Identification of a Receptor-Binding Domain of Bordetella Dermonecrotic Toxin (original) (raw)
Abstract
Bordetella dermonecrotic toxin (DNT) stimulates the assembly of actin stress fibers and focal adhesions by deamidating or polyaminating Gln63 of the small GTPase Rho. DNT is an A-B toxin which is composed of an N-terminal receptor-binding (B) domain and a C-terminal enzymatically active (A) domain. In this study, to analyze the functional and structural organization of DNT, we prepared 10 clones of hybridoma producing anti-DNT monoclonal antibodies. One of these antibodies, 2B3, neutralized the effects of DNT on target cells when mixed with the toxin. When microinjected into cells, however, 2B3 did not inhibit the intoxication by DNT. Western blot analysis revealed that 2B3 recognized the N-terminal region of DNT. To delineate the DNT-binding domain, we examined a series of truncated DNT mutants for the ability to competitively inhibit the intoxication of cells by the full-length DNT and found that a fragment consisting of the N-terminal 54 amino acids (DNT1-54) was the smallest inhibitory fragment. The radioiodinated DNT1-54 actually bound to target cells, which was inhibited by 2B3. These results suggest that the N-terminal 54 amino acids of DNT are responsible for the binding to target cells. DNT1-54 bound to none of the DNT-resistant cells, implying the presence of a cell surface receptor specific to DNT-sensitive cells.
Dermonecrotic toxin (DNT), commonly produced by Bordetella pertussis, Bordetella bronchiseptica, and Bordetella parapertussis, exerts lethal, dermonecrotic, and splenoatrophic activities in a variety of experimental animals (2, 3, 7, 8, 12, 19). B. bronchiseptica DNT is considered to be responsible for turbinate atrophy in swine atrophic rhinitis (4, 6, 9). The turbinate atrophy caused by DNT likely results from a deficiency of osteoblastic differentiation in bone tissues (9, 11). DNT also alters cell morphology, stimulating the anomalous reorganization of actin stress fibers and focal adhesions, which is elaborately regulated by the small GTPase Rho (5, 10). The small GTPases function as a molecular switch regulating various cell functions besides the reorganization of actin cytoskeletons by changing between the GDP-bound inactive and the GTP-bound active forms. The GDP-bound GTPases in resting cells exchange GDP for GTP in response to various stimulations, transduce the signals downstream by interacting with effector proteins, and thereafter revert to the GDP-bound inactive form by hydrolyzing the bound GTP. Our research group recently demonstrated that DNT was a transglutaminase catalyzing deamidation or polyamination at Gln63 of Rho and the corresponding Gln residues of the other members of the Rho family, Rac and Cdc42 (5, 18). The deamidation and polyamination result in a reduction of the GTP hydrolyzing activity. In addition, the polyaminated Rho comes to interact with a downstream effector, ROCK, in a GTP-independent manner (17). Thus, these modifications render the intracellular GTPases constitutively active, which probably mediates various effects of DNT on target cells.
DNT is a single-chain polypeptide which consists of 1,464 amino acids with a calculated molecular mass of 160,602 (13). Previously, we localized the catalytic domain of DNT to the C-terminal region from Ile1176 to the C-terminal end. We also found that the N-terminal fragment spanning Met1 to Pro531 of DNT competitively blocked the intoxication of cells by the full-length DNT (13), implying that this fragment retains the receptor-binding or internalizing property. In the present study, we attempted to define the N-terminal receptor-binding region of DNT by using a series of toxin mutants with various lengths and a monoclonal antibody (MAb) that neutralizes the toxin. The results presented here indicate that DNT binds to the cells through the N-terminal region consisting of 54 amino acids, in which the MAb recognized the region including Arg44.
MATERIALS AND METHODS
Materials.
DNT was purified from B. bronchiseptica S798 as described previously (8). The numbering of the amino acids of DNT was based on the sequence available from the DDBJ/EMBL/GenBank databases under accession no. AB020025. C3 exoenzyme was provided by S. Kozaki, University of Osaka Prefecture, Osaka, Japan. MC3T3-E1 cells were cultured at 37°C in α-minimum essential medium (α-MEM; Gibco Laboratories, Grand Island, N.Y.) supplemented with 10% fetal calf serum under 5% CO2 in air. The cells were subcultured every 3 days at a dilution of 1:10. Anti-DNT MAbs were prepared as described previously (15) with a slight modification. X63-Ag8-6.5.3 myeloma cells were used for the production of the anti-DNT MAbs.
