Genomic Analysis of Carbon Source Metabolism of Shewanella oneidensis MR-1: Predictions versus Experiments (original) (raw)
Abstract
Genomic sequences have been used to find the genetic foundation for carbon source metabolism in Shewanella oneidensis MR-1. Annotated S. oneidensis MR-1 gene products were examined for their sequence similarity to enzymes participating in pathways for utilization of carbon and energy as described in the BioCyc database (http://www.biocyc.org/) or in the primary literature. A picture emerges that relegates five- and six-carbon sugars to minor roles as carbon sources, whereas multiple pathways for utilization of up to three-carbon carbohydrates seem to be present. Capacity to utilize amino acids for carbon and energy is also present. A few contradictions emerged in which enzymes appear to be present by annotations but are not active in the cell according to physiological experiments. Annotations are based on close sequence similarity and will not reveal inactivity due to deleterious mutations or due to lack of coordination of regulation and transport. Genes for a few enzymes known by experiment to be active are not found in the genome. This may be due to extensive divergence after duplication or convergence of function in separate lines in evolution rendering activities undetectable by sequence similarity. To minimize false predictions from protein sequences, we have been conservative in predicting pathways. We did not predict any pathway when, although a partial pathway was seen it was composed largely of enzymes already accounted for in any other complete pathway. This is an example of how a biochemically oriented sequence analysis can generate questions and direct further experimental investigation.
Tens of Shewanella species have been isolated and named, and their phylogenetic relationships have been determined. Shewanella bacteria are aquatic organisms found in nature in marine ocean settings, in marshes, and in riverine and lake settings. They are gammaproteobacteria, mesophilic heterotrophic facultative anaerobes (22, 25). Although rich medium promotes faster growth, Shewanella species grow on minimal medium supplemented with a few nutrients, with lactate as a carbon source and any of a number of electron acceptors such as fumarate (25). Shewanellae are particularly distinguished by their ability to use many compounds as terminal electron acceptors in anaerobic respiration. The species whose genome we are studying is Shewanella oneidensis, and the strain is MR-1. It is one of a collection of bacteria isolated from sediment of Lake Oneida, N.Y. (18).
The taxonomic position of Shewanella bacteria has been refined over time. Early names applied to the organism were _Pseudomonas putrefacien_s and Alteromonas putrefaciens. Shewanella was recognized as a species in 1985 (15). Classification by 16S RNA sequences revealed its relationship to other aquatic organisms (25). Most recently, among _Alberomonas_-like bacteria, the genus Shewanella was placed within a new family, Shewanellaceae (10, 11).
Some of the interest in the organism stems from its ability to reduce metals and metal oxides in the environment. This has raised the possibility that shewanellae could serve as decontaminating agents in the environment (2, 18, 19, 24). Also, these bacteria can cause food spoilage and can act as opportunistic pathogens (3, 12) and thus are of some interest to the food industry and medicine.
Recently the S. oneidensis MR-1 genome was fully sequenced and its gene products were annotated (9). The early annotation has been extended since (6) and continues to be studied (13). In this work, we have placed predicted enzymes in pathways of intermediary metabolism for carbon and energy utilization in order to gain a picture from the genetic point of view of the metabolic capacities of the bacterium and to relate them to current experimental knowledge (22).
Information from the genome sequence was used to predict the presence of enzymes of carbon source metabolism. Before doing sequence comparisons, we identified fused genes in the S. oneidensis MR-1 genome and divided them so all gene sequences encode single proteins only (23). The list of all S. oneidensis MR-1 unimodular protein sequences has been compared to the protein sequences of 107 other organisms using the Darwin AllAllDb program (7). We have described previously the particular suitability of the Darwin analysis for sequence annotation (14).
Unlike the situation for most microorganisms whose genomes have been sequenced, there is a modest body of experimental information on some of the phenotypic characteristics of S. oneidensis MR-1. Thus, we have the opportunity to relate sequence-based predictions for Shewanella to existing experimental information about the organism. Do the sequence annotations for enzymes reflect experimentally known metabolic characteristics? In a few cases, experimentally derived information does not agree with the predictions. In other cases, information on expression would be required to relate phenotypic information to genomic results.
Some cautions and modest improvements of methods for pathway prediction have emerged in the process of genomic annotation of central metabolism of a Shewanella strain. Annotations of protein sequences are only predictions. We realize that there are limits to the accuracy of predicting metabolic properties by sequence comparisons. We describe steps taken to eliminate some sources of artifact from the analysis. We minimize predictions of pathways when evidence is weakened by the multiple uses of some enzymes in a cell.
Relating gene and protein sequences of any one organism to proteins of other organisms can only tell us about similarities or variations on metabolic themes already known to us through experimental work in other organisms. No completely new enzymatic functions or pathways will be revealed by current methods of gene annotation.
MATERIALS AND METHODS
Analysis of protein sequence similarities.
