identical, with the exception of the mock-transfected cells, where the percentage of unpaired chromatids increased from 11 to 23% (our unpublished data). Only the analyses of the cells without nocodazole treatment were presented here. Surprisingly, a significant increase in mitotic cells with unpaired chromatids was observed in either EFO1 or EFO2 depleted cells (67 and 65%, respectively; Figure 3B), suggesting that both EFO1 and EFO2 were required for stable cohesion between sister-chromatids. Furthermore, we observed a further increase in unpaired chromatids (93%) when both the EFO1- and EFO2-siRNA oligonucleotides were simultaneously introduced into HeLa cells. Figure 3C showed two examples of chromosome spreads containing paired and unpaired chromatids prepared from the mock and double-depletion cells. Therefore, we concluded that EFO1 and EFO2 are indeed Eco1/Ctf7 orthologues required for stable sister-chromatid cohesion and they are not functionally redundant.
Depletion of EFO1 and/or EFO2 Led to Defective Chromosome Congregation or Segregation
To determine whether the cell cycle distribution was affected by the depletion, we analyzed the siRNA cells by FACS. As shown in Figure 3D, the EFO1 and EFO2 depletion cells were enriched in G2/M (∼30 and 40%, respectively, compared with ∼15% in wild-type cells). A further increase in G2/M cells was detected in double-depletion cells (∼60% G2/M cells). It seemed that these cells had difficulty to progress through G2/M phases of the cell cycle, presumably due to the activation of the mitotic checkpoint. The mitotic delay also was reported in yeast Eco1/Ctf7 mutants, where the delay was dependent on Mad2 (Skibbens et al., 1999). To dissect the defects in more details, we visualized the mitotic chromosomes by 4,6-diamidino-2-phenylindole (DAPI) staining. We detected a marked increase in the cells with their chromosomes scattering along the spindles (Figure 3E, a and b). This abnormality was found in ∼0, 7, 5, and 15% of the mitotic cells treated with mock-, EFO1-, EFO2-, or double-siRNA, respectively. There was also a significant increase in cells with multipole spindles (Figure 3E, c and d), presumably caused by missegregation during the previous cell cycle. This abnormality was found in ∼8, 18, 27, and 53% of the mitotic cells treated with mock-, EFO1-, EFO2-, and double-siRNA, respectively. The percentage of abnormal mitotic cells was significantly smaller than the percentage of the cells with unpaired chromatids as revealed by examining the chromosome spreads. It is possible that the harsher manipulations during the preparation of the spreads broke some weaker cohesion, which would have survived the milder treatment during immunofluorescence staining. All these phenotypes could be a result of defective chromatid cohesion, which led to misalignment of chromatids at the metaphase plate, random segregation of chromosomes, and aneuploidy.
Depletion of EFO1 and/or EFO2 Did Not Reduce the Binding of Cohesin with Chromosomes in Interphase Cells
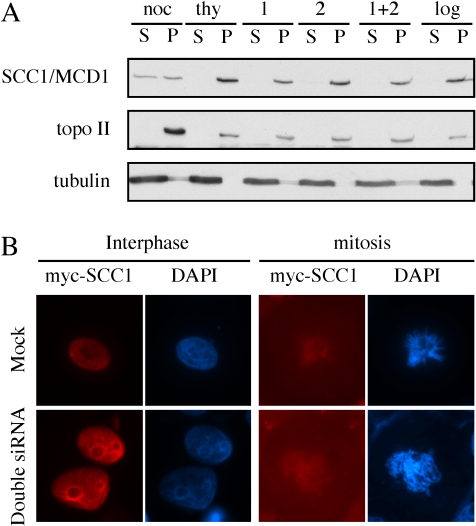
Because sister-chromatid cohesion is mediated by the cohesin complex, we next analyzed the overall binding of cohesin with chromosomes in the cells depleted of EFO1 and/or EFO2. To this end, the chromosomes were separated from the soluble fraction, and the amount of cohesin was examined in both fractions. Using a protocol described previously (Mendez and Stillman, 2000), the cytoplasm fraction (C) was separated from the nuclei, and later, the nucleoplasm fraction (N) was separated from the chromosomes (P). The soluble fraction (S) contained both the cytoplasm fraction and the nucleoplasm fraction from the same sample. P were resuspended in the same volume as the combined S fraction, and the distribution of the concerned proteins was analyzed by immunoblot. In every cellular fractionation experiment described here, we also blotted the cytoplasm and the nucleoplasm fractions separately (our unpublished data). In all the cases, the proteins that were found in the S fractions were in fact in the C fractions. Consistent with the previous report (Sumara et al., 2002), cohesin was detected mostly on chromosomes in thymidine-arrested G1/S cells. In nocodazole-arrested metaphase cells, ∼50% of the cohesin complexes dissociated with chromosomes and were detected in the soluble fractions (Figure 4A). The siRNA cells described in Figure 3 were subjected to the same analysis. We did not observe any decrease in the amount of chromosomal cohesin in these cells, suggesting that the depletion of EFO1 and/or EFO2 did not reduce the gross localization of cohesin to chromosomes.
