Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses (original) (raw)
Abstract
Double-stranded RNA (dsRNA) longer than 30 bp is a key activator of the innate immune response against viral infections. It is widely assumed that the generation of dsRNA during genome replication is a trait shared by all viruses. However, to our knowledge, no study exists in which the production of dsRNA by different viruses is systematically investigated. Here, we investigated the presence and localization of dsRNA in cells infected with a range of viruses, employing a dsRNA-specific antibody for immunofluorescence analysis. Our data revealed that, as predicted, significant amounts of dsRNA can be detected for viruses with a genome consisting of positive-strand RNA, dsRNA, or DNA. Surprisingly, however, no dsRNA signals were detected for negative-strand RNA viruses. Thus, dsRNA is indeed a general feature of most virus groups, but negative-strand RNA viruses appear to be an exception to that rule.
Double-stranded RNA (dsRNA) of more than 30-bp length is a key activator of the innate immune response against viral infections (2, 8, 25, 50, 61, 66). The interaction of the host cell with dsRNA occurs by several mechanisms. Specific receptors activate the synthesis of antiviral type I interferons (IFN-α and IFN-β) and antiviral proteins (20), and dsRNA-activated enzymes can directly inhibit viral replication (50, 55, 70). The RNA helicases RIG-I (72) and MDA-5 (4) as well as the protein kinase PKR (70) bind to intracellular dsRNA and lead to the activation of the transcription factors interferon regulatory factor 3 (IRF-3) (51, 67, 73) and NF-κB (31, 74), respectively, which are important for IFN synthesis. Toll-like receptor 3 (TLR3) binds to extracellular and endosomal dsRNA and also activates IFN transcription via IRF-3 and NF-κB (3, 46, 61). In addition, IFN effector enzymes, such as PKR (70), 2′-5′-oligoadenylate synthetase (2-5-OAS) (55) and the RNA-specific adenosine deaminase (50), need to be activated by dsRNA and inhibit viral replication at various levels. Viruses, in turn, escape this immune response either by expressing dsRNA-binding proteins or by other strategies to inhibit the dsRNA-induced pathways (18, 25, 68).
Clearly, both host and viral pathogens apply a range of measures to deal with dsRNA, indicating that this molecule represents a danger signal of central importance for the innate immune response. It is widely assumed that dsRNA is generated by viral RNA polymerases either as an intermediate in genome replication (RNA viruses) or as an erroneous product due to converging bidirectional transcription (DNA viruses) (25, 32). However, to our knowledge, this has been directly shown for only a few viruses (33, 57, 69), whereas in most cases, only indirect evidence, such as activation of the dsRNA-dependent enzymes PKR, 2-5-OAS, and RNA-specific adenosine deaminase (28, 50, 70), activation of TLR3 (60), and investigation of the dsRNA content of cellular lysates (25, 32, 40), is available.
In this study, we attempted to find evidence for virally produced dsRNA in situ, using a nondestructive method. By employing a dsRNA-specific antibody for immunofluorescence analysis, we investigated the presence and localization of dsRNA in cells infected with a range of viruses. Indeed, we detected significant amounts of dsRNA for viruses with a positive-strand RNA or a dsRNA genome as well as for DNA viruses. Surprisingly, however, no significant dsRNA signals were detected for negative-strand RNA viruses, suggesting that other viral components are more important in triggering the host's antiviral response.
MATERIALS AND METHODS
Cells and viruses.
Vero cells, HeLa cells, and BHK-1 cells were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The viruses used were adenovirus type 5 (AdV), encephalomyocarditis virus strain FA (EMCV), influenza A virus strain PR8 (FLUAV), herpes simplex virus 1 (HSV), La Crosse virus (LACV), modified vaccinia virus strain Ankara (Vac), reovirus strain Lang (ReoV), and severe acute respiratory syndrome coronavirus strain FFM (SARS-CoV). AdV and Vac were propagated in HeLa and BHK cells, respectively, while the other viruses were propagated in Vero cells.
