Genetic analysis reveals demographic fragmentation of grizzly bears yielding vulnerably small populations (original) (raw)
Abstract
Ecosystem conservation requires the presence of native carnivores, yet in North America, the distributions of many larger carnivores have contracted. Large carnivores live at low densities and require large areas to thrive at the population level. Therefore, if human-dominated landscapes fragment remaining carnivore populations, small and demographically vulnerable populations may result. Grizzly bear range contraction in the conterminous USA has left four fragmented populations, three of which remain along the Canada–USA border. A tenet of grizzly bear conservation is that the viability of these populations requires demographic linkage (i.e. inter-population movement of both sexes) to Canadian bears. Using individual-based genetic analysis, our results suggest this demographic connection has been severed across their entire range in southern Canada by a highway and associated settlements, limiting female and reducing male movement. Two resulting populations are vulnerably small (≤100 animals) and one of these is completely isolated. Our results suggest that these trans-border bear populations may be more threatened than previously thought and that conservation efforts must expand to include international connectivity management. They also demonstrate the ability of genetic analysis to detect gender-specific demographic population fragmentation in recently disturbed systems, a traditionally intractable yet increasingly important ecological measurement worldwide.
Keywords: population fragmentation, microsatellites, Ursus arctos, carnivores, genetic analysis
1. Introduction
Habitat fragmentation is a serious threat to biological diversity and is at the root of the present extinction crisis (Wilcox & Murphy 1985). Fragmented systems yielding small isolated populations suffer increased extirpation or extinction probabilities primarily from demographic processes (Lande 1988; Woodroffe & Ginsberg 1998), and secondarily from more gradual genetic processes (Frankham et al. 2002). In fragmented ecosystems, inter-population dispersal of both sexes may be important for population augmentation, rescue or re-colonization within natural and human-caused meta-populations (Hanski & Gilpin 1997). In species with sex-biased dispersal the movement of one sex, often males in mammals (Greenwood 1980; Pusey 1987), may mediate genetic connectivity, while females, the reproductive component of the population, improve a recipient population's demographics. Furthermore, species that display sex-biased dispersal may experience gender-specific fragmentation. Therefore, to be useful, any connectivity measure (or study) should have the ability to distinguish gender-specific inter-population movement.
Several large carnivores such as grizzly bear (Ursus arctos), wolf (Canis lupus) and lynx (Lynx canadensis) are threatened in the conterminous USA (Novak et al. 1987). Viability of these carnivores may require immigration from Canada. For example, human persecution led to the extirpation of wolves from the western USA in the last century. Tolerant attitudes toward carnivores, however, allowed natural re-colonization of the wolf into the USA from source populations in southern Canada (Mech 1995; Forbes & Boyd 1996).
Continental range contraction of grizzly bears over the past two centuries has resulted in four sub-populations within the conterminous USA. All four are isolated from one another, but three of these are assumed to be connected to Canadian populations (Servheen et al. 1999). To resist further range contraction and reduce the risk of eventual extirpation from the USA, maintaining connections to Canadian populations is thought essential (U.S.F.W.S. 1993). However, factors that caused historic declines of grizzly bears in the USA are also active in southern Canada and, in particular, there is a major east–west transportation corridor that bisects the entire range of grizzly bears just north of the Canada–USA border. Grizzly bear connectivity across this corridor is an important but untested assumption in grizzly bear conservation (McLellan 1998). Our objective was to quantify the extent to which grizzly bears in the USA and Canada are genetically and demographically connected. We also wanted to test the hypothesis that it is the transportation corridor and associated human settlements, and not simply major valleys, that have caused any reduction in connectivity. Measuring animal movement with radio-telemetry was impractical for this study due to its spatial and temporal limitations, and traditional genetic methods provide unreliable estimates of inter-population migration in recently disturbed systems (Whitlock & McCauley 1999). Instead, we estimated gender-specific movement rates in a recently disturbed system and identified inter-population migrants by using individual-based genetic analyses.
To quantify sex-specific connectivity of grizzly bears, we used regional-scale, non-invasive sampling (Woods et al. 1999) in three mountain ranges across the trans-border area that included both sides of the human transportation and settlement corridor that is a potential fracture for bears (figure 1). We also sampled both sides of a wide, but uninhabited valley that did not contain a transportation corridor. The broad-based sampling was followed by 15-locus microsatellite genotyping and analyses that measured inter-population movements of individuals (Waser & Strobeck 1998; Pearse & Crandall 2004; Manel et al. 2005).
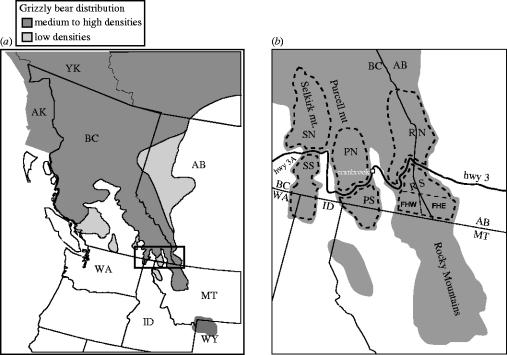
Figure 1.