Neutralization assay.
MC3T3-E1 cells were plated in wells of 24-well plates or in 35-mm culture dishes at an initial density of 1,300 cells per cm2. The cells were grown for 24 h, washed three times with serum-free α-MEM, and incubated in the same medium for an additional 24 h. DNT was added to the culture at given concentrations after preincubation with or without MAb in serum-free α-MEM at 37°C for 30 min. After incubation for 12 h, the cells were examined for the formation of actin stress fibers and the modification of intracellular Rho by a method reported previously (10). The DNT-induced polynucleation of the cells was assessed as described elsewhere (13).
Construction of expression vectors for DNT mutants.
The expression vectors for DNT and its mutants were constructed as follows. pBSDNT, pUCDNT3′, pUCSTOP, pETDNTwt, pBSSTOP, pGEXDNT1-531, pETDNT1-531-glutathione _S_-transferase (GST), and pGEXGST-DNT523-1464 were constructed as previously described (13).
(i) Series of 3′-end truncation and internal deletion mutants of DNT.
The _Eco_RI-_Apa_LI (blunted), _Eco_RI-_Eco_RV, or _Eco_RI-_Hin_cII fragment of pUCDNT3′ was inserted into the _Sma_I-_Eco_RI site of pUCSTOP. The fragments obtained by treatment of these plasmids with _Eco_RI and _Hin_dIII were separately introduced into the _Eco_RI-_Hin_dIII site of pETDNTwt. The resultant plasmids were designated psApaLI encoding DNT Met1-Cys1305, psRV encoding DNT Met1-Asp1101, and psHinc encoding DNT Met1-Val889. Expression vectors for mutants of DNT with internal deletions, dRV (_Eco_RV), dSP (_Sma_I-_Pvu_II), dSph (_Sph_I), dEM (_Eco_RI-_Mfe_I), and dSX (_Sal_I-_Xho_I) were constructed by treatment of pETDNTwt with the restriction enzymes shown in parentheses and recircularization.
(ii) pBSGST.
The GST gene was amplified by PCR with the primers 5′-AATATGCGGCCGCTCATGTCCCCTATACTAGG-3′ (the underline indicates an _Not_I site) and 5′-GGCAGATCGTCAGTCAGTCACG-3′, with pGEX4T3 (Amersham Pharmacia Biotech) as a template. The amplified DNA was digested with _Not_I and _Eco_RI and inserted into the _Not_I-_Eco_RI site of pBSSTOP. The resultant plasmid was designated pBSGST and used for the preparation of DNT mutant genes with C terminally-tagged GST.
(iii) pETDNT1-344-GST.
pETDNTwt was digested with _Stu_I, ligated with a phosphorylated 10-mer _Not_I linker, and digested with _Nco_I and _Not_I. The resultant DNA fragment encoding DNT from Met1-Glu344 and the _Not_I-_Hin_dIII fragment of pBSGST were ligated with pET21d (Novagen, Inc., Madison, Wis.) treated with _Nco_I and _Hin_dIII.
(iv) pETDNT1-244-GST.
The _Not_I site of pBSGST was replaced with an _Nde_I site by treatment with _Not_I and T4 DNA polymerase and ligation with a phosphorylated 12-mer _Nde_I linker. The GST gene was excised by treatment with _Nde_I and _Hin_dIII and inserted into the _Nde_I-_Hin_dIII site of pETDNT1-531-GST.
(v) pETDNT1-94-GST.
pETDNT1-244-GST was digested with _Xho_I, and an _Nde_I site was added by ligation with a 10-mer _Nde_I linker. A DNA fragment encoding DNT Met1-Leu94 was excised from the plasmid by digestion with _Nco_I and _Nde_I and inserted into the _Nco_I-_Nde_I site of pETDNT1-244-GST.
(vi) pETDNT1-54-GST.