Figure 4.
Chromosome localization of SCC1/MCD1 in interphase cells was not affected by EFO1 and/or EFO2 depletion. (A) The same samples described in Figure 2A were fractionated into S and P fractions. As controls, thymidine-nocodazole arrested prometaphase cells (noc), double-thymidine arrested G1/S cells (thy), and log phase cells (log) were fractionated together with HeLa cells transfected with EFO1 (1), EFO2 (2), and EFO1 and 2 (1 + 2) siRNA oligonucleotides. The distribution of SCC1/MCD1 was analyzed. Topo IIα and α-tubulin, which localized to chromosomes and the cytoplasm, respectively, were used as controls for the cellular fractionation. (B) Immunofluorescence images of HeLa cells mock-treated or double-depleted of EFO1 and EFO2. Extraction with 0.1% Triton X was performed to remove soluble proteins. DNA (blue) was stained DAPI, and SCC1/MCD1-myc9 (red) was detected with a monoclonal anti-myc (9E10).
It was somewhat unexpected that no increase in the amount of the soluble cohesin was detected even in the double-depletion cells despite that ∼60% of them were in G2/M as determined by FACS analysis (Figure 3D) and that cohesins dissociated from the chromosome arms in mitosis (Figure 4B). The enrichment of mitotic cells was further confirmed qualitatively by immunoblot with anti-phospho-H3 (our unpublished data). The inability to detect any increases in soluble cohesin might be due to the intrinsic less-quantitative nature of this assay because we also had difficulty to determine the difference of cohesin distribution between the fractions prepared from log phase cells and the fractions prepared from a mixture of 50% mitotic cells and 50% interphase cells (our unpublished data). Alternatively, EFO1 and EFO2 depletion might somehow prevent or reduce the mitotic dissociation of cohesins from the chromosome arms. To directly test the latter possibility, we performed immunofluorescence microscopy analysis of HeLa cells transiently expressing a myc-tagged SCC1/MCD1 (Waizenegger et al., 2000). A brief 0.1% Triton X-100 extraction was performed to remove any soluble cohesin. In both the mock and double-depletion cells, cohesin localized to the nucleus in interphase (Figure 4B), suggesting that the overexpressed SCC1/MCD1 was capable of localizing to chromosomes, presumably by incorporating themselves into the cohesin complexes. It also confirmed the result of the cellular fractionation showing that the gross localization of cohesins was not affected by the depletion (Figure 4A). As expected, after the extraction, little cohesin was detected in mitotic cells because most of the cohesins became soluble and were removed by the extraction (Waizenegger et al., 2000). Consequently, we could not confidently decide which mitotic cells were expressing the myc-tagged SCC1/MCD1. Nonetheless, the transfection efficiency was ∼40% as determined by counting the interphase cells, and it was reasonable to assume that the same percentage of mitotic cells expressed the myc-tagged SCC1/MCD1. We examined >20 mitotic cells from each of the two samples, and we did not detect any difference between the mock-treated cells and the siRNA-depletion cells. Examples of the mitotic cells were shown in Figure 4B. Although the data did not allow us to conclude whether the depletion of EFO1 and EFO2 reduced the mitotic localization of cohesins to the centromeres, it indicated that the depletion did not affect the mitotic dissociation of cohesins from chromosome arms. Together, these results indicated that the cohesion defects in the absence of EFO1 and EFO2 were unlikely due to a global dissociation of cohesins from chromosomes before mitosis.
