Molecular Diversity of Denitrifying Genes in Continental Margin Sediments within the Oxygen-Deficient Zone off the Pacific Coast of Mexico (original) (raw)
Abstract
To understand the composition and structure of denitrifying communities in the oxygen-deficient zone off the Pacific coast of Mexico, the molecular diversity of nir genes from sediments obtained at four stations was examined by using a PCR-based cloning approach. A total of 50 operational taxonomic units (OTUs) for nirK and 82 OTUs for nirS were obtained from all samples. Forty-four of the nirS clones and 31 of the nirK clones were sequenced; the levels of similarity of the nirS clones were 52 to 92%, and the levels of similarity of the nirS clones were 50 to 99%. The percentages of overlapping OTUs between stations were 18 to 30% for nirS and 5 to 8% for nirK. Sequence analysis revealed that 26% of the nirS clones were related to the nirS genes of Alcaligenes faecalis (80 to 94% similar) and Pseudomonas stutzeri (80 to 99%), whereas 3 to 31% of the nirK clones were closely related to the nirK genes of Pseudomonas sp. strain G-179 (98 to 99%), Bradyrhizobium japonicum (91%), Blastobacter denitrificans (83%), and Alcaligenes xylosoxidans (96%). The rest of the clones, however, were less than 80% similar to nirS and nirK sequences available in sequence databases. The results of a principal-component analysis (PCA) based on the percentage of OTUs and biogeochemical data indicated that the nitrate concentration and oxygen have an effect on the denitrifying communities. The communities at the stations in oxygen-deficient zones were more similar than the communities at the stations in the oxygenated zone. The denitrifying communities were more similar at the stations that were closer together and had similar nitrate levels. Also, the results of PCA based on biogeochemical properties suggest that geographic location and biogeochemical conditions, especially the nitrate and oxygen levels, appear to be the key factors that control the structure of denitrifying communities.
The continental margin occupies only a fraction of the total ocean, but it is critical for carbon and nutrient cycling. This margin contributes 30 to 50% of the total marine primary productivity (28, 43). The high productivity results in high carbon input into margin sediments that stimulates rapid benthic carbon and nutrient cycling (12, 18). Nearly 90% of marine carbon burial (permanent sequestration) occurs in margin sediments (5, 24). Recent studies have suggested that the present-day oceanic nitrogen budget is unbalanced (15), although some evidence suggests that it may be balanced (11, 22). In the unbalanced view of the N budget, the rate of supply of nitrogen to the ocean is much lower than the removal rate, primarily due to denitrification (1, 14, 20, 31). Denitrification contributes both indirectly and directly to decreasing carbon sequestration. It decreases the amount of nitrogen available to phytoplankton, thus affecting primary productivity. It also produces greenhouse gases, such as nitric (NO) and nitrous oxide (N2O), which contribute to global warming and the destruction of the ozone layer (16, 27).
The uncertainty about estimating marine denitrification is a problem in understanding global N dynamics. Marine denitrification occurs in sediments, primarily continental shelf and slope sediments, as well as in oxygen-deficient water columns (13, 17, 18). Not only is sedimentary denitrification the largest sink in the N budget, it is also one of the most poorly quantified (10, 14, 17). This is partially due to the fact that prediction of denitrification is difficult, and estimates of oceanic sedimentary denitrification vary severalfold (19, 31). The difficulty is compounded by considerable uncertainty about the organisms and control of the dynamics of denitrification in the marine environment. Thus, understanding the diversity of denitrifying bacterial populations in marine environments, the responses of microbial communities to environmental factors (e.g., O2, carbon, and NO3−), and the impact of changes in microbial community structure and composition on the rate of denitrification is critical in understanding global N dynamics and how it might be altered with global change.
The genetic diversity of denitrifiers in marine sediments has been explored by using specific genes as functional markers. Braker et al. (7, 8) used the nitrite reductase genes (nirK and nirS), and the nitrous oxide reductase gene, nosZ, has also been used (34, 36). In one study, Braker et al. (7) found that distinct denitrifier communities were correlated with the oxygen exposure time of the carbon in the overlying water. Later, Braker et al. (8) observed that the denitrifying community structures were very similar at different depths of sediments, although the oxidant profiles were different. Scala and Kerkhof (36) observed horizontal heterogeneity of denitrifying bacterial communities in marine sediments.
Oxygen-deficient zones are considered major sites for water column denitrification and combined nitrogen loss. No studies of the genetic diversity of denitrifiers in marine sediments have been conducted in an oxygen-deficient zone, such as the one off western Mexico. To understand the composition and structure of denitrifying communities in an oxygen-deficient zone and to compare the composition and structure in such a zone to the composition and structure in more typical oceanic regions, the molecular diversity of nirS and nirK genes was investigated in this study by using a PCR-based cloning approach. Samples were obtained from the oxygen-deficient zone off the western coast of Mexico, one of the three major oxygen-deficient zones in the world. The Mexican margin is characterized by a strong oxygen-deficient zone in the water column at depths between about 150 and 1,000 m. The primary productivity was, at most, 100 g of C m−2 year−1 (29). Our results suggest that geographic location, biogeochemical properties (especially nitrate levels), and the oxygen profile affect the structures of the denitrifier communities in marine sediments.
MATERIALS AND METHODS
Study area and sediment sampling.