A DNA fragment encoding DNT Met1-Glu54 was amplified by PCR with the primers 5′-GGGCCATGGATAAAGATGAATCGGCATTGC-3′, where the underline indicates an _Nco_I site, and 5′-GGCATATGTTTCGCCAAACAGCGCGAATTCGGCCTTC-3′, where the underline indicates an _Nde_I site and with pETDNTwt as a template. The DNA fragment was digested with _Nco_I and _Nde_I and inserted into the _Nco_I-_Nde_I site of pETDNT1-243-GST.
(vii) pETDNT47-244-GST.
The _Eco_RI site of pETDNTwt was replaced with _Nco_I by digestion with _Eco_RI and ligation with a 12-mer _Nco_I linker. A DNA fragment encoding DNT Ala47-Gly244 was excised by digestion with _Nco_I and _Nde_I and ligated with pETDNT1-244-GST digested with the same enzymes.
(viii) pGEXDNT1-54-hexahistidine (His6).
A gene was amplified by PCR with the primers 5′-GGATCCGATAAAGATGAATCGGCATTGC-3′, where the underline indicates a _Bam_HI site, and 5′-CCGAGCTCTCGCCAAACAGCGCGAATTCGG-3′, where the underline indicates an _Sac_I site, and pETDNTwt as a template DNA. The amplified DNA was digested with _Bam_HI and _Sac_I and ligated with pGST-CAN4 (14) treated with the same enzymes.
(viii) pQEDNTwt.
The _Bam_HI-_Nde_I fragment of pGEXDNT1-531 was ligated with the _Nde_I-_Hin_dIII fragment of pETDNTwt and pQE40 (Qiagen) digested with _Bam_HI and _Hin_dIII.
(ix) pQEDNT(R44G), pQEDNT(R44S), and pQEDNT(R44K).
pQEDNT(R44G), pQEDNT(R44S), and pQEDNT(R44K) were prepared from pQEDNTwt by site-directed mutagenesis with a QuickChange kit (Stratagene, La Jolla, Calif.).
Expression and purification of recombinant proteins.
The expression vectors for DNT mutants were introduced into Escherichia coli BL21(DE3). The bacteria were cultivated in Luria-Bertani broth containing 50 μg of ampicillin/ml and induced to produce the proteins by using 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The GST fusion proteins were purified with glutathione-Sepharose 4B (Amersham Pharmacia Biotech). The His6-tagged proteins were purified with His-Bind Resin (Novagen).
Microinjection of anti-DNT antibodies.
MC3T3-E1 cells were seeded at an initial density of 520 cells per cm2 on coverslips. The cells were grown for 24 h, washed three times with serum-free Dulbecco modified Eagle medium (Gibco Laboratories), and incubated in the same medium for 48 h at 37°C. The cells were injected with 50 μg of anti-DNT polyclonal antibody or 1 mg of anti-DNT MAb 2B3/ml. Rabbit immunoglobulin G (IgG; ICN Pharmaceuticals, Inc., Aurora, Ohio) was also microinjected, together with the samples, as an indicator for the injected cells. Microinjection was performed with an Eppendorf micromanipulator (Eppendorf, Humburg, Germany). The cells were incubated for 1 h at 37°C, washed three times with prewarmed Dulbecco modified Eagle medium, and further incubated for 2 h. After the incubation, 5 ng of DNT/ml was added to the culture and incubated for 20 h. The cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, washed three times with PBS, and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. The cells were then incubated with 2.5 U of Alexa 586-phalloidin (Molecular Probes, Inc., Eugene, Oreg.)/ml and 4 μg of Alexa 488-goat anti-rabbit IgG (Molecular Probes)/ml at room temperature for 1 h. The cells were washed three times with PBS and mounted in PermaFlour aqueous mounting medium (Shandon/Lipshaw Co., Pittsburgh, Pa.).
Binding assay.
DNT1-54 was iodinated with Na125I by the chloramine-T method. The specific activity of 125I-DNT1-54 was 0.42 to 0.72 mCi/mg of protein. MC3T3-E1 cells suspended in PBS were incubated with 125I-DNT1-54. The 125I-DNT1-54 bound to the cells was separated by filtration through a Millititer-GV plate (Millipore). The filters were washed five times with 200 μl of chilled PBS. The radioactivity retained on the filter was determined by using a gamma counter. The specific binding amount was evaluated from the difference between the means of the radioactive counts in the absence and presence of a 100-fold excess of unlabeled DNT1-54.