Enzymes.
RNase III was supplied by Ambion and RNase A by Sigma-Aldrich. Both enzymes were incubated for 2 h at 37°C in their respective reaction buffers provided by the manufacturers.
Poly(I:C) transfection.
For transfection of cells with synthetic dsRNA, 10 μg of poly(I:C) (Sigma) was prepared with 10 μl of Metafectene (Biontex) in 200 μl of serum-free medium according to the manufacturers' instructions. After 15 min of incubation, the dsRNA-liposome mixture was dropped onto cells using the same medium.
Immunofluorescence analysis.
Cells were grown on coverslips to 30 to 50% confluence and transfected or infected as indicated. Cells were fixed with 3% paraformaldehyde and permeabilized with 0.5% Triton X-100 dissolved in phosphate-buffered saline (PBS). For dsRNA immunofluorescence, the mouse monoclonal antibody J2 (Scicons, Hungary) was diluted 1:200 in PBS, and the fluorophore signal was visualized using the tyramide signal amplification (TSA) cyanine 3 system (Perkin plus Elmer). For viral immunofluorescence, cells were incubated with a rabbit polyclonal anti-FLUAV N (1:200) or a rabbit polyclonal rabbit anti-LACV N (1:500). For nuclear counterstain, the rabbit anti-acetyl histone H4 antiserum (Upstate) was used at a dilution of 1:500. After incubation at room temperature for 1 h, the coverslips were washed three times in PBS and then treated with the secondary antibody goat anti-mouse or anti-rabbit at a dilution of 1:200 each. Cells were again washed three times in PBS and mounted using Fluorsave solution (Calbiochem). Stained cell samples were examined using a Leica confocal laser scanning microscope with a 63× NA-1.4 objective. The confocal pinhole was set to 1 Airy unit, and pictures were digitally magnified twofold. The same microscope settings and exposure times were used within each set of experiments.
RESULTS
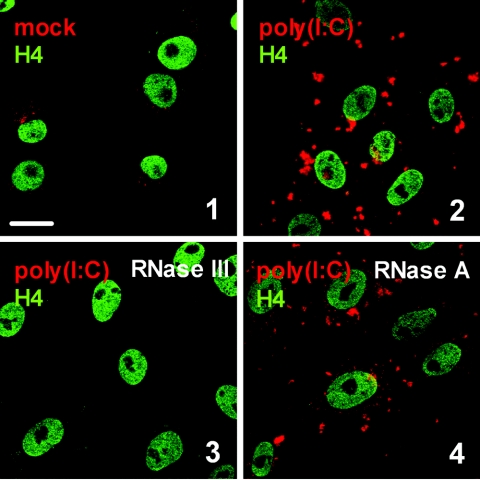
The monoclonal dsRNA-specific mouse antibody J2 specifically recognizes dsRNA of more than 40-bp length (52). It was previously used for an enzyme-linked immunosorbent assay and immunoblot analysis of virus-infected plants (37, 52) but, to our knowledge, not for immunohistochemistry of animal cells. We tested the suitability of J2 to detect transfected poly(I:C), a synthetic dsRNA, in Vero cells in situ. Figure 1 (panel 2) shows that dsRNA-transfected cells indeed give rise to a strong signal, whereas in nontransfected cells, a much weaker immunofluorescence is present (Fig. 1, panel 1). Importantly, the fluorescence signal was sensitive to the dsRNA-specific RNase III (Fig. 1, panel 3) but not to the single-stranded RNA (ssRNA)-specific RNase A (Fig. 1, panel 4) or to DNase I (data not shown), indicating that the target structure detected by J2 was authentic dsRNA. RNase III treatment also reduced the background fluorescence (data not shown), confirming previous findings of endogenous cellular dsRNA (32, 66).
FIG. 1.