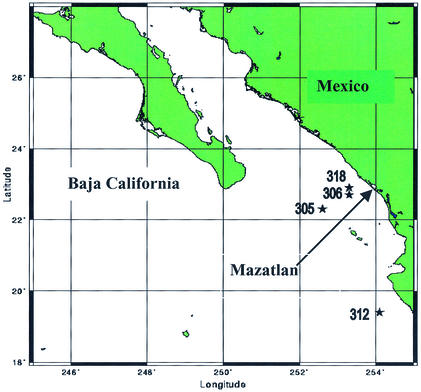
Marine sediment samples were collected from the continental margin of northwestern Mexico along a transect consisting of four stations (stations 305, 306, 312, and 318) (Fig. 1) during a cruise of the R/V New Horizon in fall 1999 (Table 1). Sediment cores with overlying water were collected at all stations with a Soutar box core, and subcores were obtained by using 7.5- and 10-cm cast acrylic tubes (28). Bulk sediment samples from selected depths within the subcores were obtained by extruding the sediments from the subcores and collecting the desired depth intervals, and samples for DNA analysis were kept at −20°C. All stations except station 305 were within the oxygen-deficient zone.
FIG. 1.
Locations of sampling stations in the continental margin off the Pacific coast of Mexico. The map was created by Online Map Creation (http://www.aquarius.geomar.de/omc/).
TABLE 1.
Locations (latitude and longitude), depth, and biogeochemical properties for the four stations (stations 305, 306, 312, and 318) in the continental margin off Mexico
| Station | Location | Depth (m) | Organic carbon (%) | Total nitrogen (%) | C/N ratio | NH4+ (μM) | NO3− (μM) | Denitrification rate (mmole m−1 day−1) | PO4+ (μM) | Fe(II) (μM) | Temp (°C) | No.of unique clonesa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nirS | nirK | ||||||||||||
| 305 | 22°11.41′N, 107°10.08′W | 2,946 | 2.9 | 0.29 | 10.0 | 17.8 | 6.9 | 0.23 | 1.5 | 0.2 | 1.6 | 12 | 15 |
| 306b | 22°41.54′N, 106°28.57′W | 340 | 11.2 | 1.20 | 9.3 | 125.6 ± 30 | 0.5 ± 0.15 | 0.85 ± 0.26 | 130.5 ± 27 | 5.4 ± 2.0 | 9.5 | 12 | 12 |
| 312 | 19°36.14′N, 105°31.12′W | 860 | 9.8 | 1.01 | 9.7 | 43.8 | 2.3 | 1.17 | 18.2 | 1.5 | 5.3 | 27 | 11 |
| 318 | 22°43.04′N, 106°23.32′W | 179 | 4.8 | 0.46 | 10.5 | 67.5 | 0.0 | 1.51 | 87.8 | 18.0 | 12.1 | 31 | 12 |
Oxygen profiles and chemical analysis of sediment samples.
Whole-core squeezing at in situ temperatures by a technique similar to that of Bender et al. (3) was used to obtain high-resolution pore water profiles of oxygen and nitrate as described by Brandes and Devol (9). Pore water was separated from the sediments by centrifugation at 1,000 × g for 20 min.
Nutrient (NH4+, NO3−, and PO4−) contents of the sediments were measured by the methods of Strickland and Parson (40). Nitrate samples from the whole-core squeezer were analyzed by using a small-volume flow injection analysis technique based on the cadmium reduction method described by Anderson (2). Dissolved iron was analyzed colorimetrically by using the ferrozine method outlined by Stookey (39). The percentages of organic carbon and total nitrogen were determined with freeze-dried, ground sediment samples by the method of Hedges and Stern (25) by using either a Carlo-Erba model 1106 CHN elemental analyzer or a Leeman Laboratories CHNS elemental analyzer. Sedimentary denitrification rates were modeled from the nitrate profiles by assuming that there was a simple, steady-state diffusion reaction system in which the downward diffusion of nitrate balanced the removal rate due to denitrification: _d_NO3/dt = 0 = Ds · δ2NO3/δ_z_2 + R, where Ds is the tortuosity-corrected sediment diffusion coefficient, z is depth from the sediment-water interface, t is time, and R is the denitrification rate. This model lacks any term for bioturbation and does not include any denitrification that may be coupled to nitrification. Thus, it provides only a minimum estimate of the denitrification rate. However, all samples except the station 305 sample were obtained from the oxygen-deficient zone or below, where bioturbation and coupled nitrification-denitrification should be minimal (23); also, due to the depth at station 305 these processes should have been insignificant (33). The equation was solved by using the Profiler algorithm (4).
DNA extraction, primer design, PCR amplification, and restriction fragment length polymorphism (RFLP) analysis.
For all four stations, DNA was extracted only from the top sediment layer, as follows: for station 305, 0.55 to 1.0 cm; for station 306, 0.0 to 0.5 cm; for station 312, 0 to 0.5 cm; and for station 318, 0 to 0.5 cm. The bulk community DNA was directly extracted from 2-g sediment samples by using combined methods that included grinding, freezing and thawing, and treatment with sodium dodecyl sulfate for cell lysis (26, 46). The crude DNA was purified by the minicolumn purification method (46), except that the DNA was eluted twice from the resin column with 50 μl of hot water (80°C) each time.
Several denitrifying bacteria belonging to subclasses α, β, and γ of the class Proteobacteria that carry nirS genes, including Paracoccus denitrificans Pd1222 (α subclass), Alcaligenes eutrophus H16 (β subclass), Pseudomonas aeruginosa NCTC 6750 (γ subclass), Pseudomonas stutzeri JM300 (γ subclass), and Pseudomonas stutzeri ZoBell (ATCC 14405) (γ subclass), and denitrifying bacteria that carry nirK genes, such as Rhizobium hedysari (α subclass), Rhodobacter sphaeroides (α subclass), Pseudomonas sp. strain G-179 (α subclass), Achromobacter cycloclastes (α subclass), Alcaligenes faecalis S-6 (β subclass), and Pseudomonas aureofaciens (γ subclass), were used to test the specificity of the primer sets. Burkholderia cepacia G4, a nondenitrifying strain belonging to the β subclass of the Proteobacteria, was used as a negative control. These bacteria were grown aerobically overnight in Luria broth, and the genomic DNA was extracted as described previously (45).