Other methods.
Protein concentrations were determined by the method of Lowry et al. (16) or Bradford (1). Rabbit anti-DNT serum was obtained as reported previously (8). Anti-DNT IgG or anti-DNT MAbs were purified with an Affi-Gel Protein A MAPS II kit (Bio-Rad, Richmond, Calif.). Expression of the DNT mutants was confirmed by Western blot analysis with the anti-DNT IgG. In the analysis, immunoreactive products were detected with a substrate mixture of 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium (Promega, Madison, Wis.) or an enhanced chemiluminescence system (Amersham Pharmacia Biotech). The [32P]ADP-ribosylation of Rho was achieved with C3 exoenzyme as described previously (10) and detected by autoradiography with a Fuji BAS 1500 image analyzer (Fuji Film Co., Tokyo, Japan).
RESULTS
MAb neutralizing DNT.
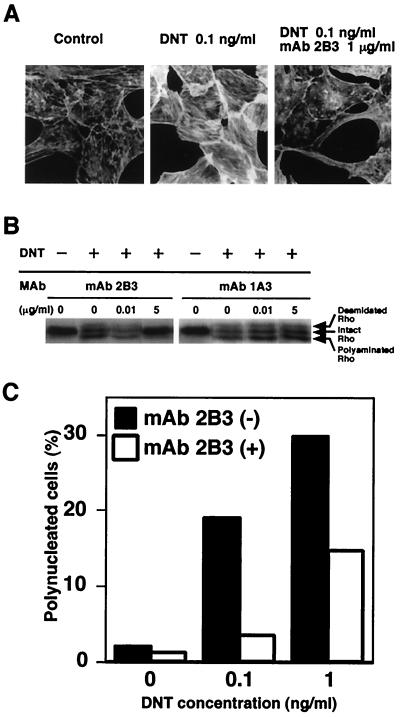
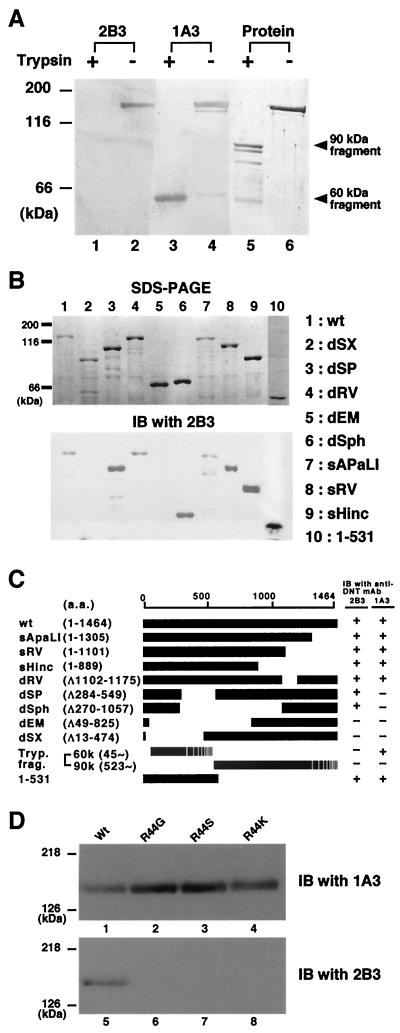
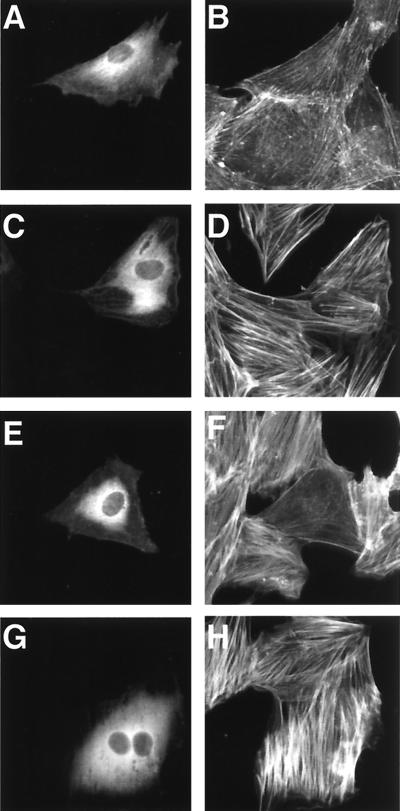
We produced 10 clones of hybridoma secreting MAbs against DNT to analyze functional domains. One of these MAbs, 2B3, was found to inhibit the formation of stress fibers caused by DNT (Fig. 1A). 2B3 also inhibited the mobility shifts in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of intracellular Rho that results from the polyamination and deamidation by the toxin, whereas another MAb, 1A3, did not (Fig. 1B). Moreover, 2B3 protected the cells from the polynucleation that results from the inhibition of cytokinesis by DNT (11) (Fig. 1C). We carried out Western blot analysis with 2B3 against tryptic fragments of DNT with apparent molecular sizes of 60 and 90 kDa (13). 2B3 bound to neither one (Fig. 2A), which was not surprising because these two fragments do not cover the overall molecule of DNT (13). It was reported that the N termini of the 60- and 90-kDa fragments were Glu45 and Gly523, respectively. Next, to localize an epitope recognized by 2B3, we subjected a series of deletion mutants of DNT to Western blot analysis (Fig. 2B and C). 2B3 bound to dSP, dSph, and DNT1-531, but not to dEM, which lacks the region from amino acids 49 to 825. These results suggest that 2B3 recognizes an epitope including amino acids 45 to 49. We previously demonstrated that the C-terminal side of Arg44 was susceptible to protease digestion (13; T. Matsuzawa et al., unpublished data), suggesting that this residue may reside on the surface of the toxin molecule and participate in a loop conformation, which readily serves as an epitope for antibody. Therefore, we prepared DNT mutants in which Arg44 was replaced with Gly, Ser, or Lys and examined whether 2B3 binds to them. Western blot analysis revealed that 2B3 recognized none of the mutants, whereas 1A3 bound to all of them (Fig. 2D). These results suggest that 2B3 recognizes the putative loop region containing amino acids 44 to 49. 