EFO1 and EFO2 Are Chromosome Binding Proteins
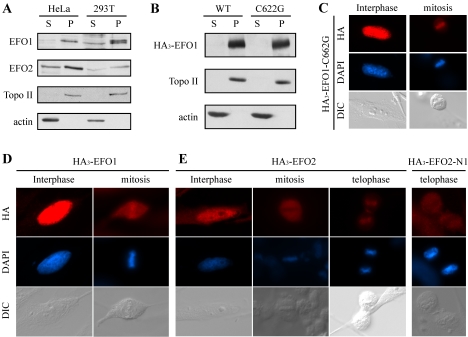
Eco1/Ctf7 in budding yeast localizes to chromosomes throughout the cell cycle (Toth et al., 1999), whereas the localization of Eso1 in fission yeast remains to be determined. To examine the localization of EFO1 and EFO2, we performed cellular fractionation to separate the chromosomes from the soluble fractions. As shown in Figure 5A, in both HeLa cells and 293T cells, ∼70% of the endogenous EFO1 and EFO2 associated with chromosomes. The rest of them were found in the S fractions. The signal in the soluble fractions was contributed by the cytoplamic EFO1 and EFO2 because little, if any, of them was detected in the N fractions when analyzed separately (our unpublished data). Furthermore, the chromosome association of EFO1 and EFO2 was stable in wash buffers containing up to 250 mM NaCl (our unpublished data). The double bands detected in the chromosome fraction of 293T cells reflected a phosphorylation modification,and the upper band collapsed into the lower band upon phosphatase treatment (our unpublished data; see below for the mitotic phosphorylation of EFO1). It was unclear why the upper band was not detected in HeLa cells. It was possible that this protein behaved somehow differently in these two cell lines. Nonetheless, similar to Eco1/Ctf7, both EFO1 and EFO2 associated with chromosomes.
Figure 5.
EFO1 and EFO2 localized to chromosomes. (A) The localization of the endogenous EFO1 and EFO2 in HeLa and 293T cells were analyzed by cellular fractionation. (B) HA3-tagged wild-type and C622G mutant EFO1 were expressed in 293T cells, and their localization was analyzed by cellular fractionation. The localization of HA3-EFO1-C622G (C), HA3-EFO1 (D), HA3-EFO2 (E), and HA3-EFO2-N1 (E) also was analyzed by immunofluorescence microscopy. HA-tagged EFO1 and EFO2 fragments (red) were detected with anti-HA, and DNA (blue) was detected with DAPI staining. Differential interference contrast (DIC) images also were presented. The seemingly midbody staining of HA3-EFO2-N1 was not consistently observed.
Many DNA binding proteins interact with DNA through their zinc finger motifs. To test whether the conserved zinc finger is required for the chromosome localization of EFO1, we substituted the cysteine 622 at the zinc finger motif of EFO1 to glycine (C662G). The mutant was tagged with HA epitope and expressed in 293T cells via transient transfection. Similar to the wild-type EFO1, the HA3-tagged EFO1-C662G mutant remained associated with chromosomes (Finger 5B), suggesting that the zinc finger is dispensable for EFO1 to bind to chromosomes. This localization also was confirmed by immunofluorescence microscopy (Figure 5C; see below)
To support the cellular fractionation results, we also performed immunofluorescence microscopy to detect the localization of HA3-tagged EFO1 and EFO2. We did not use the antibodies to the native proteins because they also cross-reacted with other proteins. Overexpressed HA3-EFO1 (Figure 5D) and HA3-EFO2 (Figure 5E) were detected inside the nucleus in interphase cells. In mitotic cells, however, EFO1 was enriched on chromosomes aligned at the metaphase plate, whereas the EFO2 signal was excluded by the metaphase chromosomes. In telophase, EFO2 reassociated with chromosomes. Therefore, the localization of EFO2 was regulated by the cell cycle. Furthermore, identical to the wild-type EFO1, EFO1-C662G localized to the interphase nucleus and the mitotic chromosomes (Figure 5C), confirming the result of cellular fractionation. Together, we concluded that both EFO1 and EFO2 associated with chromosomes in interphase cells. EFO1 remained on chromosomes in mitosis, whereas EFO2 dissociated from chromosomes and/or was degraded (see below). The overall chromosome localization of EFO1 was not affected by a mutation in the zinc finger motif.