Pairwise sequence alignments and scores were generated using the AllAllDb program of Darwin (Data Analysis and Retrieval With Indexed Nucleotide/peptide sequence package), version 2.0, developed at the ETHZ in Zurich, Switzerland (http://cbrg.inf.ethz.ch) (7). Maximum likelihood alignments are generated with an initial global alignment by dynamic programming (Smith and Waterman algorithm) followed by dynamic local alignments (Needleman and Wunsch algorithm). A single scoring matrix is used for these steps. After the initial alignment, the scoring matrix is adjusted to fit the approximate distance between each protein pair to produce the minimum Pam value. Pam units are defined as the numbers of point mutations (base pair differences) per 100 residues. The final report includes Pam distances and variances. Darwin's ability to apply scoring matrixes according to the distance between each protein pair ensures a data set of highly accurate similarity calculations (Pam scores) for distantly as well as closely related protein pairs. While the closely related homologs are mainly used for annotation purposes, the identification of distantly related proteins are valuable in finding divergent but related protein functions. The Darwin algorithms and use of multiple substitution matrices have been evaluated in relation to other sequence analysis approaches and have been given high credit for sensitivity and performance (20).
Genomic sequences for 107 microorganisms were obtained from the NCBI RefSeq web site (www.ncbi.nih.gov/RefSeq/) by ftp (ftp.ncbi.nih.gov/genomes/Bacteria/). Sequences of all proteins of the 107 genomes, predicted and known, were compared to sequences of all S. oneidensis MR-1 proteins. Data for S. oneidensis MR-1 genes were from the NCBI RefSeq database NC_004347.1 except that with further study some annotations have changed to greater specificity (unpublished data). A tab-delimited text table listing all genes for predicted enzymes of S. oneidensis MR-1 is available (see Table S1 in the supplemental material). For enzymes having sequence-similar alignments with homologs of at least 83 residues and occupying more than 45% of the sequence of both proteins, Pam values are reported. Table columns give the gene identification (ID), gene name, enzyme name or partial information, GenBank ID for the best sequence match, organism with best match, and Pam value of best match. EC numbers are included for the enzymes addressed in the paper as well as PubMed IDs for experimentally verified functions.
For the work reported here, sequence pairs were extracted from the totality of less stringent data collected: those pairs that had alignment lengths of at least 83 amino acids and distances of 125 Pam units or less. We chose the length requirement of 83 residues as it improves the significance of the sequence alignments for the more distantly related protein pairs (1). The requirement for at least 83 residues also avoids a class of commonly occurring protein domains smaller than 83 residues that appear widely in many otherwise unrelated proteins (such as small binding sites for a type of substrate, cofactor, or regulator). In addition, for this study we removed proteins directly involved in horizontal gene transfer (IS proteins, transposases, and known prophage components) from the data set.
Pathway reconstruction and vetting incomplete pathways.
S. oneidensis MR-1 proteins annotated as enzymes were examined one by one and placed in known microbial pathways. Pathways that were experimentally determined in another organism were deemed to be present in S. oneidensis MR-1 by the presence of sequence similarity for all component enzymes. Pathways for which not all enzymes were found required careful inspection. It is a fact that some enzymes occur in more than one pathway. In the cases of pathways only partially filled with homologs, we noted which of the enzymes present were already being used in other complete pathways in the cell. These have little or no significance in establishing the presence of the second pathway. Evidence for the presence of enzymes unique to the second pathway is necessary before designating them as present. We did not lower thresholds of similarity in an extended attempt to find enzymes for the holes in such tenuous pathways. Following this rule eliminated many seemingly partial pathways that we believe are not in fact present in S. oneidensis MR-1.
Comparison with experimental information.
We have compared experimental results on central metabolism of S. oneidensis MR-1 with the predictions based on sequence similarity to known enzymes. Besides published work, unpublished results have kindly been made available to us as a private communication from K. E. Nealson (noted in the text as K.N.) and extensive Biolog data have been shared by J. Klappenbach and J. Tiedje. Phenotypic results, published and unpublished, are not completely consistent for this organism from different laboratories at different times, but there is agreement on the major characteristics.
RESULTS AND DISCUSSION
Sequence similarities.
We identified the coding sequences (CDS) of S. oneidensis MR-1 that are similar in sequence to proteins in any one of the 107 genomes searched using Darwin analysis (7). A complete list of the organisms searched can be found in Table S1 in the supplemental material. Of a total of 4,325 genes coding for proteins in the S. oneidensis MR-1 genome, 3,382 CDS for proteins have matches by sequence similarity with Pam values up to 125 to at least one ortholog in the 107 genomes examined. The data collected are as follows: the ID of each S. oneidensis MR-1 protein, an abbreviation of the name of the organism, and the IDs of orthologous proteins of similar sequence, the length of all proteins, the start and end residues of both members of each alignment, the percentage of total protein aligned for each member of a pair, and the name of the orthologous protein. For this study, data for the category of enzymes were extracted.
Most closely related organisms.
To identify the organisms most closely related to S. oneidensis MR-1 among those tested, we counted the number of proteins having matches with a Pam value less than 75 (a stringent threshold). The results, listed in column 1 of Table 1, identify Yersinia and Vibrio spp. as most similar to S. oneidensis MR-1. Both Yersinia and Vibrio spp. are aquatic organisms like Shewanella. Both are major pathogens, the meaning of which is not clear at present as shewanellae are known only to be opportunistic pathogens. In this context, the proteins of the opportunistic pathogen Pseudomonas aeruginosa and the pathogen Pasteurella multocida have many similarities to those in S. oneidensis MR-1. Nevertheless, it seems clear that the aqueous habitats (marine, riverine, and estuararian) of the high-ranking Vibrio and Yersinia spp. may confer the most important physiological similarities that are reflected in the level of genome sequence similarity.
TABLE 1.