(a) Grizzly bear distribution and inter-mountain study area in western North America. (b) Southern Rocky, Purcell and Selkirk Mountain study area. Dotted lines outline geographic areas where bears were genetically sampled on both sides of Highways 3 and 3A. SN and SS are the Selkirks North and South of Highway 3A, PN and PS are the Purcells North and South of Hwy 3 and RN and RS are the Rockies North and South of Hwy 3. FHW and FHE are sampled areas west and east of the Flathead River and constitute the ‘control’ area ecologically within and similar to the southern Rocky system. Map 1_a_ adapted from McLellan (1998). YK, Yukon; AK, Alaska; BC, British Columbia; AB, Alberta; MT, Montana; ID, Idaho; WA, Washington; WY, Wyoming.
2. Methods
We genetically sampled wild grizzly bears on both sides of Canadian highways 3 and 3A as they traverse the Rocky, Purcell and Selkirk mountain ranges in southern British Columbia (BC) and Alberta just north of the Canada–USA border (figure 1). Average summer traffic volume is 7000 vehicles per day in the Rockies (Highway 3) and 4000 in the Purcells (Highway 3) and Selkirks (Highway 3A; B.C Ministry of Transportation). A railway with 8–16 trains per day parallels highways 3 and 3A. Communities along this route range from 500 to 10 000 inhabitants. The Rocky and Purcell routes have a discontinuous set of rural enclaves between towns, while the Selkirk route has a narrow yet continuous human settlement (figure 1). For a control, we also quantified connectivity across the large north fork of the Flathead River valley in the Rocky Mountains, which does not have a major transportation corridor or settlements. We quantified the topographic similarities between the control valley and our test valleys.
In our 32 000 km2 study area, between 1996 and 2001, we sampled bears using hair-traps that consisted of a single strand, barbed-wire corral surrounding a scent lure (Woods et al. 1999). Hair traps were placed within 1 km and as far away as 110 km from the highway resulting in a continuous sampling distribution across highways and the control area. Biopsy samples from hunter kills and radio-telemetry bears from other studies were also used. Total cell DNA from 2–10 hair follicles or a tissue biopsy was extracted using Qiagen kits (Qiagen, Inc., Mississauga, Ontario, Canada). All samples were initially genotyped at six microsatellite loci (Paetkau et al. 1998_a_). Because we sampled individuals multiple times, individuals were determined by unique 6-locus genotypes and one sample from each individual was sexed (Ennis & Gallagher 1994) and genotyped further to 15 loci (Paetkau et al. 1998_a_). To minimize genotyping errors we followed protocols detailed in Woods et al. (1999) and Paetkau (2003). Our sample sizes ranged from 27 to 122 (table 1).
Table 1.
Grizzly bear sample sizes, study area sizes, heterozygosity and STRUCTURE self-assignment percentages for three mountain ranges north and south of BC Highways 3 and 3A in SW Canada and the NW USA.
| sampling area | N | females | males | unknown | area (km2) | _H_E | STRUCTURE % self-assigned |
|---|---|---|---|---|---|---|---|
| Rockies South | 99 | 37 | 54 | 8 | 4695 | 0.67 | 98.3 |
| Rockies North | 122 | 57 | 58 | 7 | 6268 | 0.66 | 94.7 |
| Purcell South | 27 | 13 | 14 | 0 | 4693 | 0.64 | 99.8 |
| Purcell North | 75 | 38 | 35 | 2 | 3117 | 0.66 | 84.7 |
| Selkirk South | 43 | 20 | 20 | 3 | 5500 | 0.54 | 100 |
| Selkirk North | 104 | 45 | 40 | 19 | 9582 | 0.68 | 100 |
| E Flathead (control) | 38 | 16 | 19 | 3 | 1500 | 0.67 | 44.0 |
| W Flathead (control) | 42 | 16 | 22 | 4 | 1580 | 0.67 | 42.6 |
We used two methods to test for individual migrants across the transportation corridor and river valleys, each side of which is herein referred to as an adjacent area. First, we used area-specific allele frequencies in a likelihood-based assignment test (Paetkau et al. 1995) that calculates the probability of each individual's assignment to a particular area as the cumulative products of each allele's frequency of occurrence in any of several areas being examined. The individual is assigned to the area with the highest probability of occurrence. Because the areas we are comparing have recent ancestry, genotypes between adjacent areas may be similar (remnant similar genotypes). It is possible that cross-assigned individuals (assigned to an area other than that of their capture) are not real migrants, but appear as such due to this recent ancestry. To examine our power to distinguish true from statistical migrants we generated significance levels for individuals that cross-assigned to a neighbouring area using the simulation routine within GeneClass 2.0 software (Paetkau et al. 2004; Piry et al. 2004). Significance levels were determined by comparing the individual genotypes of cross-assigned individuals with a simulated set of 10 000 genotypes that were generated using area-specific allele frequencies. While several other assignment methods determine migrant significance based on simulations, we chose the Paetkau et al. (2004) routine because they demonstrated accurate type I error rates, a direct result of their improved simulation process; their simulation routine mimics natural population processes by generating individuals through uniting gametes. For our pool of migrant candidates, we identified individuals in the distribution tails beyond the _α_0.01 or _α_0.05 thresholds. This pool contains putative migrants which can be explained by chance (type I error rate). Any individuals in excess of this number of chance migrants are likely to be true migrants. We identified those migrants by using the Dunn–Sidak adjusted α value (_α_adj; Sokal & Rohlf 1995). This assignment test assumes that all loci in each area are in Hardy–Weinberg and linkage equilibria, which we verified using GENEPOP 3.1.