The monoclonal antibody J2 specifically recognizes dsRNA. Vero cells were transfected with poly(I:C) as indicated in Materials and Methods. After an incubation period of 6 h, the cells were fixed and analyzed for dsRNA using the mouse monoclonal antibody J2 (red). To visualize the cell nuclei, histone H4 was stained using a specific rabbit antiserum (green). Shown are untransfected cells (1), poly(I:C)-transfected cells (2), and poly(I:C)-transfected cells treated with either 2 U of RNase III (3) or 2 U of RNase A (4). Bar, 20 μm. All pictures were taken with the same magnification.
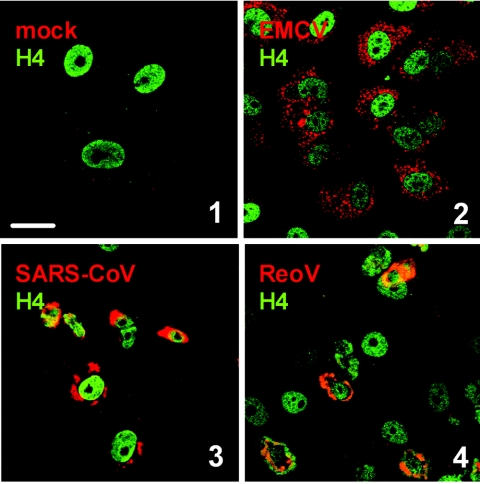
Using the J2 antibody, we analyzed the dsRNA content of cells infected with positive-strand RNA viruses and dsRNA viruses. For members of the togaviruses (57) and the flaviviruses (33, 69), in situ detection of dsRNA has been performed previously. We therefore restricted our analysis to the remaining groups, namely, the picornaviruses represented by EMCV, the coronaviruses represented by SARS-CoV, and ReoV. As shown in Fig. 2, strong dsRNA signals were present after infection with all these viruses. Note that for this and all other studies, time points were chosen in which a peak signal for dsRNA was detected. Taking this together with data from the literature (33, 57, 69), we can conclude that infection with both positive-strand RNA and dsRNA viruses results in the production of significant amounts of dsRNA. This dsRNA is indeed of viral and not of cellular origin, since the dsRNA signal was also detected under treatment with the DNA-dependent RNA polymerase inhibitor actinomycin D (data not shown). Also, similar to the situation with synthetic dsRNA (Fig. 1), the viral dsRNA signal appears to consist of ssRNA hybrids, since RNase A treatment did not alter the immunofluorescence signal (data not shown).
FIG. 2.
dsRNA in cells infected with positive-strand RNA and dsRNA viruses. Vero cells were infected at a multiplicity of infection of 5 with EMCV (2), SARS-CoV (3), or ReoV (4) or left uninfected (1). At 5 h (EMCV), 16 h (SARS-CoV), or 48 h (ReoV) postinfection, cells were fixed and analyzed by immunofluorescence as indicated in the legend to Fig. 1. Bar, 20 μm. All pictures were taken with the same magnification. The differences in size and morphology of the nuclei are most probably caused by initiation of apoptosis or by disturbances in nuclear-cytoplasmic transport, as has been described for several viruses (6, 21, 23, 27).
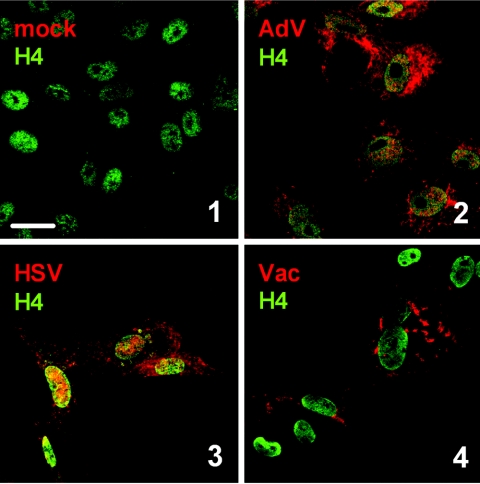
Next, we monitored dsRNA synthesis in cells infected with DNA viruses. As shown in Fig. 3, all viruses tested, namely, AdV, HSV, and Vac, were invariably positive for dsRNA. Thus, although the nature of their genome is different from RNA viruses, DNA viruses apparently also produce dsRNA during their replication cycle.