Similarity of genes in other genomes to those of Shewanella oneidensis MR-1
| Organism | No. of similar genesa | No. of best hitsb |
|---|---|---|
| Yersinia pestis | 1,943 | 189 |
| Vibrio cholerae | 1,423 | 522 |
| Shigella flexneri | 1,342 | 83 |
| Escherichia coli | ||
| CFT073 | 1,204 | 42 |
| O157:H7 | 1,173 | 43 |
| K-12 | 1,135 | 16 |
| Erwinia carotovora | 1,118 | 103 |
| Salmonella enterica | ||
| Serovar Typhimurium | 1,115 | 109 |
| Serovar Typhi | 1,104 | 16 |
| Pseudomonas aeruginosa | 1,087 | 192 |
| Pseudomonas syringae | 976 | 101 |
| Chromobacterium violaceum | 776 | 62 |
| Pasteurella multocida | 764 | 45 |
As another measure of degree of relatedness, the proteins having the closest similarity to each of the S. oneidensis MR-1 proteins were listed by organism and counted. The result, column 2 of Table 1, shows the number of “best hits” to S. oneidensis MR-1 proteins. Again Vibrio and Yersinia spp. ranked higher and Escherichia coli species ranked lower than they did when ranked by total number of closely related homologs. Note, however, that when orthologs in closely related strains of a species are counted—in this case, E. coli and Salmonella enterica serovars—one must be aware that proteins closely related in all strains will be counted only once as the best match to a Shewanella protein, yet all are highly significant. Thus, low scores by close relatives are not indicative of genetic distance as the marginally lower similarities are not counted in the ranking. Nevertheless, Vibrio and Yersinia species far outstrip the other organisms tested.
Although E. coli strains ranked low in numbers of best matches to S. oneidensis MR-1 coding sequences, nevertheless the E. coli data were vital to interpretation of the results. E. coli among all organisms tested had the most information on gene products that have been experimentally characterized, not simply predicted by sequence similarity
Metabolism pathways—overview.
The subset of orthologs with the best Pam scores from any of the genomes as well as all significant orthologs in E. coli K-12, whether the best or not, were collected for all the enzymes of central metabolism. We grouped enzymes by pathway, some enzymes appearing more than once if present in more than one pathway. We found, based on sequence similarity, that S. oneidensis MR-1 like E. coli has complete pathways for the biosynthesis of all amino acids, nucleotides, and cofactors. However, utilization of compounds as carbon and energy sources was more restricted than in E. coli, and in some instances the pathways were not those of E. coli but of other organisms such as P. aeruginosa.
Carbon source and energy metabolism by respiration.
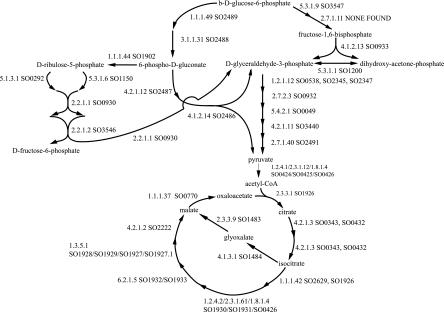
Homologous genes for enzymes of main carbon utilization and central metabolism are shown in Fig. 1. Convincing support for the major pathways of carbon utilization and multipurpose enzymes of central metabolism is present. Sequences are present for all enzymes of the pentose pathway, the pyruvate dehydrogenase complex, all enzymes of the tricarboxylic acid cycle, the glyoxylate bypass, the Entner-Douderoff pathway, major anaplerotic reactions, and other enzymes of central carbohydrate metabolism. Note that by sequence similarity an essential enzyme of glycolysis, 6-phosphofructokinase, is not found. In agreement, it is known experimentally that S. oneidensis MR-1 does not use glucose as a carbon and energy source (Biolog; K.N.). The critical glycolytic enzyme 6-phosphofructokinase was not found in cell extracts (22). Consistent with the inability to use glucose for growth, we find that the sequences for enzymes for the feeder reactions that convert six-carbon carbohydrates to glucose that are present in E. coli are not found in S. oneidensis MR-1 (see Table S2 in the supplemental material). Among E. coli enzymes used in the conversion of 15 compounds, we only detected S. oneidensis MR-1 homologs for galactokinase and ribokinase. The presence of these two kinases does not reflect utilization of galactose or ribose for carbon and energy; rather they are conversions in the course of intermediary metabolism. The predicted enzyme composition of S. oneidensis MR-1 in this respect—shy on enzymes for utilization of five- and six-carbon carbohydrates—is consistent with known phenotypic characterization (25).
FIG. 1.
Carbon source metabolism in S. oneidensis MR-1. The intermediary metabolic steps of glycolysis, tricarboxylic acid (TCA) cycle, glyoxalate bypass, the pentose phosphate, and the Entner-Doudoroff pathways are shown. Enzymes are shown by their EC number followed by the S. oneidensis locus tag (SO number) of the gene predicted to encode the respective activity. The function predictions were based on sequence similarity to proteins with experimentally verified functions. Predicted isozymes are shown as SO numbers separated by a comma. Enzyme complexes are indicated by SO numbers separated by a forward slash. Predictions were made for all steps, except for that of 6-phosphofructokinase, EC 2.7.1.11.