We independently tested for migrants in a model-based clustering method using a Monte Carlo Markov Chain (MCMC) algorithm (STRUCTURE; Pritchard et al. 2000). STRUCTURE clusters individuals into groups through iterative assignments and develops probabilities of area origin for each individual through the cumulative results of those assignments. Individuals that repeatedly assign to a group other than that of their capture are considered putative migrants from their ‘source’ area. The strength of their migrant status is reflected in the resulting probability of their cross-assignment. An index to the genetic separation of two compared areas can be seen in the percentage of self-assigned individuals that cluster to a group reflecting their area of capture. We assumed that adjacent areas had correlated allele frequencies because bears in this region were recently one continuous population. We used a migration rate of 0.01; varying this rate did not change the results.
We also used genetic distances, _D_LR (Paetkau et al. 1997) and _F_ST (Weir & Cockerham 1984), to compare levels of genetic separation between sampling areas. To control for geographic distance as a variable influencing genetic distance, in each study area only bears within the same geographic distance as our control area (80 km) were used to measure genetic distance in our test areas. Unbiased estimates of mean expected heterozygosity (_H_e) were calculated as an index of relative genetic variability (Nei & Roychoudury 1974).
Fragmentation is the reduction of movement between two areas and can occur at varying intensities. For the purposes of this study, we defined genetic fragmentation as being when movement of both sexes between geographic areas has been limited and in extreme cases can result in an isolated population (where no evidence of interchange of either sex exists). Demographic fragmentation occurs when one sex's movement has been restricted. For example, restricted female movement would limit natural augmentation of a population's reproductive capacity through immigration of females.
3. Results
We identified 470 individual grizzly bears (table 1) and found limited evidence of female movement across the southern distribution of grizzly bears just north of the US–Canada border. Furthermore, we found one isolated small population in the southern Selkirk Mountains. The resulting sample intensity was approximately 50% of the bears living in our study areas. We know this because population estimates were carried out in all six areas (Wielgus et al. 1994; Mowat & Strobeck 2000; Boulanger 2001; M. Proctor, unpublished data; W. Kasworm, unpublished data). The relative large sample sizes and high percentage of the estimated population size enhance our analytical power.
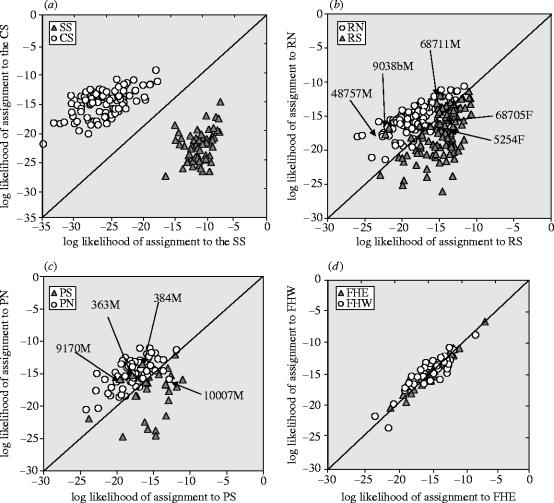
The Selkirk Mountain system showed the greatest degree of fragmentation, with genetic distances across the transportation corridor of _D_LR=14.4 and _F_ST=0.23. We found no evidence of movement for either gender between the Selkirks South and Selkirks North or the Purcells to the east. The assignment plot (figure 2a) suggests that all bears were captured in their population of birth. GeneClass2 and STRUCTURE produced the same result. All individuals in both areas had a resident probability of 1.0 according to GeneClass2 and STRUCTURE yielded 100% self-assignments for both areas (table 1). This population is fragmented demographically and genetically and appears isolated.
Figure 2.
Population assignments of grizzly bears in SW, Canada and NW, US across BC/Alberta Highways 3 and 3A in three mountain ranges and one control system. Individuals highlighted by an arrow are likely migrants across Highway 3 as determined by STRUCTURE and GeneClass2 (see text and table 2). M, male and F, female. (a) Selkirk Mountains, (b) Rocky Mountains, (c) Purcell Mountains and (d) Flathead River (control system) population. Abbreviations as defined in figure 1.