Conserved primers were designed by comparing nir sequences by using the ARB probe program (41). To achieve specificity, mismatches near the 3′ ends of the primers were designed to be minimal for the organisms of the target groups and maximal for the reference strains. The nirS primers (Heme 832F [5′ TAC CAC CCC GAG CCG CGC GT 3′] and Heme 1606R [5′ AGK CGT TGA ACT TKC CGG TCG G 3′]) were designed to amplify an approximately 800-bp region of the nirS gene by comparing the available sequences of P. denitrificans Pd1222, A. eutrophus H16, P. aeruginosa NCTC 6750, and P. stutzeri JM300 and ZoBell (ATCC 14405) (K = G, T, or U). The nirK primers (Copper 583F [5′ TCA TGG TGC TGC CGC GKG ACG G 3′] and Copper 909R [5′ GAA CTT GCC GGT PGC CCA GAC 3′]) were designed to amplify an approximately 348-bp region based on the previously published sequences of R. hedysari, A. cycloclastes, A. faecalis S-6, P. aureofaciens, R. sphaeroides, and Pseudomonas sp. strain G-179.
PCRs were performed with a Gene Amp PCR System 9700 thermal cycler (Applied Biosystems, Norwalk, Conn.) by using a 20-μl (total volume) reaction mixture containing 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, 0.1% Triton X-100; pH 9.0), each of the deoxyribonucleotide triphosphates (dTTP, dCTP, dGTP, and dATP) at a concentration of 1 mM, 1.5 mM MgCl2, each primer at a concentration of 1 μM, 4 μg of bovine serum albumin (Roche Diagnostics Corp., Indianapolis, Ind.), and 2.5 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). To minimize PCR artifacts, the PCR amplification conditions were optimized based on the suggestions described previously (32). The amplification conditions were one cycle of 80°C for 30 s and 94°C for 2 min, followed by 25 cycles of 94°C for 30 s, 60°C (for nirK) or 65°C (for nirS) for 1 min, and 72°C for 1 min, with a final extension step of 72°C for 7 min. To avoid potential sample biases and to obtain enough PCR products for cloning, five replicate amplifications were carried out for each sample. The samples were then pooled and dried to a volume of about 15 μl. The PCR products were quantified and used for cloning and sequencing. Two-microliter aliquots of the PCR products were analyzed on 1.5% agarose gels. The amounts of the PCR-amplified nirS and nirK gene products were estimated by comparing the band intensities on agarose gels to the band intensities of known concentrations of standard lambda DNA. The amplified PCR products were directly ligated to the pCR II vector obtained from Invitrogen (San Diego, Calif.). Ligation and transformation were carried out as described previously (48). All white colonies were picked and screened for desired gene inserts, which were detected with primers specific for the polylinker of the vector pCR II (47).
A total of 392 nirS clones and 378 nirK clones were screened from the four samples. Unique nirS and nirK clones were detected by RFLP analysis with two tetrametric enzymes (_Msp_I and _Rsa_I). Enzyme digestion and gel electrophoresis of the digested products were performed as described previously (48). The RFLP patterns were analyzed and clustered with the Molecular Analyst 1.6 software (Applied Math, Kortrijk, Belgium) by using the unweighted pair group method with arithmetic averages and the Jaccard algorithm. The resulting clusters were validated visually by comparing the clusters with gel images.
Sequencing and phylogenetic analysis.
Ward (44) investigated sequence divergence in ribosomal genes of known strains and isolates of aquatic denitrifying bacteria using RFLP analysis. In her study, RFLP analysis clustered most of P. stutzeri strains together but detected a considerable degree of diversity within this group. To understand phylogenetic diversity, representative nirK and nirS clones that occurred more than once in a given library, as well as representatives of some of the unique OTUs as determined by cluster analysis based on RFLP patterns, were partially sequenced. A total of 82 nirS clones and 50 nirK clones were partially sequenced, but only 44 nirS clones and 31 nirK clones were used in the phylogenetic tree analyses. Amplified double-stranded DNA templates were purified for DNA sequencing by using an ArrayIt PCR purification kit (TeleChem International Inc., Sunnyvale, Calif.). DNA sequencing was performed with an ABI PRISM BigDye terminator cycle sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.) and an ABI PRISM 3700 DNA analyzer (Applied Biosystems). One microliter (about 30 ng) of purified DNA was used for each sequencing reaction. The vector-specific primers TAF (5′-GCCGCCAGTGTGCTGGAATT-3′) and TAR (5′-TAGATGCATGCTCGAGCGGC-3′) were then used for sequencing (29). DNA sequences were assembled and edited by using the Sequencher program, version 4.0 (Gene Codes Corporation, Ann Arbor, Mich.).
Preliminarily analysis of the sequences was carried out by searching the current databases by using the program FASTA in the Genetics Computer Group software package (21). The nirS or nirK sequences obtained in this study were aligned with all of those available in current databases by using the PILEUP program in the Genetics Computer Group software package. The alignments were edited by using the genetic data environment (38). The initial phylogenetic trees were based on all available sequences and were constructed by using the DNA distance program Neighbor-Joining with Felsenstein correction in ARB (38). Based on the initial phylogenetic results, appropriate subsets of nirS or nirK sequences were selected and subjected to a final phylogenetic analysis by using the maximum-likelihood method with the program fastDNAml in the Ribosomal Database Project (30). Final phylogenetic trees were constructed with a transition/transversion ratio of 2.0 by using jumbled orders of 10 for the addition of taxa. The accession numbers of the sequences have been deposited in the GenBank database (see below).