The existence of multiple copies of genes or genes for isozymes might reflect a gene dosage effect supporting heavy use of these particular enzymes. We note that isozymes are not present in S. oneidensis MR-1 for five- and six-carbon metabolism as they are in E. coli, but do exist for glyceraldehyde-3-phosphate dehydrogenase, an enzyme that is vital to central metabolism at the three-carbon level (see Table S3 in the supplemental material). Also in S. oneidensis MR-1, different from E. coli, large numbers of phosphotransferase system (PTS) genes for transport of simple sugars are not present. Only one set of PTS genes is found in S. oneidensis MR-1, whereas E. coli has 41 genes encoding PTS proteins for a variety of four-, five-, and six-carbon carbohydrates.
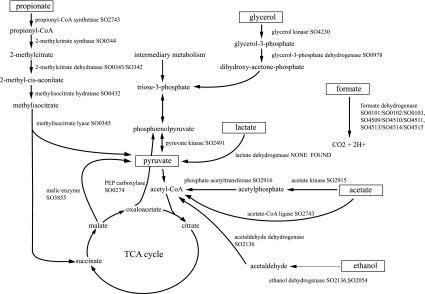
Instead of six-carbon sugars, S. oneidensis MR-1 prefers three-carbon carbohydrates for growth. Experimentally, l-lactate, pyruvate, and acetate are among compounds utilized as sources of carbon and energy (22, 25; Biolog data). Agreeing with the experiment, genes are present for the enzymes for metabolism of pyruvate and acetate and other two- and three-carbon molecules (Fig. 2). However, the genetic picture seemingly does not agree for l-lactate, which does in fact support growth, but no homolog for a currently sequenced bacterial l-lactate dehydrogenase is found in S. oneidensis MR-1. A homolog for an unrelated fermentative d-lactate dehydrogenase gene is present, although S. oneidensis MR-1 is not considered a fermentative organism and is unable to ferment glucose (22, 25). Since S. oneidensis MR-1 is usually grown with l-lactate as a carbon and energy source, the absence of a homolog for l-lactate dehydrogenase is puzzling. Perhaps there is a different kind of lactate dehydrogenase not represented in current sequence databases. We note that two genes, designated by locus tags SO1520 and SO1521, are located adjacent to the lactate permease gene. One codes for an iron-sulfur protein, and the product of the other contains a domain for binding flavin adenine dinucleotide and has similarity to a glycolate dehydrogenase. Could these CDS represent a different kind of prokaryotic l-lactate dehydrogenase, a dimer that contains iron sulfur center(s) and flavin adenine dinucleotide bound to separate subunits? Experimental exploration of the possibility could be fruitful.
FIG. 2.
Enzymes in S. oneidensis MR-1 for utilization of one-, two-, and three-carbon compounds. Enzymes involved in the degradation of compounds with one carbon (formate), two carbons (acetate, ethanol), and three carbons (propionate, glycerol, lactate, and pyruvate) are shown by their names and predicted S. oneidensis locus tags (SO number). The function predictions were based on sequence similarity to proteins with experimentally verified functions. Predicted isozymes are shown as SO numbers separated by a comma. Enzyme complexes are indicated by SO numbers separated by a forward slash. TCA, tricarboxylic acid.
Although reports of ability to grow on propionate are mixed (22; K.N.; Biolog), on the basis of sequence similarity, support is found. Propionate could be metabolized by a variation on the 2-methyl citrate pathway in which 2-methyl citrate is converted to 2-methyl-_cis_-aconitate by the AcnD-PrpF combination, a complex 2-methyl citrate dehydratase. In this respect, S. oneidensis MR-1 is like P. aeruginosa and Vibrio cholerae, not like E. coli, which has the PrpD version of the enzyme (8).
S. oneidensis MR-1 uses a C-1 compound, formate, as an energy source (22). Standing out from a plethora of genes concerning formate are genes for three formate dehydrogenase operons and one hydrogenase operon found in the genome. The three clusters are similar to those known for three-subunit formate dehydrogenase enzymes that participate in respiration, using either oxygen as an electron acceptor or nitrate. The three clusters are SO0101 to -0103 plus the accessory protein product of SO0107; SO4509 to -4511, and SO4513 to -4515 plus the accessory protein product of SO4503. The hydrogenase SO2097, -2098, and -2099 genes code for the subunits of a quinone-reactive type of hydrogenase known to work together with respiratory formate dehydrogeases. Adjacent to the hydrogenase operon is a contiguous set of genes coding for hydrogenase accessory proteins (SO2089 to SO2096). Absence of the fermentative type of formate dehydrogenase in complex with hydrogenase to make the formate hydrogen lyase complex is consistent with the observation that S. oneidensis MR-1 is not a fermentative organism. The genes encoding for pyruvate formate-lyase and its activator were found (SO2912 and -2913). Although this enzyme usually is associated with fermentative metabolism, it may be supplying S. oneidensis with C-1 (formate) units.
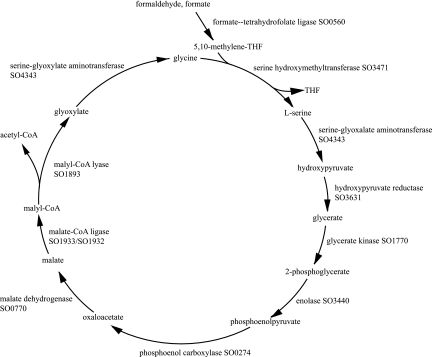
The CO2 produced by formate oxidation cannot be fixed by S. oneidensis MR-1 as no homologs are found for any known enzymes for CO2 fixation. Consideration has been given to the possibility S. oneidensis MR-1 is a facultative methylotroph with the cyclic C-1 serine pathway. Some of the enzymes for the cyclic serine pathway are present but, by stringent sequence comparisons, not all. An alternative to the classical serine pathway that uses only enzymes of general metabolism has been proposed (22). However, this version may not be able to effect net incorporation of C-1 moieties into metabolism. Since all of the enzymes in this scheme are also required for other pathways, their presence in MR-1 is ambiguous.