In the Rocky Mountains the genetic distances across the transportation corridor were _D_LR=2.97 and _F_ST=0.035. STRUCTURE clustered individuals into two distinct groups with self-assignment percentages of 94.7 and 98.3 for the Rocky North and South areas, respectively (table 1) suggesting reasonable power to detect migrants. Our migrant results are consistent across α levels and statistical tests. In GeneClass2, at _α_0.05, 15 putative migrants (ten males, five females) between North and South areas were detected (table 2). Eleven of these would be expected by chance due to type I errors (5% of 220) and can be explained as remnant similar genotypes due to recent ancestry (table 3). At _α_0.01, GeneClass2 identified five putative migrants, two of which can be explained by chance (1% of 220). Adjustment of α by the Dunn–Sidak method produced four migrants that cannot be explained by chance and are therefore likely migrants (table 3, figure 2b). STRUCTURE identified the same four individuals as migrants with probabilities >85% (table 2). One _α_0.01 GeneClass2 putative migrant, female 68705, was not selected by STRUCTURE to be a migrant (0.189; table 2). The bimodal distribution of migrant probabilities produced by STRUCTURE is remarkable; the top four migrant candidates are >0.85 and all others are <0.36, quickly diminishing to <0.1 (table 2). Female 5254 had in fact, been previously translocated across the corridor by wildlife managers (figure 2b). Two additional males were DNA-sampled on both sides of the corridor, for a total of five true migrants. Overall, we found possibly one (as female 68705 remains a questionable migrant; table 2) out of 94 females and five out of 112 males (4.4%) have moved across the highway corridor in the Rocky Mountains. The Rocky Mountain system is not genetically fragmented (male interchange occurs) but appears demographically fragmented evidenced by the dearth of female movement.
Table 2.
GeneClass2 and STRUCTURE putative migrants are listed for the Purcell and Rocky study systems.
| bear ID | sex | capture population | assigned population | GeneClass cross-assign log ratio | resident probability | STRUCTURE migrant probability | |
|---|---|---|---|---|---|---|---|
| RN versus RS | |||||||
| _D_LR=2.97 | 9038ba | M | RS | RN | 5.078 | 0.000** | 0.999 |
| 48757a | M | RS | RN | 4.631 | 0.000** | 0.998 | |
| 5254b | F | RN | RS | 3.227 | 0.000** | 0.933 | |
| 68711a | M | RS | RN | 2.933 | 0.004** | 0.876 | |
| 60104a | M | RS | RN | 1.219 | 0.037 | 0.362 | |
| 68705a | F | RN | RS | 1.556 | 0.005* | 0.189 | |
| 1153 | M | RN | RS | 0.934 | 0.015 | 0.121 | |
| 1331 | F | RN | RS | 1.087 | 0.011 | 0.105 | |
| 60108a | RN | RS | 0.731 | 0.013 | <0.0.1 | ||
| 5238 | M | RN | RS | 0.484 | 0.028 | <0.0.1 | |
| 60104a | M | RS | RN | 1.219 | 0.037 | <0.0.1 | |
| 1182 | M | RS | RN | 1.299 | 0.038 | <0.0.1 | |
| 1143 | M | RS | RN | 1.087 | 0.041 | <0.0.1 | |
| 48756a | M | RN | RS | 0.299 | 0.042 | <0.0.1 | |
| 1148 | F | RN | RS | 0.224 | 0.047 | <0.0.1 | |
| PN versus PS | |||||||
| _D_LR=2.04 | 9170 | M | PS | PN | 3.687 | 0.007* | 0.995 |
| 384 | M | PS | PN | 2.759 | 0.017 | 0.991 | |
| 363 | M | PS | PN | 3.146 | 0.012 | 0.985 | |
| 128 | M | PS | PN | 2.278 | 0.024 | 0.945 | |
| 358 | M | PS | PN | 0.000 | 1.000 | 0.149 | |
| 72832a | M | PS | PN | 0.000 | 1.000 | 0.123 | |
| 10007 | M | PN | PS | 3.020 | 0.000** | 0.099 | |
| 355 | M | PS | PN | 0.882 | 0.013 | <0.0.1 | |
| 9104 | M | PN | PS | 0.166 | 0.024 | <0.0.1 | |
| 10005 | M | PN | PS | 1.914 | 0.025 | <0.0.1 |
Table 3.
Summary of migrant decisions in the Purcell and Rocky Mountains relative to analytical method, type I errors and various α thresholds for significance.
| area | alpha | GeneClass bears | GeneClass expected type I errors | GeneClass expected migrants | D–S Adj observed migrants | STRUCTURE observed migrants |
|---|---|---|---|---|---|---|
| Rockies | <0.05 | 15 | 11 | 4 | ||
| _N_=220 | 4 | 4 | ||||
| <0.01 | 5 | 1 | 4 | |||
| Purcells | <0.05 | 8 | 5 | 3 | ||
| _N_=104 | 1 | 4 | ||||
| <0.01 | 2 | 1 | 1 |
In the Purcell Mountains the genetic distances across the transportation corridor were _D_LR=2.04 and _F_ST=0.024. STRUCTURE clustered individuals into two distinct groups with self-assignment percentages of 84.7 and 99.8 for the Purcells North and South areas, respectively (table 1). GeneClass2 identified eight putative migrants at _α_0.05 (expected type I error of five bears, 5% of 104), two at _α_0.01 (expected type 1 error of one bear, 1% of 104) and one using _α_adj (tables 2 and 3; figure 2c). STRUCTURE's bimodal distribution yielded four individuals with high probabilities of being migrants (>94%; table 2) and the remaining bears with migrant probabilities <15%. Male 128 was known to cross from radio-telemetry data (Kasworm et al. 2000). There is inconsistency with the status of one bear, 10007 (table 2); GeneClass2 yielded a relatively high log ratio (3.02) and low resident probability, while STRUCTURE determined its migrant probability to be only 9.9%. Unlike in the Rockies, no evidence of any female movement was detected. One additional male was DNA-sampled on both sides of the highway corridor (and known from radio telemetry), for a total of five migrants between the Purcell areas. The Purcell system is demographically fragmented, as evidenced by the complete lack of female interchange, but appears not to be genetically connected due to male movement.