Statistical methods.
Principal-component analysis (PCA) was performed by using the SYSTAT statistical computing package (version 10.0; SPSS, Inc., Chicago, Ill.). PCA could provide a means to separate and group sediment samples based on their complex biogeochemical profiles and denitrifying community patterns, since it simultaneously considers many correlated variables and then identifies the lowest number to accurately represent the structure of the data (37). In the present study, PCA was used to group or separate stations, which were similar or different, based on the percentages of operational taxonomic units (OTUs) (unique RFLP patterns) obtained from nirS, nirK, _nirS_-nirK, and biogeochemical data [organic carbon, total nitrogen, C/N ratio, NH4+, NO3−, denitrification rate, PO4−3, Fe(II), and temperature] for each station. For PCA based on the percentages of OTUs, the relative amounts of the unique clones for each station were used as variables. In contrast, the biogeochemical parameters were selected as variables for PCA based on biogeochemical properties. To determine which biogeochemical parameters contributed to the differences among stations, PCA results were also computed based on water depth, oxygen, and biogeochemical data [organic carbon, total nitrogen, C/N ratio, NH4+, NO3−, denitrification rate, PO4−3, Fe(II), and temperature] for all four stations. In this analysis, the stations (stations 305, 306, 312, and 318) were used as variables.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 44 nirS clones are as follows: AY195897 (M318a45), AY195898 (M318b36), AY195899 (M312b96), AY195900 (M318b85), AY195901 (M305015), AY195902 (M312b01), AY195903 (M305029), AY195904 (M312b67), AY195905 (M312b48), AY195906 (M306b04), AY195907 (M312b20), AY195908 (M312a19), AY195909 (M318a21), AY195910 (M318b27), AY195911 (M312a24), AY195912 (M305027), AY195913 (M318a58), AY195914 (M306a68), AY195915 (M318a19), AY195916 (M312a89), AY195917 (M306a26), AY195918 (M312a38), AY195919 (M318a96), AY195920 (M318b18), AY195921 (M318a50), AY195922 (M318a07), AY195923 (M305044), AY195924 (M318a86), AY195925 (M305010), AY195926 (M312b45), AY195927 (M318a38), AY195928 (M318a42), AY195929 (M318b04), AY195930 (M318b12), AY195931 (M318a25), AY195932 (M318a80), AY195933 (M305059), AY195934 (M318a36), AY195935 (M306b03), AY195936 (M312a29), AY195937 (M312a93), AY195938 (M306b38), AY195939 (M318b28), and AY195940 (M318b34).
The GenBank accession numbers for the 31 nirK clones are as follows: AY195804 (M306051), AY195805 (M306066), AY195806 (M312079), AY195807 (M305100), AY195808 (M318006), AY195809 (M312087), AY195810 (M318049), AY195811 (M306034), AY195812 (M318095), AY195813 (M306013), AY195814 (M305073), AY195815 (M306026), AY195816 (M312084), AY195817 (M318061), AY195818 (M318029), AY195819 (M318014), AY195820 (M305039), AY195821 (M312053), AY195822 (M306071), AY195823 (M312010), AY195824 (M305088), AY195825 (M306027), AY195826 (M306061), AY195827 (M306012), AY195828 (M306069), AY195829 (M305109), AY195830 (M305044), AY195831 (M312045), AY195832 (M318015), AY195833 (M312068), and AY195834 (M305054).
RESULTS
Biogeochemical properties of sediments.
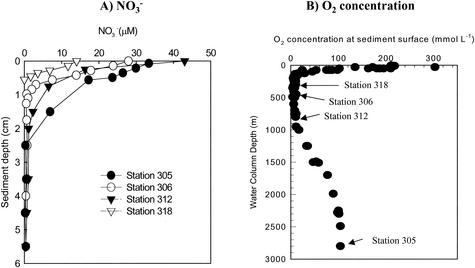
The sediments in the Mexican margin region were rich in organic carbon, and the concentration increased from 4% at 150 m to almost 10% at 1,000 m (23). Station 305, the deepest station, was outside the oxygen-deficient zone and was the least similar station in terms of biogeochemical properties compared to the other stations; it had the highest oxygen and NO3− concentrations and the lowest denitrification rate. The water column oxygen distribution was typical of that in the permanent oxygen-deficient zone of the eastern tropical North Pacific Ocean (Fig. 2B) (9, 15, 20, 22, 23, 42). In this zone, benthic macrofauna were virtually absent, and none of the cores showed that there was detectable oxygen in the overlying water or within the sediments. The station 305 sample had a bottom water oxygen concentration of 104 μM and a shallow penetration depth, less than 1 cm (three replicate profiles); this penetration depth fits well with the general trend of increasing oxygen penetration depths reported by Hartnett and Devol (23) for the Mexican continental margin. Nitrate had a low penetration depth and was depleted within the upper 3.5 cm in all samples (Fig. 2A). As noted by Hartnett and Devol (23), the NO3 penetration depth also increased with increasing depth, which is characteristic of western North American continental shelves. At a depth of 1.5 cm, where the samples for molecular analysis were collected, the NO3− contents at the stations were in the following order: station 305> station 312 > station 306 > station 318. The corresponding denitrification rates ranged from 0.23 mmol m−2 day−1 at station 305 (the deepest station) to 1.51 mmol m−2 day−1 at station 318 (the shallowest station) (Table 1). The four sediment samples also varied considerably in terms of the organic C and total N contents, C/N ratio, and temperature (Table 1); again, station 305 was the most divergent. Samples from stations 306 and 312 had the highest concentrations of organic C (9.8 to 11.2%) and total N (1.01 to 1.20%), and samples from stations 305 and 318 had the lowest concentrations of organic C (2.9 to 4.8%) and total N (0.29 to 0.46%). Among the four stations, samples from station 318 had the highest C/N ratio (10.5). The in situ temperature and Fe2+ content at this station were also the highest (Table 1). Station 305, the deepest station and the station that was farthest offshore, had the lowest temperature (1.6°C), and the samples from this station had lowest NH4+, PO4−3, and Fe2+ contents. Overall, both the solute and solid-phase data fit well with the depth trends and overall values (within about ±30%) that were obtained on three previous cruises in the same area (23).