EFO1 and EFO2 Bind to Chromosomes via Their Diverse N Termini
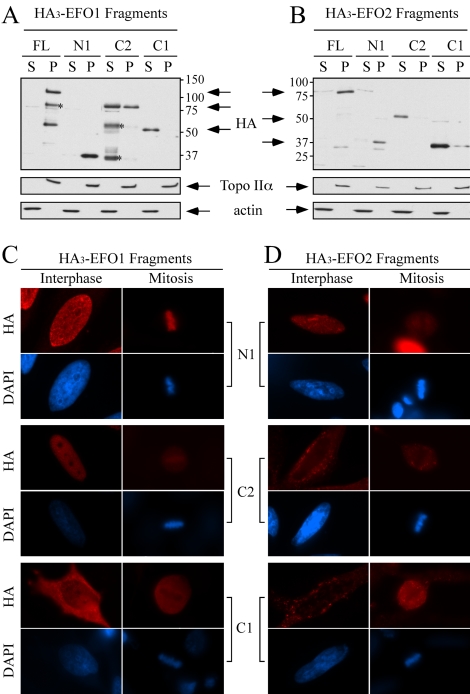
To determine the domains of EFO1 and EFO2 that mediated chromosome association, we performed a series of deletion analyses. First, we demonstrated that the overexpressed EFO1 and EFO2 localized to chromosomes. To this end, HA3-tagged EFO1 and EFO2 were expressed in 293T cells by transient transfection. As shown in Figure 6, A and B, full-length (FL) EFO1 and EFO2 were detected almost exclusively in the P fractions. For unknown reasons, this distribution was different from that of their endogenous counterparts, of which ∼30% also were found in the soluble fractions prepared from 293T (Figure 5A). Regardless, this assay provided an approach to map the chromosome binding domains. The fragments used in the mapping experiment were summarized in Figure 1B. For EFO1, the C1 fragment, which includes both the conserved Eco1/Ctf7 domain and the zinc finger motif, failed to bind to chromosomes (Figure 6A). The longer C2 fragment was detected in both the S fraction and the P fraction. On the other hand, the N1 fragment (a.a. 1–203) was detected exclusively in the P fraction. For EFO2, both the C1 and C2 fragments were detected in the soluble fractions, whereas the N1 fragment was found only in the chromosome fraction (Figure 6B). These results suggested that the N-terminal diverse region was responsible for the chromosome binding of both EFO1 and EFO2. It should be noted that all the fragments that were found in the S fractions were in fact in the C fractions as revealed by analyzing the C and the N fractions separately (our unpublished data).
Figure 6.
The N-terminal domains of EFO1 and EFO2 mediate their chromosome association. Various fragments of EFO1 (A) and EFO2 (B) were fused with HA3-tag and introduced into 293T cells. Their localizations were determined by cellular fractionation followed by Western blot with anti-HA antibody. The molecular weight (kilodaltons) of the size markers is indicated. The asterisks indicate the degradation fragments. The random weak signals in B are cross-reacting bands. The localization of the EFO1 (C) and EFO2 (D) fragments also was analyzed in HeLa cells by immunofluorescence microscopy.
We also performed immunofluorescence staining to determine the localization of these fragments in HeLa cells. As shown in Figure 6C, the localization of the overexpressed HA3-tagged EFO1-N1 and -C1 fragments was consistent with what was found using the cellular fractionation method. The EFO1-C2 fragment was found mostly in the nucleus in interphase cells and was enriched, but not as intense, as the EFO1-N1 fragment, on the chromosomes. This was different from the result of cellular fractionation, which demonstrated that more than half of EFO1-C2 was in the cytoplasm. This could be due to the difference of the cell lines used in these two experiments. Alternatively, EFO1 might contain two chromosome binding domains, and the interaction between EFO1-C2 and chromosomes might be relatively weak and failed to survive the buffers used in the cellular fractionation procedure. Nonetheless, the additional sequence of EFO1-C2 over EFO1-C1 is outside of the conserved region, indicating that the diverse N-terminal domain mediated the chromosome binding. As for EFO2, the result from the immunofluorescence staining of the C1, C2, and N1 fragments was in complete agreement with that from the cellular fractionation method (Figure 6D). Furthermore, unlike the full-length EFO2, the N1 fragment did not reassociate with chromosomes in telophase cells (Figure 5E), suggesting that additional sequence is needed to fully reproduce the cell cycle regulation. Together, we concluded that the conserved Eco/Ctf7 domains of EFO1 and EFO2 did not support their chromosome association. On the other hand, it was the diverse N-terminal domains that were both necessary and sufficient to mediate their chromosome localization. It is worth noting that overexpressing these fragments did not affect the localization of the endogenous proteins, nor did it cause any detectable defects in cell cycle progression and chromatid cohesion as determined by FACS and analyzing metaphase chromosome spreads, respectively (our unpublished data).
EFO1 and EFO2 Are Differentially Regulated during the Cell Cycle
To further investigate the differences between EFO1 and EFO2, we analyzed their status at different stages of the cell cycle. HeLa cells were arrested at G1/S transition with double-thymidine treatments and synchronously released into S phase (Fang et al., 1998). To determine the cell cycle stages of the harvested samples, both phospho-histone H3 and securin were analyzed by immunoblot (Figure 7A). Specifically, histone H3 was phosphorylated only in mitosis (8–11 h), whereas as securin was phosphorylated in S phase (0–6 h), hyperphosphorylated in mitosis, and degraded at the onset of anaphase (10 h). The reappearance of securin after 18 h indicated that the culture had reentered later G1 or S phase because degradation of securin persisted throughout early G1.
Figure 7.