The results along the Highway 3/3A corridor in all three mountain ranges contrast sharply with the control area in the Flathead River system. Comparing 38 bears captured on the east with 42 on the west side, we found the areas were genetically identical, suggesting that individuals mix freely across the river valley (figure 2d). This result is corroborated by the ∼50% self-assignment rates by STRUCTURE for each of the control units (table 1). The genetic distances between the bears captured on each side of the centre of the control valley were _D_LR=0.15 and _F_ST=0.001, an order of magnitude below the Rocky and Purcell systems and two orders of magnitude below the Selkirk system. Comparing several topographic characteristics of the control and test valleys, the Flathead is the widest valley (100 m from the bottom, control=2–7 km, test=0.5–3.0 km) with a greater distance between the delineating peaks (control=8–20 km, test=4–12 km). Furthermore, no difference exists in vegetation or ecology across the river valleys in all four systems (Medinger & Pojar 1991).
4. Discussion
The ecological and biophysical environments across the four mountain valley systems in this study are essentially identical (Medinger & Pojar 1991). Because the control valley is larger than the test valleys, it should show the largest genetic separation. However it has the smallest genetic distance (a compete mixing), suggesting that factors present in the test valleys are mediating fragmentation. The terrain within each test valley would be considered continuous grizzly bear habitat, just as in the Flathead valley, were it not for the presence of the transportation corridor and associated human settlements. The distance across each test corridor is well within the average daily movement distance of a grizzly bear (2.4 km day−1; B. McLellan, unpublished data). Because of the short spatial distances in each system, the wide-ranging movement capability of grizzly bears and the very low genetic distance with thoroughly integrated assignment plots of the control area (figure 2d), we expected to see similar connectivity in all river valleys sampled. Instead, we found much larger genetic distances across the transportation corridors. In the undisturbed Canadian north, these same genetic distances (Purcells _D_LR=2.04; Rockies _D_LR=2.97) correspond to groups of bears separated by 650 and 1000 km, respectively (Paetkau et al. 1998_b_). Our evidence strongly suggests that the transportation corridor and associated human settlements are fragmenting grizzly bears.
The power to detect the number and sex of migrants moving between geographic areas varies between systems. In the control area we have no power to detect individual movements across the landscape because no genetic separation has occurred. Many animals move across this valley, as determined by 28 years of radio-telemetry data (B. McLellan, unpublished data). Conversely, in the Selkirk system we have excellent power to detect migrants (figure 2a) because of distinct genetic separation; no migrants were detected in this study. In the Rocky and Purcell Mountains we had limited but adequate power to detect individual migrants as genetic separation had occurred, but to a lesser degree than in the Selkirks. This power is demonstrated by the ability of STRUCTURE to separate the groups into relatively distinct groups based on the dynamic iterative assignment cluster analysis and to separate migrants from residents so clearly with bimodal migrant probability distributions (tables 1 and 2). We found that the allele frequencies in the Purcells and Rockies were sufficiently distinct to detect more individuals in the tails of the migrant probability distributions than can be explained by chance (GeneClass2) and these migrant choices were corroborated by STRUCTURE. We chose the GeneClass2 method because of its use of an improved simulation routine (Paetkau et al. 2004) that more closely mimics natural population processes by developing individuals through united gametes. We found that this process results in accurate type I error rates as theoretically demonstrated in Paetkau et al. (2004). STRUCTURE may be more sensitive than GeneClass2 in detecting migrants between areas that share recent ancestry and have only moderate genetic structure, as suggested by the bimodal probability distribution evident in the Purcell system (tables 2 and 3).