FIG. 3.
dsRNA in cells infected with DNA viruses. Vero cells were infected at a multiplicity of infection of 5 with AdV (2), HSV (3), or Vac (4) or left uninfected (1). At 7 h (AdV, HSV) or 5 h (Vac) postinfection, cells were fixed and analyzed by immunofluorescence as indicated in the legend to Fig. 1. Bar, 20 μm. All pictures were taken with the same magnification.
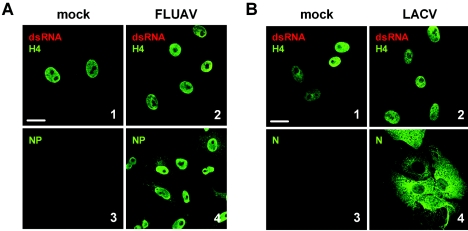
The third large group of viruses comprises the negative-strand RNA viruses. As examples, we chose FLUAV, which replicates in the nucleus, and LACV, which replicates in the cytoplasm as most other RNA viruses do. Much to our surprise, when we investigated dsRNA in infected cells, we detected hardly any signals above background levels (Fig. 4A and B, panels 1 and 2). Occasionally, single cells exhibited a cytoplasmic dsRNA signal (curiously, also for the nucleus-borne FLUAV), but these were always exceptions. Nevertheless, the viruses replicated efficiently, as is demonstrated by the synthesis of viral nucleocapsid proteins (Fig. 4A and B, panels 3 and 4). To further strengthen this last point, we measured infectious particle production and viral RNA levels and did not find significant differences between positive-strand RNA viruses (exemplified by SARS-CoV) and negative-strand RNA viruses (exemplified by LACV) growing in Vero cells (data not shown). As with FLUAV and LACV, we were also unable to detect significant dsRNA signals for Sendai virus and Newcastle disease virus, two members of the Paramyxoviridae family with a strong ability to induce IFN (data not shown). Thus, despite having comparable efficiencies of growth and RNA synthesis, negative-strand RNA viruses appear to produce much less dsRNA than positive-strand RNA viruses and DNA viruses.
FIG. 4.
Negative-strand RNA viruses. Vero cells were infected at a multiplicity of infection of 5 with FLUAV (A, panels 2 and 4) or LACV (B, panels 2 and 4) or left uninfected (A and B, panels 1 and 3). At 5 h postinfection, cells were fixed and analyzed by immunofluorescence either for dsRNA as indicated in the legend to Fig. 1 (A and B, panels 1 and 2) or for viral antigens (A and B, panels 3 and 4). Bar, 20 μm. All pictures were taken with the same magnification.
DISCUSSION
To our knowledge, no study was hitherto performed in which the production of intracellular dsRNA by different viruses was systematically investigated. The formation of dsRNA was thought to be a general feature of all viruses (25, 32), although direct in situ proof for this was provided only for a few viruses, notably ones with a positive-strand RNA genome (33, 57, 69). Using a specific mouse monoclonal antibody, we were able to confirm the production of dsRNA by positive-strand RNA viruses, dsRNA viruses, and DNA viruses.