FIG. 2.
Bottom-water oxygen concentrations and typical sediment nitrate profiles off the Pacific coast of Mexico. At each station triplicate nitrate profiles were determined, and all profiles were similar. The nitrate profiles shown are a subset of the profiles for the entire section of 24 stations collected during four cruises between 1990 and 1999 described by Hartnett and Devol (23) and are characteristic of the environment along the western Mexican margin.
RFLP analysis of nirK and nirS clone libraries.
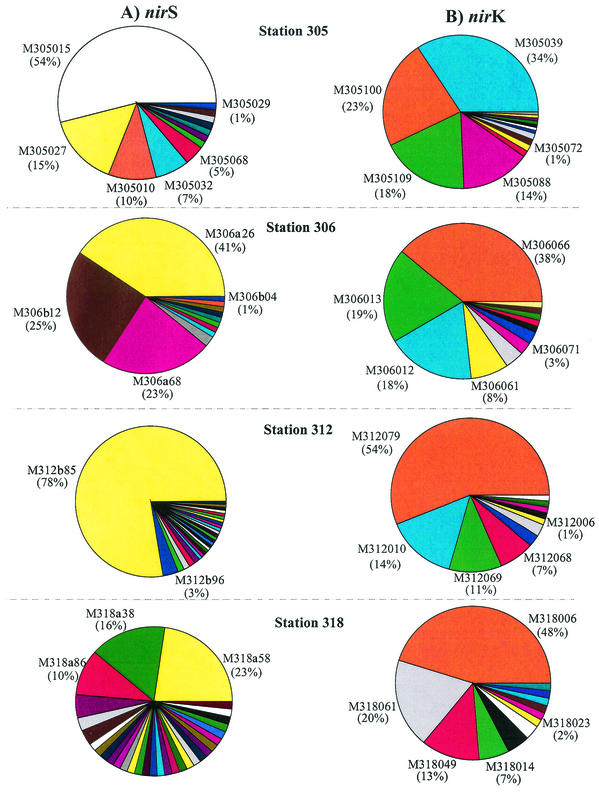
Different diversity patterns were observed for the 392 nirS clones and 378 nirK clones from the four samples screened by RFLP analysis. A total of 82 nirS OTUs and 50 nirK OTUs were obtained. The greatest number of unique nirS clones was found in the sample from station 318 (31 clones), whereas the lowest number of unique clones was found in samples from stations 305and 306 (12 clones each). Although the sample from station 305 had the lowest number of nirS clones, it had the highest number of unique nirK clones (15 clones). Samples from stations 306 and 318 had the same number of unique nirK clones (12 clones) and were similar in that regard to the sample from station 312 (11 clones) (Table 1).
Two to five dominant nirS clones were detected for each station (Fig. 3A). The RFLP patterns of clone M305027 from station 305, clone M306A26 from station 306, clone M312B85 from station 312, and clone M318a58 from station 318 were identical. These clones represented 15, 41, 78, and 18% of the total clone populations recovered, respectively (Fig. 3A). Although clone M318a58 was the most dominant clone at station 318, it represented only 23% of the clone population present (Fig. 3A). Thus, the nirS clones at this station were diverse.
FIG. 3.
Distribution of sequences in clone libraries for nirK and nirS constructed with nirS (A) and nirK (B) fragments amplified from genomic DNA in sediment samples. Clones having the same OTU are represented by the same color.
At least four dominant nirK clones were observed in each station (Fig. 3B). Three of the most abundant nirK clones from station 305 had the same RFLP pattern as the most dominant clones from stations 306 and 312. Two of the most abundant clones (M318006 and M318014, which represented 48 and 7% of the total clone population, respectively) had the same RFLP pattern as clones from stations 305, 306, and 312 (Fig. 3B).
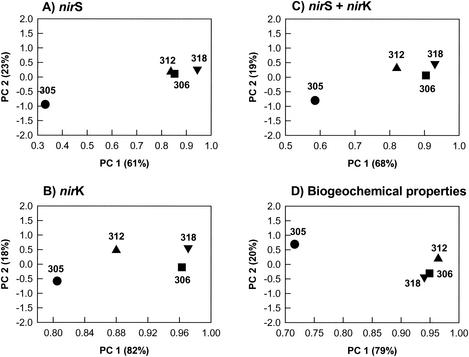
PCA of the nirS data (Fig. 4A), the nirK data (Fig. 4B), and the combined data (nirS plus nirK) (Fig. 4C), which represent 84, 97, and 87% of the total variance of the clone distributions, respectively, revealed some consistent differences among the four stations examined. nirS and nirK had somewhat different distribution patterns at the stations, as revealed by PCA. For nirS only stations 306 and 312 grouped together in principal component 1 (PC 1), but for PC 2 stations 306, 312, and 318 were close together. These three stations were distantly separated from station 305 (Fig. 4A). For nirS plus nirK station 305 was a great distance from stations 306 and 318 in PC 1, and station 312 was intermediate (Fig. 4C). Thus, station 305 was separated from all the other stations on the basis of the nirS community, and the other stations appeared to be similar (Fig. 4B). However, with regard to the nirK community, although station 305 was still separated from all the other stations, station 312 was also separated from stations 306 and 318 and from station 305 (Fig. 4B). Thus, nirS and nirK differed both in terms of diversity patterns (see above) and in terms of distribution among stations.