As to the classical C-1 cyclic serine pathway, almost all needed genes are present in S. oneidensis MR-1. It appears to be able to activate both formate and formaldehyde substrates by enzymes of tetrahydrofolate metabolism, converting them into the derivative _N-_5,10-methylenetetrahydrofolate (methyleneTHF) (4, 16). The methyleneTHF enters the cyclic C-1 serine pathway by donating the C-1 moiety to combine with glycine producing serine, resulting ultimately in capture of carbon as acetyl-coenzyme A (CoA) and regeneration of the C-1 acceptor molecule glycine for the next cycle of capture. There are two unique enzymes that could identify the pathway: malate-CoA ligase and malyl-CoA lyase (Fig. 3). Searching for sequences for the two unique enzymes in this pathway did not reveal homologs in S. oneidensis MR-1 within the threshold of significance applied in this work. However, we note that enzymes are present for similar reactions such that broad substrate specificity could enable the enzymes to encompass the activities of the two missing reactions. The succinyl-CoA ligase (SO1932 and SO1933) might serve as the malyl-CoA ligase if it also had the ability to bind malate. As a possible substitute for the malyl-CoA lyase, the S. oneidensis MR-1 genome has a close homolog of the gene for 3-hydroxy-3-methylglutaryl-CoA lyase (SO1893). Could this enzyme, if it has broad substrate specificity, also serve as the malyl-CoA lyase? 3-Hydroxy-3-methylglutarate and malate are chemically related compounds. Experimental characterization of the proteins of SO1893 and SO1932 to -1933 could establish whether either possibility is the case. Otherwise Shewanella spp. could employ an as yet unknown pathway as a recycling mode for capturing carbon when growing on formate or formaldehyde as a carbon source (5). Biochemical investigation is needed.
FIG. 3.
Classical C-1 serine pathway enzymes in S. oneidensis MR-1. Prediction of S. oneidensis MR-1 enzymes involved in the growth and assimilation of one-carbon compounds via the serine pathway are shown. Function predictions were made based on sequence matches to proteins with experimentally verified functions. Predicted enzyme complexes are indicated by SO numbers separated by a forward slash. Significant sequence matches to the two key enzymes in the pathway, malyl-CoA lyase (EC 4.1.3.24) and malate-CoA ligase (EC 6.2.1.9), were not found. Tentative assignments for these reactions were made to S. oneidensis MR-1 gene products catalyzing similar reactions.
Given its evident preference for C-3 and C-1 carbon sources, S. oneidensis MR-1 will depend on anaplerotic reactions, the glyoxylate pathway, gluconeogenesis, and the nonoxidative pentose pathway reactions to generate the intermediates required for biosynthetic metabolism. Not mentioned above are the critical anaplerotic enzymes, all of which seem to be present: phosphoenolpyruvate carboxykinase, phosphoenolpyruvate carboxylase, and malic enzyme.
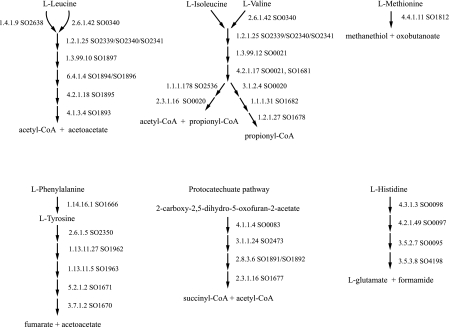
Ability to utilize amino acids as carbon sources is also widespread in S. oneidensis MR-1 (21). Sequence similarity was found to the genes coding for the enzymes of the pathways for utilization of amino acids as they are known in E. coli. In addition, in S. oneidensis MR-1, some of the amino acids are broken down by pathways found in other organisms. These are shown in Fig. 4. The ability to utilize histidine is not by the pathway found in E. coli K-12 but is like the transformations in Bacillus subtilis. The ability to utilize aromatic amino acids is not by the pathway in E. coli, but has similarities to part of the protocatechuate pathway as found in pseudomonads. Genes for the enzymes of the latter steps of the protocatechuate meta-pathway are present in S. oneidensis MR-1, but only in the steps after the intermediate γ-carboxymuconolactone. Utilization of aromatic compounds may have unique early steps in S. oneidensis MR-1 that join the known pathway at the carboxymuconolactone step. S. oneidensis MR-1 also degrades leucine, isoleucine, and valine not simply by transamination as in E. coli but rather by pathways used in pseudomonads. Methionine is converted to 2-oxobutanoate as in pseudomonads.
FIG. 4.
Amino acid utilization in S. oneidensis MR-1 by other than E. coli pathways. S. oneidensis MR-1 enzymes predicted to be involved in the degradation of amino acids are shown by their locus tags (SO numbers) and the EC number of the reaction. Gene predictions are based on sequence similarities to known enzymes. Predicted isozymes are shown as SO numbers separated by a comma. Enzyme complexes are indicated by SO numbers separated by a forward slash. Only pathways not utilized by E. coli are included in the figure.