Cell cycle regulation of EFO1 and EFO2. (A) Expression levels of EFO1 and EFO2 in cells released from double-thymidine arrest. (B) The expression level and cellular localization of EFO1 and EFO2 in cells released from nocodazole arrest. The time points were indicated above the panels. Phosphatase treatment indicated that the chromosomal EFO1 is specifically phosphorylated during mitosis. Securin and phospho-histone H3 were used as mitotic markers (see text). Topoisomerase IIα (Topo II) and α-actin are used as the marker for the P fractions and the S fractions, respectively.
In this series of samples, we found that the level of EFO2 was significantly down-regulated in mitosis presumably due to protein degradation, and during the same period, the gel mobility of some EFO1 was retarded and formed up-shifting smears in mitosis (Figure 7, A and B). The down-regulation of EFO2 might start after 6 h postrelease when some cells entered M phase as indicated by weak phosphohistone H3 signal. EFO2 reappeared after 18 h, released from double-thymidine arrest when the cells entered S phase of the next cell cycle (Figure 7A). To further confirm this, we synchronized HeLa cells at prometaphase with nocodazole and released them into anaphase. Consistent with the result in Figure 7A, EFO2 was absent in mitosis and reappeared 8 h after releasing from prometaphase arrest (Figure 7B). The faint signals of EFO2 detected in the chromosome fractions were most likely due to cells that were not synchronized in mitosis. The down-regulation of EFO2 did not seem to be regulated by the anaphase-prompting complex or cyclosome because in vitro translated EFO2 was stable in both Xenopus mitotic extract and interphase extract supplemented with recombinant CDH1 (our unpublished data).
To confirm that the slower mobility of EFO1 in mitosis was due to phosphorylation, we also analyzed EFO1 in cells released from nocodazole arrest. Because EFO1 was detected both in the soluble form and on chromosomes, we performed cellular fractionation to determine which pools of EFO1 were modified. As shown in Figure 7B, the up-shift of EFO1 became more prominent in these samples, presumably due to the prolonged arrest at prometaphase. Interestingly, the up-shift was only detected in the chromosome fractions. Furthermore, phosphatase treatment collapsed the smear signals into sharp bands, indicating that the modification was indeed due to phosphorylation.
DISCUSSION
The mechanism that establishes sister-chromatid cohesion in S phase has been elusive despite many attentions. Among many factors that have been implicated in this process, the members of the Eco1/Ctf7 acetyltransferase family were demonstrated to be important in different organisms, including budding yeast (Skibbens et al., 1999; Toth et al., 1999), fission yeast (Tanaka et al., 2001), Drosophila (Williams et al., 2003), and, in this report, humans. This highlights the importance of these acetyltransferases in this critical process.
Two Eco1/Ctf7 Acetyltransferase Orthologues in Human Cells
We independently identified two human homologues of Eco_1_/Ctf7 by sequence homology search. Although, one of the homologues was later reported as EFO1, its functional relevance in sister-chromatid cohesion was not established (Bellows et al., 2003). Here, we demonstrated that both EFO1 and EFO2 are functional orthologues of Eco1/Ctf7, based on the following observations. First, their C-terminal domains exhibited strong sequence homology with other established members of the Eco1/Ctf7 family. The homology included the acetyltransferase domain and the C2H2 zinc finger, organized in a similar manner as other members of the family. Second, recombinant EFO1 and EFO2 containing the putative acetyltransferase domain were capable of autoacetylation (Figure 2). Similar observations also were reported for Eco1/Ctf7 and EFO1 (Ivanov et al., 2002; Bellows et al., 2003). It is unclear whether the acetyltransferase activity is necessary for sister-chromatid cohesion (Brands and Skibbens, 2005). The conservation of the enzymatic activity itself from yeast to human speaks toward its significance, and this issue merits more scrutiny. Third, siRNA depletion in HeLa cells directly demonstrated that both EFO1 and EFO2 were necessary for proper sister-chromatid cohesion. The depletion caused a mitotic delay, scattering chromosomes in mitotic cells, and consequently aneuploidy in cultured cells (Figure 3). Furthermore, precocious chromatid separation at the centromeres and heterochromatin regions also was documented in cells isolated from patients of Roberts syndrome with mutations in EFO2 or ESCO2 (Jabs et al., 1989, 1991). Fourth, similar to Eco1/Ctf7, EFO1 and EFO2 bind to chromosomes. Together, we concluded that EFO1 and EFO2 are orthologues of Eco1/Ctf7 and are necessary for stable sister-chromatid cohesion in human cells.