FIG. 4.
Ordinate plots from PCA of RFLP profiles of nirS (A), nirK (B), and _nirS_-nirK (C) clones and biogeochemical properties [organic carbon, total nitrogen, C/N ratio, NH4+, NO3−, denitrification rate, PO4+, Fe(II), and temperature] (D) for four marine samples. The values in parentheses are percentages of the total variances of PCA derived from nirS, nirK, _nirS_-nirK, and biogeochemical data.
The results of the PCA of biogeochemical data, which represented 99% of the total variance, revealed a pattern similar to the pattern obtained by PCA of nirS (Fig. 4D). Stations 306 and 318 were close together and were distant from station 305. Thus, the biogeochemical properties at stations 306 and 318 were similar but different from those at station 305. Station 312 was somewhat different but was much more similar to stations 306 and 318 than to station 305. The overall patterns showed that the denitrifying community and biogeochemical properties were more similar at the stations that were closer together, as well as at the stations in the oxygen-deficient zone.
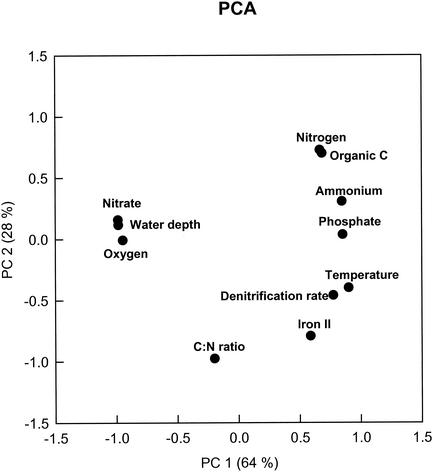
The PCA of the physical and chemical parameters reduced the data to two principal components that explained a large amount (89%) of the variation in the geochemical parameters. Oxygen, water depth, and nitrate were separated from other biogeochemical parameters with regard to PC 1, which explained 64% of the variation. PC 1 also separated the C/N ratio from the other variables. PC 2 separated the other variables, such as total and organic nitrogen, NH4+, PO4−3, temperature, denitrification rate, Fe2+, and C/N ratio (Fig. 5). Some of these variables (e.g., nitrogen and organic C; temperature and denitrification rate) were not separated with these two principal components and the small number of samples. These results suggested that several of the parameters tended to vary together in the sample set.
FIG. 5.
Ordinate plot from PCA based on water depth, oxygen, and biogeochemical data from four different sites. The values in parentheses are percentages of the total variances of PCA derived from water depth, oxygen, and biogeochemical data.
Sequence data and phylogenetic analysis of nirS and nirK sequences.
A wide range of sequence divergence was observed in the 44 nirS clones and 31 nirK clones that were sequenced from all samples. Sequence comparison showed that the nirS clones were 52 to 92% similar and that the nirK clones were 50 to 99% similar.
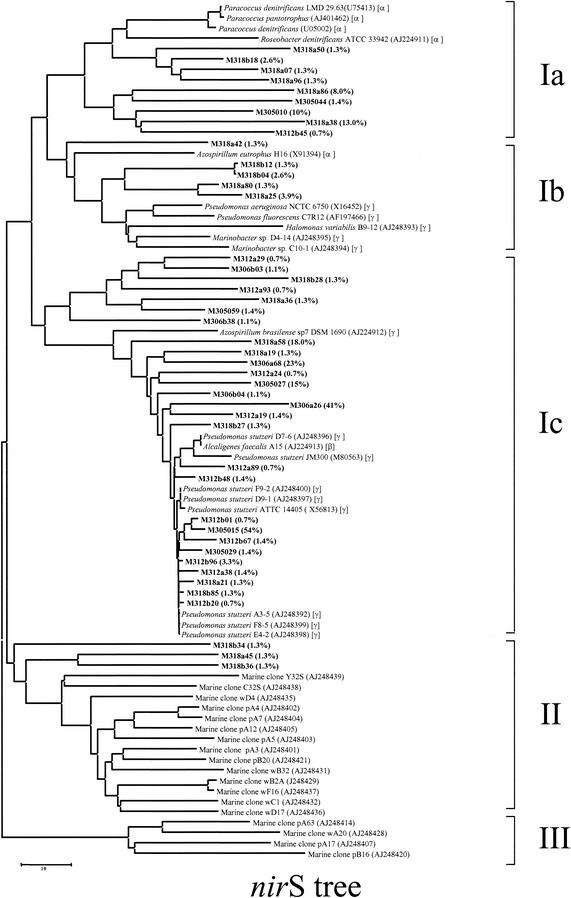
The phylogenetic tree constructed by the DNA maximum-likelihood method showed that there were three major clusters for the nirS clones (Fig. 6). Most of the nirS clones were closely related to the nirS clones belonging to three phylogenetic subdivisions (α, β, and γ subclasses of the Proteobacteria) (Fig. 6). Many of the clones (26% of the nirS clones) (group Ib-c) were associated with the nirS genes of A. faecalis (β subclass) (80 to 94% similar) and P. stutzeri (γ subclass) (80 to 99%). The most abundant clones in the samples (Fig. 3A) fell into the β and γ subclasses of the Proteobacteria and were closely related to the nirS genes of A. faecalis (78 to 84%) and P. stutzeri (85 to 91%) (group Ic) (Fig. 6). Most of the rest of the clones were less than 80% similar to nirS sequences of known denitrifying bacteria in the database; the only exceptions were clones M318A45, M318B36, and M318B34 (group II), which represented 4% of the total nirS gene population, were grouped into a distinct cluster, and were different from the rest of the nirS clones in this study. These clones also clustered with clones from Washington margin and Puget Sound sediments (oxygenated zones) (group II) (Fig. 6), but the levels of similarity were low (26 to 47%).