For negative-strand RNA viruses, however, we were unable to detect dsRNA in infected cells. In line with this finding, we have previously shown that expression of the dsRNA-binding domains of RIG-I or PKR in myeloid cells could abrogate the cytokine response only to the positive-strand RNA virus EMCV but not to the negative-strand RNA virus Sendai virus (45). This again indicates fundamental differences between different virus groups with respect to dsRNA formation. Nevertheless, it is well known that negative-strand RNA viruses can strongly induce IFN synthesis (18, 68), activate PKR or be sensitive to it (7, 56, 58), or express a dsRNA-binding protein, such as FLUAV NS1 (22, 36). However, since as little as one molecule of dsRNA per cell can be effective in triggering an antiviral response (42), it is possible that the amounts of dsRNA produced by negative-sense RNA viruses are below our detection limit. Alternatively, other viral structures may take the role of dsRNA as a danger signal for the host cell. Indeed, it was previously shown that the ribonucleoprotein particles (RNPs) of the negative-strand vesicular stomatitis virus (VSV) and measles virus are capable of triggering IFN induction (62, 63). In addition, ssRNAs of FLUAV and VSV are capable of triggering IFN induction via TLR7 (13, 38), and the FLUAV NS1 protein also binds to viral ssRNA (22). Moreover, PKR can also be activated independently of dsRNA by cellular proteins, such as PACT/RAX (24, 47). Since these proteins are stress activated, it is conceivable that PKR activation in response to negative-strand RNA virus infections occurs mostly via PACT/RAX and not via dsRNA. Also, the importance of dsRNA binding for the anti-IFN activity of FLUAV NS1 is still a matter of debate (19, 30). Indeed, the dsRNA-binding activity is not absolutely required for the inhibition of IFN induction (14), and NS1 confers a host of other dsRNA-independent activities (15, 30). Interestingly, to our knowledge, FLUAV NS1 is the only anti-IFN protein of a negative-strand RNA virus known to directly bind dsRNA. All other members of this taxonomic group rely on inhibiting other downstream parts of the IFN induction signaling chain, such as MDA-5 (4), the IRF-3 kinase TBK-1 (53, 65), IRF-3 (5, 11, 12, 26, 29), the RNA polymerase II complex (9, 10, 17, 34, 64), nuclear export of RNAs (16), and translation (1). On the other hand, however, the dsRNA-binding proteins MDA-5 (which is inhibited by paramyxoviruses) (4) and RIG-I both are important sensor molecules for negative-strand RNA viruses (45, 48, 71). Also, expression of a dsRNA-binding protein can rescue VSV from the antiviral effects of IFN (54). Thus, we cannot exclude the presence of small amounts of dsRNA produced by negative-strand RNA viruses, but we suppose that in the case of this virus group, other signals, such as ssRNA and RNPs, may be equally important for the host as danger signals in triggering an antiviral response. This view is not contradicted by previous reports about biologically active dsRNA derived from FLUAV-infected cells (41), since those studies involved the extraction of RNA from infected tissue and the removal of all proteins, thus allowing hybridization of RNAs which may have been well separated from each other before disruption (25).
It is tempting to speculate that negative-strand RNA viruses avoid the formation of dsRNA by packaging the genomic and antigenomic RNAs into RNPs. For most other viruses, hiding or sequestering of once-formed dsRNA is the strategy for circumventing activation of the innate immune system (68). The dsRNA of ReoV, for example, remains within the inner capsid throughout the viral replication cycle (25), and positive-strand RNA viruses replicate their genome enclosed in membrane vesicles (49). Many viruses, e.g., poxviruses, also express dsRNA-binding proteins (25). One may therefore wonder why dsRNA can nevertheless be detected in cells infected with ReoV (Fig. 2, panel 4) or Vac (Fig. 3, panel 4). Most likely, either the monoclonal antibody J2 can access dsRNA even when it is bound by a protein or the amount of dsRNA can exceed that of the virus-expressed proteins. Also, it was suspected that ReoV subviral particles only imperfectly cover the dsRNA (25). This view of leaky anti-dsRNA mechanisms is supported by the fact that ReoV can partly activate PKR (35) and that Vac needs to express the PKR decoy substrate K3L in addition to the dsRNA-binding E3L to overcome IFN sensitivity (25).