Depletion of EFO1 and EFO2 Did Not Grossly Affect the Cohesin Binding to Chromosomes
The Eco1/Ctf7 family proteins clearly play a critical role in sister chromatid cohesion. However, their upstream regulators and downstream targets remain elusive. In Drosophila, mutations in deco destabilized the centromeric localization of the cohesin complex in prometaphase cells, suggesting a maintenance role for deco (Williams et al., 2003). However, this defect might be an indirect result of poor cohesion manifested from improper establishment. On the other hand, in yeast, both Eco1/Ctf7 and Eso1 are dispensable after S phase (Toth et al., 1999; Tanaka et al., 2000). This has been interpreted as they play a role in cohesion establishment. However, they might be required for cohesion maintenance in S phase. Regardless the exact function of the Eco1/Ctf7 family proteins, they do not affect the gross localization of cohesins to chromosomes. This is true in budding yeast where cohesin was found on chromosomes in Eco1/Ctf7 mutants (Toth et al., 1999). Similarly, we did not detect any change in the expression levels of cohesin subunits SCC1/MCD1 and SMC3 in EFO1- and/or EFO2-depleted cells (Figure 3A; our unpublished data). Nor did the depletion reduce the amount of cohesin on chromosomes (Figure 4). These results indicated that the cohesin binding to chromosomes alone is not sufficient for stable cohesion. Additional regulation must exist and identification of the substrates will shed light on this mechanism.
EFO1 and EFO2 Are Not Functionally Redundant
Remarkably, two orthologues of Eco1/Ctf7 were identified in humans, and both were expressed in HeLa and other cultured cells (Figure 1C). This seems to be unique to higher eukaryotes such as human because only one such protein was found in insect and fungus. More importantly, EFO1 and EFO2 are not functionally redundant as suggested by the siRNA depletion experiment. When the expression of either one was depleted, a marked increase in defective cohesion was observed (Figure 3B). This result indicated that EFO1 and EFO2 could not replace each other. Furthermore, double-depletion of EFO1 and EFO2 led to a further increase in unpaired chromatids from ∼65 to 93%. This can be explained in four scenarios. First, EFO1 and EFO2 perform the same functions but are present in limited amount. Depleting one of the two partially impaired the pathway and depleting both led to a more severe phenotype. However, it was unlikely that either EFO1 or EFO2 was limited because both likely functioned as enzymes, and both also were detected in the soluble forms, which presumably could serve as backups (Figure 5A). Second, EFO1 and EFO2 belong to a protein complex. Lacking of either of the two compromised the function of the complex. Third, EFO1 and EFO2 might interact with different specific cofactors to be recruited to or stabilized at different chromosome structures. Consistent with this scenario, their chromosome localization depends on their diverse N-terminal domains (Figure 6). Therefore, it is possible that defects in one of the pathways caused by the depletion leads to a less stable cohesion, which has a higher tendency to be separated before metaphase. Fourth, EFO1 and EFO2 played distinct functions that could be efficiently replaced by each other. However, in the absence of one, the remaining acetyltransferase was able to partially perform the function of the missing one, resulting in a partial cohesion defect. The third and fourth scenarios are not mutually exclusive.
EFO1 and EFO2 Associated with Chromosomes through Their N-Terminal Domains
Both EFO1 and EFO2 localized to chromosomes in interphase (Figure 5). This was demonstrated by cellular fractionation and immunofluorescence microscopy. The interaction between EFO1/EFO2 and chromosomes survived stringent buffers containing up to 250 mM NaCl (our unpublished data). In mitosis, EFO1 remained on chromosomes and was phosphorylated. On the other hand, EFO2, when overexpressed, did not associate with chromosomes until telophase.
Surprisingly, we found that the N-terminal domains of EFO1 and EFO2 were both necessary and sufficient for chromosome association (Figure 6). This finding is unexpected because the N termini of their yeast counterparts are dispensable for chromatid cohesion. For example, the budding yeast Eco1/Ctf7 lacks the N-terminal domain, yet retains it ability to associate with chromosomes (Toth et al., 1999). Similarly, deletion of the N-terminal domain of fission yeast Eso1 had no effect on sister-chromatid cohesion (Tanaka et al., 2000). One possibility is that the conserved zinc fingers could mediate chromosome binding if EFO1 and EFO2 were expressed in their full lengths. To test this, we constructed a point mutation (cysteine622 to glycine) in the zinc finger motif of EFO1. Consistent with our deletion mapping result, this mutation did not affect the chromosome binding of EFO1 (Figure 5B). Therefore, the zinc finger motif was not necessary for EFO1 and, perhaps, EFO2 to bind to chromosomes. However, it remains possible that the zinc finger, although dispensable for chromosome binding, contributes to the functional localization of these proteins. Alternatively, the function of the N-terminal domains was merely to bring the C-terminal conserved domains into the nucleus. Once the C-terminal domains were inside the nucleus, they would bind to chromosomes even in the absence of the N-terminal domains. Although we cannot rule out this possibility, the observation that the N1, but C1, fragment of EFO1 localized to chromosome in mitosis (Figure 6B) indicated that, during this phase of the cell cycle, the accessibility was not sufficient for chromosome binding. Interestingly, overexpression of the N-terminal fragments of EFO1 and EFO2 did not grossly affect the chromosome binding of the endogenous EFO1 and EFO2, respectively, as determined by cellular fractionation (our unpublished data). Nor does it caused any dominant-negative effect on sister chromatid cohesion as detected by examining chromosome spreads (our unpublished data). Perhaps there are abundant binding sites for EFO1 and EFO2. Consistent with this notion, most of the exogenously expressed full-length proteins also bind to chromosomes (Figure 5). Based on their role in chromosome association, we propose that the diverse N termini of EFO1 and EFO2 may contribute to their functional nonredundancy.