All putative migrants are heavily skewed toward males and no female migrants were detected by either method, other than the translocated female in the Rockies. Sex-biased dispersal is widespread in mammals (Greenwood 1980; Pusey 1987), has been demonstrated in grizzly bears in North America by comparing mtDNA and nDNA (Paetkau et al. 1998_a_) and is documented in our study area (McLellan & Hovey 2001; Proctor et al. 2004). The lesser dispersing sex may be more affected by human influence and this is evident in our results. Because each test valley is <3 km wide and in these areas females disperse on average 10–14 km (McLellan & Hovey 2001; Proctor et al. 2004), the limited female movement observed across the corridors is unexpected. In contrast, both males and females in the unpopulated control valley were moving across the valley (B. McLellan, unpublished data). While male movement in the Purcells and Rockies is mediating genetic connectivity, the limited female movement is cause for demographic concern. This disruption of the female dispersal process diminishes the possibility of natural population augmentation or re-colonization (Lande 1988) and may have serious implications for the small south Purcell and Selkirk areas along the Canada–USA border. The south Purcell area has an estimated 40–50 bears (Kasworm et al. 2000; M. Proctor, unpublished data) and is declining at approximately 3.7% per year (Wakkinen & Kasworm 2004). The predominance of movement from north to south in the Purcells suggests that grizzly persistence to the south of Highway 3 into the USA may depend on connectivity to the north. The demographically and genetically isolated south Selkirk population has an estimated 70–100 bears (Wielgus et al. 1994) and although the population is reported to be stable (Wakkinen & Kasworm 2004), it should be considered threatened.
The threat of local grizzly extirpation is primarily driven by demographic forces in the form of human-induced mortality (McLellan et al. 1999). Without demographic connectivity, the two small demographically isolated populations revealed in this study are vulnerable to stochastic events and are reliant on positive fecundity rates, a challenge in a context of negative human–bear interactions. Loss of these two border populations would leave only one US border population tenuously linked to Canadian populations.
While the Rocky Mountain population south of Highway 3 is experiencing demographic fragmentation, it is still genetically connected (male mediated) to bears in the north and is relatively large (>400 animals; B. McClellan, unpublished data); as such it is not under immediate conservation risk due to fragmentation. However, consideration should be given to the paucity of female connectivity across the Highway 3 corridor where monitoring and enhanced connectivity management may be necessary.
The mechanism leading to limited movement across the transportation and settlement corridor is probably a combination of some bears avoiding human activity centres (Mattson et al. 1987) and increased grizzly bear mortality in these areas, due to bear attractants such as garbage, human food and perceived threats to human safety (McLellan et al. 1999). Dispersal of male and female grizzly bears in the southeastern BC region requires several years, resulting in newly established home ranges that usually overlap or are adjacent to the maternal home range (McLellan & Hovey 2001). When this gradual dispersal process requires bears to spend time in human-dominated landscapes, mortality rates can increase; out of the three natural migrants detected in the Rockies by both GeneClass2 and STRUCTURE, all are dead, killed either by hunters or as ‘problem wildlife’. During the past 10 years, 60 grizzly bears were removed from the Rocky Mountain study area by conservation officers because of conflicts with people, and over the past 25 years an additional 500 were harvested legally (BC Ministry of Water, Land and Air Protection files). These mortalities may also decrease density-dependent movement within the system (Swenson et al. 1998; McLellan & Hovey 2001). We suspect that human-caused mortality is a contributory factor to decreased migration (Proctor 2003) and recommend management to reduce human–bear conflict and ultimately mortality in the transportation/settlement corridors. Further, we recommend research to locate areas with landscape attributes that foster successful bear movement through these corridors. These identified linkage areas can then be managed accordingly. We also recommend further work to determine the benefits and feasibility of population augmentation in the south Purcell and Selkirk populations.
Demographic processes appear to be the dominant influence over grizzly bear persistence within North America (McLellan et al. 1999). Excessive mortality and isolation played a primary role in the extirpation of approximately 31 small isolated grizzly bear populations between 1922 and 1970 within the conterminous USA (Mattson & Merrill 2002). There is evidence that the deleterious effects of reduced genetic variation are minimal in grizzly bears. For example, Kodiak Island grizzly bears have 33% reduction in heterozygosity (_H_E) and have been thriving for centuries (Paetkau et al. 1998_a_). However, this population is large (ca 3000; L. Van Daele, personal communication) relative to the isolated southern Selkirk population of 70–100 bears (20% reduction in _H_E; table 1), and the effects of inbreeding depression tend to be more detrimental for small populations (Frankham et al. 2002).
There is a recent but growing body of evidence that anthropogenic fragmentation is influencing carnivore populations in North America, but no studies documenting sex-specific differences in fragmentation. Schwartz et al. (2002) found little genetic structure in lynx (L. canadensis) populations in western North America and recommended maintenance of connectivity. At a finer scale, Campbell (2002) found genetic structure in lynx across a major highway in Alberta, Canada, suggesting that human disturbance may be influencing connectivity. Kyle & Strobeck (2002) found increased genetic structure in southern peripheral populations relative to northern core populations in the North American wolverine (Gulo gulo), although they did not measure immediately adjacent populations within the dispersal distance of a wolverine. Cegelski et al. (2003) also found wolverines to be fragmented in the southern periphery of their western North America distribution.
Our methods demonstrate the importance of using individual-based analyses in addition to traditional population genetics techniques (e.g. _F_ST and genetic distance) to provide insight into gender-specific processes of immigration and emigration in recently disturbed systems. Our results underscore the need for connectivity management and highlight the importance of international co-operation for the management of highly vagile animals.