1A3 was considered to recognize amino acids 284 to 474, since it bound to the 60-kDa fragment but not dSP and dSX. Next, we tested whether 2B3 microinjected into cells neutralizes the action of DNT added extracellulary (Fig. 3). The microinjected 2B3 did not protect the cells from intoxication by DNT, whereas anti-DNT polyclonal antibody did (Fig. 3E to H), suggesting that 2B3 neutralizes DNT by inhibiting its binding or translocation.
FIG. 1.
Neutralization of DNT by 2B3. (A) DNT (1 ng/ml) was incubated with (10 μg/ml) or without 2B3 at 37°C for 30 min and added to cultures of MC3T3-E1 cells at a final concentration of 0.1 ng/ml. The cells were incubated for 36 h and stained for actin cytoskeletons. (B) DNT (10 ng/ml) was treated with or without 0.1 and 50 μg of 2B3 or 1A3/ml, and the mixtures were diluted 10-fold and added to cultures of MC3T3-E1 cells. After incubation for 36 h, the cells were examined for modifications of Rho. Upper and lower mobility shifts of Rho represent deamidation and polyamination caused by DNT, respectively. (C) DNT at 1 or 10 ng/ml was incubated with or without 10 μg of 2B3/ml at 37°C for 30 min. A 10-fold dilution of the mixture was applied to MC3T3-E1 cells. After incubation for 36 h, the mono- and polynucleated cells were enumerated, and the percentage of polynucleated cells in at least 400 cells was calculated as described elsewhere (13). Three independent experiments were carried out, and representative data are shown.
FIG. 2.
Localization of an epitope recognized by 2B3. (A) Trypsin-treated (lanes 1, 3, and 5) or nontreated (lanes 2, 4, and 6) DNT was sujected to Western blot analysis with 2B3 (lanes 1 and 2) or 1A3 (lanes 3 and 4). Total proteins electrotransferred on the membrane were stained with Coomassie brilliant blue R-250 (lanes 5 and 6).(B) The lysates of E. coli expressing DNT mutants were subjected to SDS-PAGE (upper panel) and Western blot analysis with 2B3 (lower panel). (C) A schematic representation of wild-type (wt) and deletion mutants of DNT. The results of the Western blot analyses are summarized on the right. The numbers in parentheses indicate the positions of amino acids covering the peptides or deletions (Δ). The C-terminal ends of the tryptic 60- and 90-kDa fragments were not identified. (D) His-DNTwt (lanes 1 and 5), His-DNT R44G (lanes 2 and 6), His-DNT R44S (lanes 3 and 7), and His-DNT R44K (lanes 4 and 8) were subjected to SDS-PAGE and immunoblotted (IB) with 1A3 (lanes 1 to 4) or 2B3 (lanes 5 to 8).