FIG.6.
Phylogenetic distribution of the unique nirS clones sequenced, as established by fastDNAml maximum-likelihood analysis. The scale bar represents 0.1 substitution per nucleotide position. The phylogenetic positions of pure cultures based on 16S ribosomal DNA genes are indicated by α, β, and γ for the α, β, and γ subclasses of the Proteobacteria, respectively. The values in parentheses are the percent distributions of the clones within the nirS population.
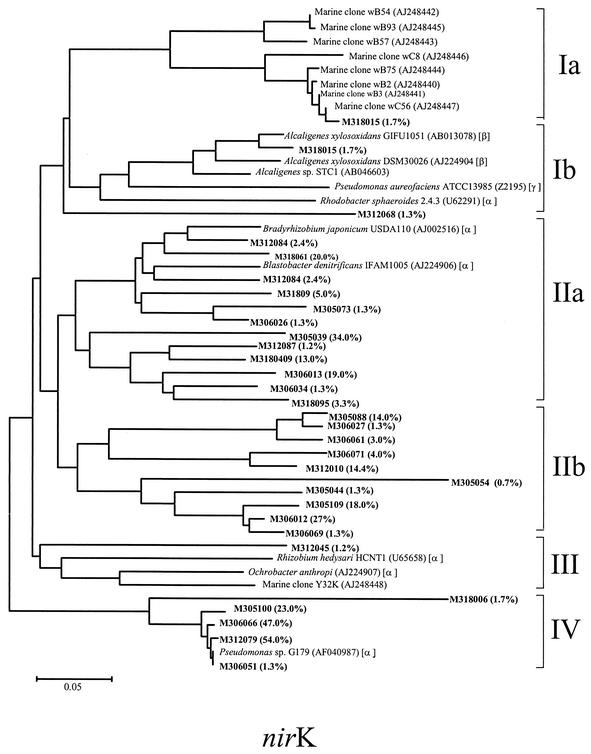
A phylogenetic analysis performed by the maximum-likelihood method for all of the nirK clones sequenced revealed the presence of four primary groups (Fig. 7). Groups Ib, IIa, and III were most closely related to the nirK genes of members of the α, β, and γ subclasses of the Proteobacteria, whereas group Ia was closely related to nirK clones found by Braker et al. (8) in Pacific Northwest sediments (Fig. 7). Sequence analysis of dominant clones revealed that the majority of the clones did not branch with any known denitrifying bacteria. The levels of similarity between dominant clones and known denitrifying bacteria were <70%. However, a few clones exhibited high levels of nucleotide identity with nirK genes. Clones M305100, M306066, M312079, and M306051 were closely related to nirK genes of Pseudomonas sp. strain G-179 (γ subclass) (98 to 99% similar); clone M312084 was closely related to nirK genes of Bradyrhizobium japonicum (α subclass) (91%); clone M318061 was closely related to nirK genes of Blastobacter denitrificans (α subclass) (83%); and M312053 was closely related to nirK genes of Alcaligenes xylosoxidans (β subclass) (96%). The rest of the clones in this study exhibited less than 80% identity to nirK sequences of known denitrifying bacteria in the database. Almost none of the nirK clones grouped closely with the clones from Washington margin sediments (group Ia); the only exception was clone M318015, which was 79 to 80% similar to clones wB75, wB2, wB23, and wC56. This clone represented 1.7% of the nirK clone population.
FIG. 7.
Phylogenetic distribution of the unique nirK clones sequenced, as established by fastDNAml maximum-likelihood analysis. The scale bar represents 0.1 substitution per nucleotide position. The phylogenetic positions of pure cultures based on 16S ribosomal DNA genes are indicated by α, β, and γ for the α, β, and γ subclasses of the Proteobacteria, respectively. The values in parentheses are the percent distributions of the clones within the nirK population.
The nirS and nirK trees did not indicate that there was a clear separation between clones from different sites. Clones from stations 305, 306, 312, and 318 were distributed throughout the phylogenetic trees for both nirS and nirK genes (Fig. 6). Although station 305 was different from stations 306, 312, and 318 in terms of the oxygen profile (Fig. 2B), biogeochemistry (Table 1), and PCA results (Fig. 4), the clones from this station were distributed throughout the phylogenetic tree and were not separated from clones from stations 306, 312, and 318 (Fig. 4A to C).
DISCUSSION
In several studies, workers have used nitrite (nir) and nitrous oxide reductase (nos) genes to monitor community differences related to geographic distance and biogeochemical properties of marine sediments (6, 8, 35, 36). Scala and Kerkhof (36) found that geographic distance (centimeters to kilometers) had a major influence on the structures of marine denitrifying communities as determined by terminal RFLP (T-RFLP) analysis of nosZ genes. Braker et al. (6) found no differences between members of the denitrifying community in spite of the dramatic changes in redox gradients in Washington margin sediments. Using T-RFLP analysis of nirS clones from a wider biogeochemical area, including Puget Sound and Washington margin sites, Braker et al. (6) observed that some dominant terminal restriction fragments occurred in all samples but differed in relative abundance. In their study, these authors found that the T-RFLP patterns within cores were more similar than those between cores. Sequence analysis (8) showed that most of the clones grouped together in accordance with the environments from which they were obtained. Braker et al. observed that all nirS gene sequences from Washington margin sediments clustered together and were separated from nirS gene sequences from the Puget Sound sediments (8).