EFO1 and EFO2 Were Regulated Differently by the Cell Cycle
Alternatively, the nonredundancy of EFO1 and EFO2 could be due to their distinct windows of function. In yeast, inactivation of Eco1/Ctf7 or Eso1 after S phase did not affect sister-chromatid cohesion (Skibbens et al., 1999; Toth et al., 1999; Tanaka et al., 2000). This result has been generally interpreted as them being cohesion factors involved in establishment. However, it remains possible that these proteins are required for the maintenance of cohesion in S phase but not in G2/M. As for EFO1 and EFO2, it is possible that each of them functions in different phases of the cell cycle to maintain proper cohesion. EFO2, whose level sharply decreases when cells enter G2/mitosis (Figure 7), must function in S phase in the establishment and/or maintenance of chromatid cohesion. As for EFO1, although its expression level is constant throughout the cell cycle, the chromosomal EFO1 is phosphorylated in mitosis (Figure 7). We do not understand the functional significance of this phosphorylation. However, we disfavor the possibility that phosphorylation is required for chromosome binding based on the following two observations. First, the unphosphorylated interphase EFO1 also localized to chromosomes (Figure 7B, 8 h). Second, the N-terminal fragment of EFO1, which did not seem to be phosphorylated, was capable of mediating chromosome binding (Figure 6, A and C). Therefore, it seems that the phosphorylation of mitotic EFO1 is the result of its chromosomal localization. We speculate that the phosphorylation may regulate the activity of EFO1. For example, chromosomal EFO1 may be up-regulated in mitosis to fill in the vacancy left by EFO2 degradation. Studies of a phosphorylation resistant mutant EFO1 should be informative in regarding this regulation.
The significance of EFO2 degradation is unclear. Its chromosome localization is also abolished in mitotic cells. One speculative scenario is that the presence of EFO2 activity after S phase may destabilize the cohesin complex, a step that, on the other hand, might be necessary during the establishment process. This notion was first proposed by Tanaka at el to explain the interaction between Eso1 and Pds5 in fission yeast (Tanaka et al., 2001). Consequently, the removal of EFO2 after S phase may therefore be necessary for the stable maintenance of sister-chromatid cohesion after the establishment. Alternatively, EFO2 might positively regulate the stability of cohesion that becomes unnecessary or even undesired for the other functions of the cohesion complex not related to chromatid cohesion. And last, the degradation of EFO2 bears no significance. To probe into these possibilities, we overexpressed untagged EFO2 in HeLa cells and were able to detect mitotic EFO2 in synchronized cultures. No defects could be detected in these cells (our unpublished data). Because the pathway involving EFO2 is not clear at all, it is possible that other factors upstream or downstream of EFO2 also are regulated. For example, EFO2 was unable to associate with chromosomes when overexpressed in mitotic cells. Therefore, further experiments are needed to elucidate the mechanism of EFO2 and significance of its G2/mitotic down-regulation.
Acknowledgments
We thank Yu Hong for excellent technical support. H. Z. is the Kenneth G. and Elaine A. Langone Scholar supported by the Damon Runyon Cancer Foundation (DRS-#35-03). This work also was supported by a Research Scholar Grant from the American Cancer Society (RSG-04-171-01-CCG) and a research grant from the Welch Foundation (I-1594) (to H. Z.).