FIG. 3.
The DNT-induced formation of actin stress fibers in MC3T3-E1 cells microinjected with antibodies. The cells were microinjected with buffer alone (A to D), anti-DNT polyclonal antibody (E and F), or 2B3 (G and H). After incubation with (C to H) or without (A and B) 5 ng of DNT/ml for 20 h, the cells were stained for actin cytoskeletons (B, D, F, and H). Microinjected cells were distinguished by staining with Alexa 488-antibody against rabbit IgG microinjected along with the test samples (A, C, E, and G).
Definition of a receptor-binding domain of DNT.
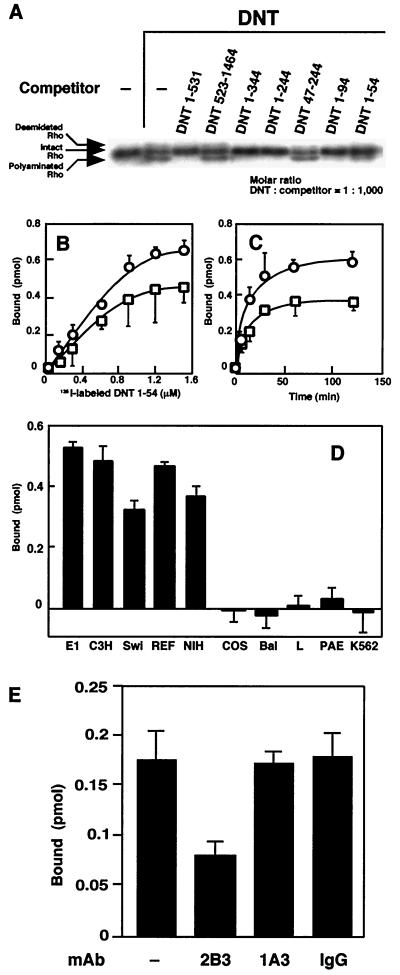
Previously, we had localized the receptor-binding domain of DNT to the N terminus composed of 531 amino acids (13). In addition, the results mentioned above raise the possibility that DNT binds to target cells via the more distal N terminus, including the 2B3-recognizing region. To test this, we attempted to further delimitate the receptor-binding region. DNT was applied to MC3T3-E1 cells, along with one of the deletion mutants of the toxin, and examined for modifications of intracellular Rho. DNT1-531, DNT1-344, DNT1-244, DNT1-94, and DNT1-54 inhibited the DNT-induced modification of Rho, whereas the N-terminally truncated mutants, DNT523-1464 and DNT47-244, did not (Fig. 4A). We speculated that these mutants inhibited the DNT action by competing with the full-length toxin for a cell surface receptor and then carried out a direct binding assay with DNT1-54, the shortest fragment inhibiting the DNT action. It was found that DNT1-54 actually bound to MC3T3-E1 cells (Fig. 4B to D). The binding of 125I-labeled DNT1-54 was saturable after 60 min of incubation and slightly attenuated at 4°C (Fig. 4B and C). Similar results were obtained with the radiolabeled DNT1-94 (data not shown). 125I-labeled DNT1-54 bound to the DNT-susceptible MC3T3-E1, C3H10T1/2, Swiss 3T3, rat embryo fibroblasts, and NIH 3T3 cells but not to the resistant COS7, BALB/3T3, L929, PAE, and K562 cells (Fig. 4D), indicating that the sensitivity of cells to DNT is attributable to the existence of an unknown receptor. The binding of 125I-labeled DNT1-54 was inhibited by preincubation with 2B3 but not 1A3 or normal mouse IgG (Fig. 4E).
FIG. 4.