Acknowledgments
We thank C. Lausen, D. Paetkau, J. Bonneville and J. Bergenske for editing, advice, lab and field assistance, as well as the BC Ministry of Water, Land and Air Protection, BC Parks, BC Forest Service, Natural Science and Engineering Research Council, Killam Foundation, Wilburforce Foundation for funding support and Alberta Sustainable Resource Development, Fish and Wildlife Division for providing samples.
References
- Boulanger J. BC Ministry of Environment, Wildlife Branch; Victoria, BC, Canada: 2001. Analysis of the 1997 Elk Valley and Flathead Valley DNA mark-recapture grizzly bear inventory projects. [Google Scholar]
- Campbell, V. A. 2002 Population genetic structure of Canada lynx (Lynx canadensis) in Alberta. M.Sc. thesis, University of Alberta, Edmonton, Alberta.
- Cegelski C, Waits L.P, Anderson N. Using assignment tests to evaluate gene flow in Montana wolverines (Gulo gulo) Mol. Ecol. 2003;12:2907–2918. doi: 10.1046/j.1365-294x.2003.01969.x. 10.1046/j.1365-294X.2003.01969.x [DOI] [PubMed] [Google Scholar]
- Ennis S, Gallagher T.F. PCR based sex determination assay in cattle based on the bovine Amelogenin locus. Anim. Genet. 1994;25:425–427. doi: 10.1111/j.1365-2052.1994.tb00533.x. [DOI] [PubMed] [Google Scholar]
- Forbes S.H, Boyd D.K. Genetic variation of naturally occurring wolves in the central rocky mountains. Conserv. Biol. 1996;10:1082–1090. 10.1046/j.1523-1739.1996.10041082.x [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Cambridge University Press; Cambridge, UK: 2002. Introduction to conservation genetics. [Google Scholar]
- Greenwood P.J. Mating systems, philopatry, and dispersal in birds and mammals. Anim. Behav. 1980;28:1140–1162. [Google Scholar]
- Hanski I.A, Gilpin M.E, editors. Metapopulation biology: ecology and evolution. Academic Press; Toronto: 1997. [Google Scholar]
- Kasworm W.F, Carriles H, Radandt T.G. United States Fish and Wildlife Service. Grizzly Bear Recovery Coordinator's Office; Missoula, Montana: 2000. Cabinet–Yaak grizzly bear recovery area 1999 research and monitoring progress report. [Google Scholar]
- Kyle C.J, Strobeck C. Connectivity of peripheral and core populations of North American Wolverines. J. Mamm. 2002;83:1141–1150. 10.1644/1545-1542(2002)083%3C1141:COPACP%3E2.0.CO;2 [Google Scholar]
- Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- Manel S, Gaggioti O.E, Waples R.S. Assignment methods: matching biological questions with appropriate methods. Trends Ecol. Evol. 2005;20:136–142. doi: 10.1016/j.tree.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Mattson D.J, Merrill T. Extirpations of grizzly bears in the contiguous United States. Conserv. Biol. 2002;16:1123–1136. 10.1046/j.1523-1739.2002.00414.x [Google Scholar]
- Mattson D.J, Knight R.R, Blanchard B. The effects of development and primary roads on grizzly bear habitat use in Yellowstone National Park, Wyoming. Int. Conf. Bear Res. Manage. 1987;7:259–274. [Google Scholar]
- McLellan B.N. Maintaining viability of brown bears along the southern fringe of their distribution. Ursus. 1998;10:607–611. [Google Scholar]
- McLellan B.N, Hovey F. Natal dispersal of grizzly bears. Can. J. Zool. 2001;79:838–844. 10.1139/cjz-79-5-838 [Google Scholar]
- McLellan B.N, Hovey F, Mace R.D, Woods J.G, Carney D.W, Gibeau M.L, Wakkinen W.L, Kasworm W.F. Rates and causes of grizzly bear mortality in the interior mountains of British Columbia, Alberta, Montana, Washington, and Idaho. J. Wildlife Manage. 1999;63:911–920. [Google Scholar]
- Mech L.D. The challenge and opportunity of recovering wolf populations. Conserv. Biol. 1995;9:270–278. 10.1046/j.1523-1739.1995.9020270.x [Google Scholar]
- Medinger D, Pojar J, editors. Ecosystems of British Columbia. British Columbia Ministry of Forests Special Report Series 6. British Columbia Ministry of Forests; Victoria, BC: 1991. [Google Scholar]
- Mowat G, Strobeck C. Estimating population size of grizzly bears using hair capture, DNA profiling, and mark–recapture analysis. J. Wildlife Manage. 2000;64:183–193. [Google Scholar]
- Nei M, Roychoudury A.K. Sampling variances of heterozygosity and genetic distance. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak M, Baker J, Obbard M, Malloch B, editors. Wildlife furbearer management and conservation in North America. Ontario Ministry of Natural Resources; Toronto, Ontario: 1987. [Google Scholar]