Even though enzymes for utilization of many amino acids seem to be present by sequence analysis, as measured by Biolog data, single amino acids do not serve as carbon sources for S. oneidensis MR-1. However, these Biolog results show that many dipeptides composed of many combinations of pairs of amino acids do register positively. The easier permeability and lower specificity of dipeptide transport systems compared with single-amino-acid transporters might account for the difference in results. Homologs for several dipeptide-specific ABC transport proteins are found in the genome, and there are numerous homologs of dipeptidases that could hydrolyze the dipeptides taken up and make available the monomeric amino acids for utilization.
Fatty acids are also used as carbon and energy sources. Homologs for genes coding for enzymes for breakdown of fatty acids are found. There are multiple copies of some of the key enzymes in these pathways. Other compounds found by Biolog testing to be utilized as carbon sources are α-ketobutyric acid, methylpyruvate, and d-lactate methyl ester, each readily convertible to mainline metabolites. Also giving positive Biolog results is _N_-acetylglucosamine, for which all sequences for the enzymes of the degradative pathway are present in the genome.
Experimental information on nucleoside utilization is interesting. Biolog and laboratory (K.N.) results for utilization of the nucleosides uridine, adenosine, 2′-deoxyadenosine, inosine, and, to a lesser extent, thymidine are positive. In contrast, the Biolog results for the bases cytosine, thymine, adenine, and guanine are poor to negative. We see that sequences for nucleoside hydrolases are present in S. oneidensis MR-1 that could split the nucleosides to their bases and ribose-5-phosphate for further utilization. However, since Biolog data indicate that the bases are not utilized, evidently it is primarily the ribose-5-phosphate moiety that is supporting growth when nucleosides are provided. Again, further experimentation is required to establish this interpretation.
Aerobic and anaerobic respiration.
With respect to respiratory electron transfer pathways, electron transfer elements seem to be emphasized in the S. oneidensis MR-1 genome. Pathways for synthesis of heme, siroheme, menaquinone, and unbiquinone are intact. S. oneidensis MR-1 is believed to contain 42 cytochrome _c_-type proteins (17), whereas by comparison there are only 5 in E. coli.
Shewanella spp. are well known for their use in anaerobic respiration of many electron acceptors other than oxygen, the most unique being metals and metallic compounds (2, 18, 25). In agreement, genes have been found by sequence similarity to code for reduction of nitrate, nitrite, sulfate, sulfur, trimethylamine _N_-oxide (TMAO), dimethyl sulfoxide, and fumarate reductase enzymes corresponding to many of the metabolites used as anaerobic respiration acceptors. Enzymes for reduction of Fe ions are known, but reductases for the numerous metallic compounds that serve as receptors have yet to be identified. However, there is no dearth of possibilities in the genome sequences. We found many candidate uncharacterized oxidoreductases.
Fermentation.
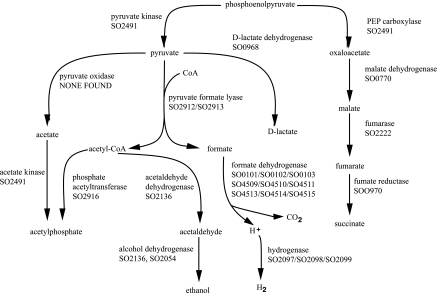
S. oneidensis MR-1 does not ferment (22, 25), yet many enzymes of mixed acid fermentation seem to be present in the genome (Fig. 5). In fermentation, choices include multiple paths from lactate or pyruvate to ethanol or acetate and conversion of acetate to acetyl-CoA or acetylphosphate. Other pathways lead from formate to production of H2 and CO2. Although all enzymes of mixed acid fermentation are present, there is no evidence it does exist. Support for a pathway does not exist if all enzymes are known to be active in other pathways (Table 2). In some cases, such as with the cyclic C-1 serine pathway, there is at least one unique enzyme in the pathway. Designation of the pathway then hangs on the presence or absence of any unique member. Table 2 gives examples of pathways that use many or all of its enzymes that are present in other pathways. Note that all of the enzymes used in fermentation pathways are also used in other known pathways. Thus, there is no indication from genomic analysis whether fermentation is used or not by this organism.
FIG. 5.
Enzymes of mixed acid fermentation in S. oneidensis MR-1. Sequence matches for S. oneidensis MR-1 gene products (SO numbers) to the enzymes used by E. coli for mixed acid fermentation are shown. Significant similarities to all enzymes except pyruvate oxidase (EC 1.2.2.2) were found. Predicted isozymes are separated by commas. Reactions predicted to be carried out by enzyme complexes are shown by SO numbers separated by forward slashes.
TABLE 2.
Some pathways that use enzymes in common with other pathways
| Pathway 1 | No. of enzymes | Pathway 2 |
|---|---|---|
| Common | Unique | |
| C-1 serine pathway | 2 | Tricarboxylic acid cycle |
| 1 | Glycolysis/gluconeogenesis | |
| 1 | Glyoxylate bypass | |
| 1 | Central metabolism | |
| 2 | ||
| Mixed acid fermentation | 3 | Tricarboxylic acid cycle |
| 3 | Glycolysis | |
| 4 | Central metabolism | |
| 0 | ||
| Valine utilization | 2 | Leucine utilization |
| 5 | ||
| Glycolysis | 8 | Gluconeogenesis |
| 1 |
Biosynthesis.