Definition of the binding domain of DNT. (A) Competitive inhibition of the Rho modifications caused by DNT with various deletion mutants. The mutant proteins were derived from pETDNT1-531-GST, pGEXGST-DNT523-1464, pETDNT1-344-GST, pETDNT1-244-GST, pETDNT47-244-GST, pETDNT1-94-GST, and pETDNT1-54-GST. MC3T3-E1 cells were incubated with DNT (5 ng/ml, ca. 31 pM) and each deletion mutant (31 nM) at 37°C for 8 h and examined for modifications of intracellular Rho. Numbers in the mutant namesindicate positions of the N-terminal and the C-terminal amino acids. (B) Dose-dependent binding of 125I-labeled DNT1-54 to the cells at 37°C (○) or 4°C (□). (C) Time course of the binding of 125I-labeled DNT1-54 to MC3T3-E1 cells at 37°C (○) or 4°C (□). (D) Binding of 125I-labeled DNT1-54 to MC3T3-E1 (E1), C3H10T1/2 (C3H), Swiss 3T3 (Swi), rat embryo fibroblasts (REF), NIH 3T3 (NIH), COS7 (COS), BALB 3T3 (Bal), L929 (L), PAE, and K562. 125I-labeled DNT1-54 (1.2 μM) was incubated with each cell line at 37°C for 1 h. (E) Inhibition of the binding of DNT1-54 to the cells by 2B3. 125I-DNT1-54 at 0.21 μM was preincubated with or without 850 μg of 2B3, 1A3, or normal IgG/ml at 37°C for 30 min and then incubated with MC3T3-E1 cells at a final concentration of 0.15 μM for 1 h. (B to E) Three independent experiments were performed, and representative data (means ± the standard deviations of three samples) are shown.
DISCUSSION
Bacterial protein toxins that enzymatically modify cytosolic substances of eukaryotic cells consist of functionally distinct domains. The designation A-B toxin refers to toxins composed of an A domain conducting enzymatic action and a B domain binding to a surface receptor on target cells. In toxins composed of a single-chain polypeptide, these domains reside in the N-terminal or C-terminal region but not the intermediate region of the molecule. Previously, we reported that the B domain of DNT to the N terminus of ca. 500 amino acids and the A domain was localized to the C terminus of ca. 300 amino acids. In the present study, we further localized the B domain to the N-terminal 54 amino acids.
To localize the functional domains of DNT, we first prepared 10 different MAbs against the toxin. Western blot analysis revealed that all of the MAbs recognized the N-terminal 600-amino-acid region of the toxin (data not shown), implying that the N-terminal region covers the surface of the toxin molecule and therefore presents epitopes. It is conceivable that the N-terminal region on the molecular surface is involved in the binding to the cell receptor. The neutralizing MAb, 2B3, was found to recognize the molecule in the vicinity of Arg44. This region seems to form a loop structure because it was also susceptible to cleavage by trypsin or furin. The results presented here indicate that 2B3 inhibits the binding of DNT to target cells by binding to this putative loop region. The DNT mutants in which Arg44 was substituted with different amino acids so that they would become resistant to furin were inactive on cells (Matsuzawa et al., unpublished). This implies that the loop structure of DNT1-54 plays an important role in the DNT action in addition to the binding to the receptor.
DNT1-54 seems to precisely recognize an unknown cell surface receptor since it strictly bound to the toxin-sensitive cells and not to resistant cells. Direct binding assays with the full-length DNT had long been unsuccessful because nonspecific binding occurred irrespective of cell type (unpublished data). This had constituted an obstacle to elucidating the nature of the binding of DNT to cells. However, we could reduce the nonspecific binding by using DNT1-54 or DNT1-94 as a probe and succeeded in detecting the specific binding, which may provide a way to identify the cell surface receptor. The receptor for DNT is probably an uncommon substance because only a few lines of cells have turned out to be sensitive to DNT. Identification of the DNT receptor is an important issue. In addition, it might be of interest to examine how a binding domain as short as 54 amino acids recognizes the receptor.
Acknowledgments
We thank S. Kozaki for the generous gift of C3 exoenzyme and K. Oka and M. Minami for critical reading of the manuscript.