- Paetkau D. An empirical exploration of data quality in DNA-based population inventories. Mol. Ecol. 2003;12:1375–1387. doi: 10.1046/j.1365-294x.2003.01820.x. 10.1046/j.1365-294X.2003.01820.x [DOI] [PubMed] [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995;3:489–495. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Paetkau D.H, Waits L.P, Clarkson P.L, Craighead L, Strobeck C. An empirical evaluation of genetic distance statistics using microsatellite data from bear (Ursidae) populations. Genetics. 1997;147:1943–1957. doi: 10.1093/genetics/147.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D.H, Shields G.F, Strobeck C. Gene flow between insular, coastal, and interior populations of brown bears in Alaska. Mol. Ecol. 1998a;7:1283–1292. doi: 10.1046/j.1365-294x.1998.00440.x. 10.1046/j.1365-294x.1998.00440.x [DOI] [PubMed] [Google Scholar]
- Paetkau D.H, Waits L.P, Waser P, Clarkson L, Craighead L, Vyse E, Ward R, Strobeck C. Variation in genetic diversity across the range of North American brown bears. Conserv. Biol. 1998b;12:418–429. 10.1046/j.1523-1739.1998.96457.x [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol. Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294x.2004.02008.x. 10.1046/j.1365-294X.2004.02008.x [DOI] [PubMed] [Google Scholar]
- Pearse D.E, Crandall K.A. Beyond FST. Analysis of population genetic data for conservation. Conserv. Genet. 2004;5:585–602. 10.1007/s10592-003-1863-4 [Google Scholar]
- Piry S, Alapetite A, Cornuet J.-M, Paetkau D, Baudouin L, Estoup A. GeneClass2: a software for genetic assignment and first generation migrants detection. J. Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donnelly P. Inferences of population structure using multilocus genotyping data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, M. F. 2003 Genetic analysis of movement, dispersal and population fragmentation of grizzly bears in southwestern Canada. Ph.D. thesis, University of Calgary, Alberta.
- Proctor M.F, McLellan B.N, Strobeck C, Barclay R.M.R. Gender specific dispersal distances for grizzly bears analysis revealed by genetic analysis. Can. J. Zool. 2004;82:1108–1118. 10.1139/z04-077 [Google Scholar]
- Pusey A.E. Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends Ecol. Evol. 1987;2:295–299. doi: 10.1016/0169-5347(87)90081-4. 10.1016/0169-5347(87)90081-4 [DOI] [PubMed] [Google Scholar]
- Schwartz M.K, Mills L.S, McKelvey K.S, Ruggiero L.F, Allendorf F.W. DNA reveals high dispersal synchronizing the population dynamics of Canada lynx. Nature. 2002;415:520–522. doi: 10.1038/415520a. 10.1038/415520a [DOI] [PubMed] [Google Scholar]
- Servheen C, Herrero S, Peyton B. IUCN/SSC Bear and Polar Bear Specialist Groups; Cambridge, UK: 1999. Bears status survey and conservation action plan. [Google Scholar]
- Sokal R.R, Rohlf F.J. 3rd edn. W. H. Freeman and Company; New York: 1995. Biometry. [Google Scholar]
- Swenson J.E, Sandergren F, Söderberg A. Geographic expansion of an increasing brown bear population: evidence for presaturation dispersal. J. Anim. Ecol. 1998;67:819–826. 10.1046/j.1365-2656.1998.00248.x [Google Scholar]
- U.S.F.W.S. U.S. Fish and Wildlife Service; Missoula, Montana: 1993. Grizzly bear recovery plan. [Google Scholar]
- Wakkinen W.L, Kasworm W.F. Demographic and population trends of grizzly bears in the Cabinet–Yaak and Selkirk ecosystems of British Columbia, Idaho, Montana, and Washington. Ursus. 2004;15:65–75. 10.2192/1537-6176(2004)015%3C0065:DAPTOG%3E2.0.CO;2 [Google Scholar]
- Waser P.M, Strobeck C. Genetic signatures of interpopulation dispersal. Trends Ecol. Evol. 1998;13:43–44. doi: 10.1016/s0169-5347(97)01255-x. 10.1016/S0169-5347(97)01255-X [DOI] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Whitlock M.C, McCauley D.E. Indirect measures of gene flow and migration. Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. 10.1038/sj.hdy.6884960 [DOI] [PubMed] [Google Scholar]
- Wielgus R.B, Bunnell F.L, Wakkinen W.L, Zager P.E. Population dynamics of Selkirk mountain grizzly bears. J. Wildlife Manage. 1994;58:266–272. [Google Scholar]
- Wilcox B.A, Murphy D.D. Conservation strategy: the effects of fragmentation on extinction. Am. Nat. 1985;125:879–887. 10.1086/284386 [Google Scholar]
- Woodroffe R, Ginsberg J.R. Edge effects and the extinction of populations inside protected areas. Science. 1998;280:2126–2128. doi: 10.1126/science.280.5372.2126. 10.1126/science.280.5372.2126 [DOI] [PubMed] [Google Scholar]
- Woods J.G, Paetkau D, Lewis D, McLellan B.N, Proctor M, Strobeck C. Genetic tagging of free-ranging black and brown bears. Wildlife Soc. Bull. 1999;27:616–627. [Google Scholar]