The emphasis in this survey is on utilization of sources of carbon and energy, yet in the course of the work, the presence of homologs in the genome of enzymes of biosynthesis of basic building blocks was noted. Although S. oneidensis MR-1 is frequently grown with supplements to increase the growth rate, the organism is able to grow on an unsupplemented minimal medium. In agreement, based on sequence similarity, S. oneidensis MR-1 has complete biosynthetic capacity for basic building blocks and cofactors. Sequences are found for enzymes similar to those in E. coli for biosynthesis of all amino acids, purines, pyrimidines, and growth factors. The essential precursor metabolites that are needed for biosynthetic activities of the cell are as follows: glucose-6-P, fructose-6-P, ribose-5-P, erythrose-4-P, triose phosphate, 3-phosphoglycerate, phosphoenolpyruvate, pyruvate, acetyl-CoA, 2-oxoglutarate, succinyl-CoA, oxalacetate, and sedoheptulose-7-phosphate. In S. oneidensis MR-1, all enzymes required to generate these precursors are supported by genomic sequences.
Summary of general considerations of pathway prediction.
We have emphasized that there are caveats for pathway prediction which we applied in this work. A common approach to connecting information about enzyme homologs with metabolic pathways is to list the enzymes that seem to be present for any and all pathways. By assessing pathways one by one, if all enzymes are present one asserts the pathway is present in the organism. If not all enzymes are present, there is a temptation to look for the missing enzymes, relaxing thresholds of sequence similarity in an effort to complete the pathway list. However, it is important first to ask whether any of the enzymes in a partial pathway are used in other pathways in the cell. If all enzymes of a pathway with “holes” are used in other pathways, then there is no evidence for existence of the partial pathway. For some pathways, there is at least one unique enzyme in the pathway that is not used in any other known pathway in the cell. If a homolog for the unique enzyme is found, then it does seem safe to assert that pathway is present, even if not all other enzymes have yet been found. Illustrations are given in Table 2. In the classical cyclic C-1 serine pathway, two enzymes are unique. However, for mixed acid fermentation, all enzymes are also used in general metabolism; none are unique to the fermentation. Therefore, one cannot know without experimentation whether any of the fermentations occur.
In asserting a metabolic capability of any kind by genomic analysis, one needs to keep in mind that not all pathways in the biological world are yet known, so failure to find standard ways to convert one substance to another need not mean the biological capability is absent in the organism. An example is evidence in S. oneidensis MR-1 for only part of the protocatechuate pathway of aromatic compound degradation.
In all predictions of enzyme activity, it goes without saying that sequence similarity does not guarantee biological activity. Mutations may be present that produce an inactive enzyme product. We have tried to demur with assertions like “appears to be present” since we cannot know whether a homolog that is not identical to an active enzyme is in fact capable of activity.
Concluding remarks.
Identification of homologs to enzymes of metabolism has allowed us to assemble a picture of central carbon metabolism in S. oneidensis MR-1. Utilization of carbohydrates larger than three carbons is not well supported by sequence similarity to appropriate genes and enzymes. Utilization of three-carbon carbohydrates and smaller has genetic support. Capability for utilization of amino acids and fatty acids as carbon sources is also present. Basic biosynthetic enzymes are present. Most pathways were similar to those in E. coli even though the closest homologs by sequence analysis were not often found in E. coli. But these pathways presumably are highly similar in the most closely related gammaproteobacteria, Vibrio spp. and Yersinia spp. Exceptions were found for some pathways for utilization of amino acids that were similar to those of pseudomonads or bacilli.
There are opportunities for connecting study of the physiology and metabolism of S. oneidensis MR-1 in the laboratory with questions of the kind raised here about operation of particular pathways and the properties of particular enzymes. Each of the questions posed here provides an opportunity for investigation that has the potential of yielding new knowledge about microbial metabolism. This would seem to be a useful outcome of sequence similarity analysis of genomic DNA.
Supplementary Material
[Supplemental material]
Acknowledgments
Patient and expert assistance from Daniella Wilmot was essential to the performance of this project. We thank John L. Ingraham for illuminating discussions. We are grateful that Biolog data were shared by J. Klappenbach and J. Tiedje and unpublished results were shared by K. E. Nealson. We acknowledge the use of BioCyc (http://www.biocyc.org/) for guidance in presenting metabolic pathways.
This work was supported by the Office of Science (BER), U.S. Department of Energy, grant no. DE-FG02-01ER63202.