This work was supported in part by Grant-in Aid for Scientific Research 11670264 from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72**:**248-254. [DOI] [PubMed] [Google Scholar]
- 2.Bruckner, I. E., and D. G. Evans. 1939. The toxin of B. parapertussis and the relationship of this organism to H. pertussis and B. bronchiseptica. J. Pathol. Bacteriol. 48**:**67-78. [Google Scholar]
- 3.Evans, D. G. 1940. The production of pertussis antitoxin in rabbits and the neutralization of pertussis, parapertussis, and bronchiseptica toxins. J. Pathol. Bacteriol. 51**:**49-58. [Google Scholar]
- 4.Hanada, M., K. Shimoda, S. Tomita, Y. Nakase, and Y. Nishiyama. 1979. Production of lesions similar to naturally occurring swine atrophic rhinitis by cell-free sonicated extract of Bordetella bronchiseptica. Jpn. J. Vet. Sci. 41**:**1-8. [DOI] [PubMed] [Google Scholar]
- 5.Horiguchi, Y., N. Inoue, M. Masuda, T. Kashimoto, J. Katahira, N. Sugimoto, and M. Matsuda. 1997. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc. Natl. Acad. Sci. USA 94**:**11623-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horiguchi, Y., T. Nakai, and K. Kume. 1991. Effects of Bordetella bronchiseptica dermonecrotic toxin on the structure and function of osteoblastic clone MC3T3-E1 cells Infect. Immun. 59**:**1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiguchi, Y., T. Nakai, and K. Kume. 1989. Purification and characterization of Bordetella bronchiseptica dermonecrotic toxin. Microb. Pathog. 6**:**361-368. [DOI] [PubMed] [Google Scholar]
- 8.Horiguchi, Y., T. Nakai, and K. Kume. 1990. Simplified procedure for purification of Bordetella bronchiseptica dermonecrotic toxin. FEMS Microbiol. Lett. 66**:**39-43. [DOI] [PubMed] [Google Scholar]
- 9.Horiguchi, Y., T. Okada, N. Sugimoto, Y. Morikawa, J. Katahira, and M. Matsuda. 1995. Effects of Bordetella bronchiseptica dermonecrotizing toxin on bone formation in calvaria of neonatal rats. FEMS Immunol. Med. Microbiol. 12**:**29-32. [DOI] [PubMed] [Google Scholar]
- 10.Horiguchi, Y., T. Senda, N. Sugimoto, J. Katahira, and M. Matsuda. 1995. Bordetella bronchiseptica dermonecrotizing toxin stimulates assembly of actin stress fibers and focal adhesions by modifying the small GTP-binding protein rho. J. Cell Sci. 108**:**3243-3251. [DOI] [PubMed] [Google Scholar]
- 11.Horiguchi, Y., N. Sugimoto, and M. Matsuda. 1993. Stimulation of DNA synthesis in osteoblast-like MC3T3-E1 cells by Bordetella bronchiseptica dermonecrotic toxin. Infect. Immun. 61**:**3611-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iida, T., and T. Okonogi. 1971. Lienotoxicity of Bordetella pertussis in mice. J. Med. Microbiol. 4**:**51-60. [DOI] [PubMed] [Google Scholar]
- 13.Kashimoto, T., J. Katahira, W. R. Cornejo, M. Masuda, A. Fukuoh, T. Matsuzawa, T. Ohnishi, and Y. Horiguchi. 1999. Identification of functional domains of Bordetella dermonecrotizing toxin. Infect. Immun. 67**:**3727-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katahira, J., K. Strasser, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18**:**2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256**:**495.. [DOI] [PubMed] [Google Scholar]
- 16.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193**:**265-275. [PubMed] [Google Scholar]
- 17.Masuda, M., L. Betancourt, T. Matsuzawa, T. Kashimoto, T. Takao, Y. Shimonishi, and Y. Horiguchi. 2000. Activation of Rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 19**:**521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda, M., M. Minami, H. Shime, T. Matsuzawa, and Y. Horiguchi. 2002. In vivo modification of small GTPase Rac and Cdc42 by Bordetella dermonecrotic toxin. Infect. Immun. 70**:**998-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardlaw, A. C., and R. Parton. 1983. Bordetella pertussis toxins, p. 327-371. In F. Dorner and J. Drews (ed.), Phamacology of bacterial toxins. Pergamon Press, Ltd., Oxford, England.