The origin of the dsRNA structures detected for positive-strand RNA viruses and for DNA viruses remains to be determined. For positive-strand RNA viruses, it could be either hybrids of cRNA strands generated during genome replication and transcription or intramolecular secondary structures within ssRNA molecules. Whereas the evidence for RNA-RNA hybrids in infected cells remains problematic due to the above-mentioned annealing artifacts during RNA extraction (25, 40), the latter view is supported by studies showing that highly structured viral ssRNAs are sufficient to trigger activation of PKR (44), 2-5-OAS (39), or RIG-I (59). For DNA viruses, dsRNA may arise as a result of overlapping converging transcription (25, 40) or highly structured ssRNAs, such as the adenovirus virus-associated RNAs, of which more than 108 copies are present in a single cell (43). The fact that the ssRNA-specific RNase A has no effect on the viral dsRNA signal does not absolutely rule out the presence of intramolecular secondary structures. Rather, it is still possible that sterical constraints do not allow the enzyme to access the single-strand regions and cut off the secondary structures.
In summary, we have demonstrated by a nondestructive in situ analysis that production of dsRNA occurs for positive-strand RNA viruses, dsRNA viruses, and DNA viruses. Negative-strand RNA viruses, however, appear to represent an exception to that rule since no significant dsRNA signals were detected. Most probably, other molecular patterns, such as RNPs and ssRNA, are more dominant as danger signals for the immune system.
Acknowledgments
We are indebted to Charles E. Samuel for his helpful comments and ideas and Otto Haller for his constant support. We thank Georg Kochs and Peter Staeheli for critically reading the manuscript and Susanne Vends for technical assistance.
Work in our laboratory was supported by grants from the Deutsche Forschungsgemeinschaft (We 2616/4), the Sino-German Center for Research Promotion [GZ Nr. 239 (202/12)], and the Danish Medical Research Council (grant no. 22-02-0144, 22-03-0183, and 22-04-0704). R.H. was supported by the Novo Nordisk Foundation senior researcher fellowship.
REFERENCES
- 1.Ahmed, M., M. O. McKenzie, S. Puckett, M. Hojnacki, L. Poliquin, and D. S. Lyles. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77**:**4646-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4**:**499-511. [DOI] [PubMed] [Google Scholar]
- 3.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413**:**732-738. [DOI] [PubMed] [Google Scholar]
- 4.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101**:**17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77**:**7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov, G. A., A. G. Evstafieva, Y. P. Rubtsov, O. V. Mikitas, A. B. Vartapetian, and V. I. Agol. 2000. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology 275**:**244-248. [DOI] [PubMed] [Google Scholar]
- 7.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74**:**6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutler, B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430**:**257-263. [DOI] [PubMed] [Google Scholar]
- 9.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78**:**9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, B. L., and D. S. Lyles. 1992. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J. Virol. 66**:**4058-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77**:**8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzózka, K., S. Finke, and K.-K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79**:**7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diebold, S. S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303**:**1529-1531. [DOI] [PubMed] [Google Scholar]
- 14.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77**:**13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falcon, A. M., R. M. Marion, T. Zurcher, P. Gomez, A. Portela, A. Nieto, and J. Ortin. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78**:**3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faria, P. A., P. Chakraborty, A. Levay, G. N. Barber, H. J. Ezelle, J. Enninga, C. Arana, J. van Deursen, and B. M. Fontoura. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17**:**93-102. [DOI] [PubMed] [Google Scholar]
- 17.Ferran, M. C., and J. M. Lucas-Lenard. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J. Virol. 71**:**371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Sastre, A. 2004. Identification and characterization of viral antagonists of type I interferon in negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283**:**249-280. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279**:**375-384. [DOI] [PubMed] [Google Scholar]