References
- Bellows, A. M., Kenna, M. A., Cassimeris, L., and Skibbens, R. V. (2003). Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res. 31, 6334-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands, A., and Skibbens, R. V. (2005). Ctf7p/Eco1p exhibits acetyltransferase activity–but does it matter? Curr. Biol. 15, R50-R51. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Shirayama, M., Shevchenko, A., Tanaka, T., Toth, A., and Nasmyth, K. (2000). Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243-254. [DOI] [PubMed] [Google Scholar]
- Edwards, S., Li, C. M., Levy, D. L., Brown, J., Snow, P. M., and Campbell, J. L. (2003). Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol. Cell. Biol. 23, 2733-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., Yu, H., and Kirschner, M. W. (1998). Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2, 163-171. [DOI] [PubMed] [Google Scholar]
- Furuya, K., Takahashi, K., and Yanagida, M. (1998). Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 12, 3408-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, P. J., and Hirano, T. (2004). Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 14, 1598-1603. [DOI] [PubMed] [Google Scholar]
- Gu, W., and Roeder, R. G. (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595-606. [DOI] [PubMed] [Google Scholar]
- Hartman, T., Stead, K., Koshland, D., and Guacci, V. (2000). Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151, 613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henegariu, O., Heerema, N. A., Lowe Wright, L., Bray-Ward, P., Ward, D. C., and Vance, G. H. (2001). Improvements in cytogenetic slide preparation: controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry 43, 101-109. [PubMed] [Google Scholar]
- Ivanov, D., Schleiffer, A., Eisenhaber, F., Mechtler, K., Haering, C. H., and Nasmyth, K. (2002). Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12, 323-328. [DOI] [PubMed] [Google Scholar]
- Jabs, E. W., Tuck-Muller, C. M., Cusano, R., and Rattner, J. B. (1989). Centromere separation and aneuploidy in human mitotic mutants: Roberts syndrome. Prog. Clin. Biol. Res. 318, 111-118. [PubMed] [Google Scholar]
- Jabs, E. W., Tuck-Muller, C. M., Cusano, R., and Rattner, J. B. (1991). Studies of mitotic and centromeric abnormalities in Roberts syndrome: implications for a defect in the mitotic mechanism. Chromosoma 100, 251-261. [DOI] [PubMed] [Google Scholar]
- Kenna, M. A., and Skibbens, R. V. (2003). Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23, 2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madril, A. C., Johnson, R. E., Washington, M. T., Prakash, L., and Prakash, S. (2001). Fidelity and damage bypass ability of Schizosaccharomyces pombe Eso1 protein, comprised of DNA polymerase eta and sister chromatid cohesion protein Ctf7. J. Biol. Chem. 276, 42857-42862. [DOI] [PubMed] [Google Scholar]
- Mendez, J., and Stillman, B. (2000). Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza, S., Tanaka, T., Hochwagen, A., Eisenhaber, F., and Nasmyth, K. (2000). Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10, 1557-1564. [DOI] [PubMed] [Google Scholar]
- Pear, W. S., Nolan, G. P., Scott, M. L., and Baltimore, D. (1993). Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90, 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, R. A., Korom, M., Aulner, N., Martens, A., and Dorsett, D. (2004). Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24, 3100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R. V., Corson, L. B., Koshland, D., and Hieter, P. (1999). Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann, O., Zou, H., Gerber, S. A., Gygi, S. P., and Kirschner, M. W. (2001). Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715-726. [DOI] [PubMed] [Google Scholar]
- Sumara, I., Vorlaufer, E., Gieffers, C., Peters, B. H., and Peters, J. M. (2000). Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151, 749-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara, I., Vorlaufer, E., Stukenberg, P. T., Kelm, O., Redemann, N., Nigg, E. A., and Peters, J. M. (2002). The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9, 515-525. [DOI] [PubMed] [Google Scholar]
- Takahashi, T. S., Yiu, P., Chou, M. F., Gygi, S., and Walter, J. C. (2004). Recruitment of Xenopus Scc2 and cohesin to chromatin requires the prereplication complex. Nat. Cell Biol. 6, 991-996. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Hao, Z., Kai, M., and Okayama, H. (2001). Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20, 5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Yonekawa, T., Kawasaki, Y., Kai, M., Furuya, K., Iwasaki, M., Murakami, H., Yanagida, M., and Okayama, H. (2000). Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20, 3459-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, E. T., Wang, T. J., Lisgo, S., Bamshad, M. J., and Strachan, T. (2004). NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36, 636-641. [DOI] [PubMed] [Google Scholar]
- Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., and Nasmyth, K. (1999). Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega, H., et al. (2005). Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 37, 468-470. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I. C., Hauf, S., Meinke, A., and Peters, J. M. (2000). Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399-410. [DOI] [PubMed] [Google Scholar]
- Williams, B. C., Garrett-Engele, C. M., Li, Z., Williams, E. V., Rosenman, E. D., and Goldberg, M. L. (2003). Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 13, 2025-2036. [DOI] [PubMed] [Google Scholar]
- Zou, H., McGarry, T. J., Bernal, T., and Kirschner, M. W. (1999). Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418-422. [DOI] [PubMed] [Google Scholar]