Footnotes
REFERENCES
- 1.Altschul, S. F. 1991. Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol. 219**:**555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, G. E., Jr., V. E. Henrich, W. H. Casey, D. L. Clark, C. Eggleston, A. Felmy, D. W. Goodman, M. Gratzel, G. Maciel, M. I. McCarthy, K. H. Nealson, D. A. Sverjensky, and M. F. Zachara. 1999. Metal oxide surfaces and their interactions with aqueous solutions and microbial organisms. Chem. Rev. 99**:**77-174. [DOI] [PubMed] [Google Scholar]
- 3.Bulut, C., G. T. Ertem, C. Gokcek, N. Tulek, M. A. Bayar, and E. Karakoc. 2004. A rare cause of wound infection: Shewanella putrefaciens. Scand. J. Infect. Dis. 36**:**692-694. [DOI] [PubMed] [Google Scholar]
- 4.Chistoserdova, L., S.-W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185**:**2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdova, L., M. Laukel, J.-C. Portais, J. A. Vorholt, and M. E. Lidstrom. 2004. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J. Bacteriol. 186**:**22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daraselia, N., D. Dernovoy, Y. Tian, M. Borodovsky, R. Tatusov, and T. Tatusova. 2003. Reannotation of Shewanella oneidensis genome. OMICS J. Integr. Biol. 7**:**171-175. [DOI] [PubMed] [Google Scholar]
- 7.Gonnet, G. H., M. T. Hallett, C. Korostensky, and L. Bernadin. 2000. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics 16**:**101-103. [DOI] [PubMed] [Google Scholar]
- 8.Grimek, T. L., and J. C. Escalante-Semerena. 2004. The acnD genes of Shewenella oneidensis and Vibrio cholerae encode a new Fe/S-dependent 2-methylcitrate dehydratase enzyme that requires prpF function in vivo. J. Bacteriol. 186**:**454-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadr, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20**:**1118-1123. [DOI] [PubMed] [Google Scholar]
- 10.Ivanova, E. P., T. Sawabe, N. V. Zhukova, N. M. Gorshkova, O. I. Nedashkovskaya, K. Hayashi, G. M. Frolova, A. F. Sergeev, K. G. Pavel, V. V. Mikhailov, and D. V. Nicolau. 2003. Occurrence and diversity of mesophilic Shewanella strains isolated from the North-West Pacific Ocean. Syst. Appl. Microbiol. 26**:**293-301. [DOI] [PubMed] [Google Scholar]
- 11.Ivanova, E. P., S. Flavier, and R. Christen. 2004. Phylogenetic relationships among marine _Alteromonas_-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54**:**1773-1788. [DOI] [PubMed] [Google Scholar]
- 12.Jorens, P. G., K. Goovaerts, and M. Ieven. 2004. Shewanella putrefaciens isolated in a case of ventilator-associated pneumonia. Respiration 71**:**199-201. [DOI] [PubMed] [Google Scholar]
- 13.Kolker, E., A. F. Picone, M. Y. Galperin, M. F. Romine, R. Higdon, K. S. Makarova, N. Kolker, G. A. Anderson, X. Oiu, K. J. Auberry, G. Babnigg, A. S. Beliav, P. Edlefsen, D. A. Elias, Y. A. Gorby, T. Holzman, J. A. Klappenbach, K. T. Konstantinidis, M. L. O. Land, M. S. Lipton, L. A. McCue, M. Monroe, L. Pasa-Tolic, G. Pinchuk, S. Purvine, M. H. Serres, S. Tsapin, B. A. Zakraisek, W. Zhu, J. Chou, F. W. Llarimer, C. E. Lawrence, M. Riley, F. R. Collart, J. R. Yates III, R. D. Smith, C. S. Giometti, K. H. Nealson, J. K. Fredrickson, and J. M. Tiedje. 2005. Global profiling of Shewanella oneidensis MR-1: expression of hypothetical genes and improved functional annotations. Proc. Natl. Acad. Sci. USA 102**:**2099-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang, P., and M. Riley. 2001. A comparative genomics approach for studying ancestral proteins and evolution. Adv. Appl. Microbiol. 50**:**39-72. [DOI] [PubMed] [Google Scholar]
- 15.MacDonell, M. T., and R. R. Colwell. 1985. Phylogeny of the Vibrionaceae, and recommendation for two new genera, Listonella and Shewanella. Syst. Appl. Microbiol. 6**:**171-182. [Google Scholar]
- 16.Marx, C. J., M. Laukel, J. A. Vorholt, and M. E. Lidstrom. 2003. Purification of the formate-tetrahydrofolate ligase from Methylobacterium extorquens AM1 and demonstration of its requirement for methylotrophic growth. J. Bacteriol. 185**:**7169-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, T. E., A. I. Tsapin, I. Vandenberghe, L. de Smet, D. Frishman, K. H. Nealson, M. A. Cusanovich, and J. J. van Beeumen. 2004. Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS J. Integr. Biol. 8**:**57-77. [DOI] [PubMed] [Google Scholar]
- 18.Nealson, K. H. 1997. Sediment bacteria: who's there, what are they doing, and what's new? Annu. Rev. Earth Planet. Sci. 25**:**403-434. [DOI] [PubMed] [Google Scholar]
- 19.Nealson, K. H., A. Belz, and B. McKee. 2002. Breathing metals as a way of life: geobiology in action. Antonie Leeuwenhoek 81**:**215-222. [DOI] [PubMed] [Google Scholar]
- 20.Pearson, W. R. 1995. Comparison of methods for searching protein sequence databases. Protein Sci. 4**:**1145-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringø, E., E. Stenberg, and A. R. Strøm. 1984. Amino acid and lactate catabolism in trimethylamine oxide respiration of Alteromonas putrefaciens NCMB 1735. Appl. Environ. Microbiol. 47**:**1084-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott, J. H., and K. H. Nealson. 1994. A biochemical study of the intermediary carbon metabolism of Shewanella putrefaciens. J. Bacteriol. 176**:**3408-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serres, M. H., and M. Riley. 2004. Structural domains, protein modules, and sequence similarities enrich our understanding of the Shewanella oneidensis MR-1 proteome. OMICS J. Integr. Biol. 8**:**306-321. [DOI] [PubMed] [Google Scholar]
- 24.Tiedje, J. M. 2002. Shewanella—the environmentally versatile genome. Nat. Biotechnol. 20**:**1093-1094. [DOI] [PubMed] [Google Scholar]
- 25.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49**:**705-724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]