- 20.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276**:**30178-30182. [DOI] [PubMed] [Google Scholar]
- 21.Gustin, K. E. 2003. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 95**:**35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatada, E., T. Takizawa, and R. Fukuda. 1992. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J. Gen. Virol. 73**:**17-25. [DOI] [PubMed] [Google Scholar]
- 23.Hoyt, C. C., R. J. Bouchard, and K. L. Tyler. 2004. Novel nuclear herniations induced by nuclear localization of a viral protein. J. Virol. 78**:**6360-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, T., M. Yang, and W. S. May. 1999. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 274**:**15427-15432. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219**:**339-349. [DOI] [PubMed] [Google Scholar]
- 26.Jennings, S., L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2005. Thogoto virus ML protein suppresses IRF3 function. Virology 331**:**63-72. [DOI] [PubMed] [Google Scholar]
- 27.Kallman, F., R. C. Williams, R. Dulbecco, and M. Vogt. 1958. Fine structure of changes produced in cultured cells sampled at specified intervals during a single growth cycle of polio virus. J. Biophys. Biochem. Cytol. 4**:**301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katze, M. G. 1995. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 3**:**75-78. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325**:**137-148. [DOI] [PubMed] [Google Scholar]
- 30.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309**:**181-189. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, A., J. Haque, J. Lacoste, J. Hiscott, and B. R. Williams. 1994. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. USA 91**:**6288-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62**:**1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200**:**307-312. [DOI] [PubMed] [Google Scholar]
- 34.Le May, N., S. Dubaele, L. P. De Santis, A. Billecocq, M. Bouloy, and J. M. Egly. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116**:**541-550. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd, R. M., and A. J. Shatkin. 1992. Translational stimulation by reovirus polypeptide σ3: substitution for VAI RNA and inhibition of phosphorylation of the α subunit of eukaryotic initiation factor 2. J. Virol. 66**:**6878-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214**:**222-228. [DOI] [PubMed] [Google Scholar]
- 37.Lukacs, N. 1994. Detection of virus infection in plants and differentiation between coexisting viruses by monoclonal antibodies to double-stranded RNA. J. Virol. Methods 47**:**255-272. [DOI] [PubMed] [Google Scholar]
- 38.Lund, J. M., L. Alexopoulou, A. Sato, M. Karow, N. C. Adams, N. W. Gale, A. Iwasaki, and R. A. Flavell. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 101**:**5598-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maitra, R. K., N. A. McMillan, S. Desai, J. McSwiggen, A. G. Hovanessian, G. Sen, B. R. Williams, and R. H. Silverman. 1994. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology 204**:**823-827. [DOI] [PubMed] [Google Scholar]
- 40.Majde, J. A. 2000. Viral double-stranded RNA, cytokines, and the flu. J. Interferon Cytokine Res. 20**:**259-272. [DOI] [PubMed] [Google Scholar]
- 41.Majde, J. A., R. K. Brown, M. W. Jones, C. W. Dieffenbach, N. Maitra, J. M. Krueger, A. B. Cady, C. W. Smitka, and H. F. Maassab. 1991. Detection of toxic viral-associated double-stranded RNA (dsRNA) in influenza-infected lung. Microb. Pathog. 10**:**105-115. [DOI] [PubMed] [Google Scholar]
- 42.Marcus, P. I., and M. J. Sekellick. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266**:**815-819. [DOI] [PubMed] [Google Scholar]
- 43.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65**:**5657-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCormack, S. J., and C. E. Samuel. 1995. Mechanism of interferon action: RNA-binding activity of full-length and R-domain forms of the RNA-dependent protein kinase PKR—determination of KD values for VAI and TAR RNAs. Virology 206**:**511-519. [DOI] [PubMed] [Google Scholar]
- 45.Melchjorsen, J., S. B. Jensen, L. Malmgaard, S. B. Rasmussen, F. Weber, A. G. Bowie, S. Matikainen, and S. R. Paludan. 2005. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR 7 and TLR 8 in a cell-type-specific manner. J. Virol. 79**:**12944-12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogensen, T. H., and S. R. Paludan. 2005. Reading the viral signature by Toll-like receptors and other pattern recognition receptors. J. Mol. Med. 83**:**180-192. [DOI] [PubMed] [Google Scholar]
- 47.Patel, R. C., and G. C. Sen. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17**:**4379-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175**:**5260-5268. [DOI] [PubMed] [Google Scholar]
- 49.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285**:**139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14**:**778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273**:**2714-2720. [DOI] [PubMed] [Google Scholar]
- 52.Schonborn, J., J. Oberstrass, E. Breyel, J. Tittgen, J. Schumacher, and N. Lukacs. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19**:**2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw, M. L., W. B. Cardenas, D. Zamarin, P. Palese, and C. F. Basler. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79**:**6078-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shors, S. T., E. Beattie, E. Paoletti, J. Tartaglia, and B. L. Jacobs. 1998. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J. Interferon Cytokine Res. 18**:**721-729. [DOI] [PubMed] [Google Scholar]
- 55.Silverman, R. H. 1994. Fascination with 2-5A-dependent RNase: a unique enzyme that functions in interferon action. J. Interferon Res. 14**:**101-104. [DOI] [PubMed] [Google Scholar]
- 56.Stojdl, D. F., B. Lichty, S. Knowles, R. Marius, H. Atkins, N. Sonenberg, and J. C. Bell. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 6**:**821-825. [DOI] [PubMed] [Google Scholar]
- 57.Stollar, B. D., and V. Stollar. 1970. Immunofluorescent demonstration of double-stranded RNA in the cytoplasm of Sindbis virus-infected cells. Virology 42**:**276-280. [DOI] [PubMed] [Google Scholar]
- 58.Streitenfeld, H., A. Boyd, J. K. Fazakerley, A. Bridgen, R. M. Elliott, and F. Weber. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77**:**5507-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79**:**2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabeta, K., P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, L. Alexopoulou, R. A. Flavell, and B. Beutler. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 101**:**3516-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeda, K., and S. Akira. 2005. Toll-like receptors in innate immunity. Int. Immunol. 17**:**1-14. [DOI] [PubMed] [Google Scholar]
- 62.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76**:**3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.tenOever, B. R., S. Sharma, W. Zou, Q. Sun, N. Grandvaux, I. Julkunen, H. Hemmi, M. Yamamoto, S. Akira, W. C. Yeh, R. Lin, and J. Hiscott. 2004. Activation of TBK1 and IKKɛ kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78**:**10636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas, D., G. Blakqori, V. Wagner, M. Banholzer, N. Kessler, R. M. Elliott, O. Haller, and F. Weber. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279**:**31471-31477. [DOI] [PubMed] [Google Scholar]
- 65.Unterstab, G., S. Ludwig, A. Anton, O. Planz, B. Dauber, D. Krappmann, G. Heins, C. Ehrhardt, and T. Wolff. 2005. Viral targeting of the interferon-beta-inducing Traf family member-associated NF-kB activator (TANK)- binding kinase-1. Proc. Natl. Acad. Sci. USA 102**:**13640-13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Q., and G. G. Carmichael. 2004. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 68**:**432-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18**:**1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber, F., G. Kochs, and O. Haller. 2004. Inverse interference: how viruses fight the interferon system. Viral Immunol. 17**:**498-515. [DOI] [PubMed] [Google Scholar]
- 69.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71**:**6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams, B. R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18**:**6112-6120. [DOI] [PubMed] [Google Scholar]
- 71.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175**:**2851-2858. [DOI] [PubMed] [Google Scholar]
- 72.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5**:**730-737. [DOI] [PubMed] [Google Scholar]
- 73.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17**:**1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zamanian-Daryoush, M., T. H. Mogensen, J. A. DiDonato, and B. R. Williams. 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell. Biol. 20**:**1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]