In our study, we found that the denitrifying community and biogeochemical properties were more similar for the stations that were geographically closer together. Stations 306 and 318 were located close together and had similar biogeochemical properties. The relative amounts of OTUs in samples from stations 306 and 318 were also similar, but they were different from the relative amounts of OTUs from station 312 and were much different from the relative amounts of OTUs from station 305. When we compared the physicochemical characteristics of samples from stations 318 and 306 with those of samples from stations 305 and 312, we found that the former samples had higher concentrations of Fe2+, NH4+, and PO4−3 and higher denitrification rates and temperatures (Table 1). Samples from stations 306 and 318 also had lower NO3− concentrations (0 and 0.5 μM) than samples from stations 305 and 312 (6.9 and 2.3 μM). The patterns obtained by PCA based on the biogeochemical data [organic carbon, total nitrogen, C/N ratio, NH4+, NO3−, denitrification rate, PO4+, Fe(II), and temperature] from each station were similar to the patterns obtained by PCA based on the percentage of nirS OTUs (Fig. 4). For nirK PC 1 separated station 305 from station 312 and separated both of these stations from stations 306 and 318 (Fig. 4). Interestingly, station 312, which is intermediate in PC 1 for nirK between station 305 and the other two stations, is also intermediate for nitrate, with station 305 having the highest level and stations 306 and 318 having the lowest levels. Thus, for nirK and nirS nitrate concentration may not have the same effect on the community since for nirS the PCA did not result in an intermediate position for station 312.
One of the key differences among the samples was oxygen level. All of samples except the sample from station 305 were from the oxygen-deficient zone. All of samples except the sample from station 305 were oxygen deficient. PCA based on water depth and various biogeochemical properties (Fig. 5) suggested that the oxygen level at the sediment surface, together with water depth and nitrate concentration, may have had a significant impact on the structures of the denitrifier communities in the marine sediments in this study. These three parameters clustered together and were separated from other biogeochemical parameters in PC 1 (Fig. 5). An examination of the PCA based on the percentage of unique nirS OTUs showed that the oxygen level may have been related to this portion of the denitrifier communities. The sample from station 305 (the deepest station and the only station outside the oxygen-deficient zone) was different from the other three samples (Fig. 4). The sample from station 312 had a similar oxygen level (Fig. 2B) but was quite different from the samples from stations 306 and 318 in terms of the water depth (Table 1). The samples from stations 306 and 318, which were much more similar to each other in terms of depth and horizontal distance and from which oxygen was absent, had similar denitrifier communities in this study. These results were quite different from the results of Braker et al. (6) obtained with Puget Sound and Washington margin sediments, in which oxygen was present in the overlying water. These authors pointed out that the denitrifier community structures were very similar at different depths, although the oxidant profiles were different.
Although the nirK and nirS trees did not show a clear division among the clones from the four samples (Fig. 5 and 6), our results showed that the frequency distribution and relative amounts of different populations of organisms containing nirS and nirK genes were affected by the selection resulting from different environmental conditions at distant geographic locations. For instance, the frequency distribution of OTUs generated for the nirK and nirS genes showed that most of the clone populations in one sample location had unique nirS and nirK sequences that were not present in the clones from other locations. Even for the majority of the clones distributed at all four sites, the relative amounts of these clones were different at different sites. These results were similar to those obtained by Braker et al. (6) by T-RFLP analysis. Braker et al. (6) found that some dominant terminal restriction fragments occurred in all samples but that the relative amounts were different. Moreover, the T-RFLP patterns within samples were more similar than those in different samples. Differences in the overall diversity of nirS and nirK clones were also found. The most obvious difference was generally observed for nirS genes. This difference was reflected in the lower percentage of overlapping OTUs of nirS clones (4 to 8%) than of overlapping OTUs of nirK clones (18 to 26%), indicating that the nirS genes were more diverse than the nirK genes.
The nir trees (Fig. 6 and 7) revealed four important patterns. (i) The majority of the clones were not closely related to any known cultivated denitrifiers or any nir clone sequences obtained from Washington margin and Puget Sound sediments. Most of the nirS and nirK clones exhibited less than 80% nucleotide identity with known denitrifiers in the database, suggesting that they are unique and may represent novel sequences of denitrifiers. Provided that the clone libraries represent the in situ microbial community structure at the functional group level, the novel groups of denitrifiers appear to be abundant in marine sediments. Real-time PCR is needed to estimate or quantify these novel groups of denitrifiers in order to verify their abundance in the sediments. In order to understand their functionality, further cultivation strategies are also desirable for recovering organisms with the novel sequences. (ii) Most of the dominant nirS clones were related to known cultivated denitrifiers, indicating that the most dominant members of the _nirS_-containing bacteria in the community might be culturable. In contrast, nirK clones that appeared to be dominant in the marine samples had little relationship (>80% similarity) with known nirK genes from cultivated denitrifiers. (iii) The majority of the nirS and nirK clones from the Pacific Northwest marine sediments (oxygenated zone) were distantly related to the clones in this study (oxygen-deficient zone), indicating that there are distinct populations of denitrifying bacteria within the oxygen-deficient zone, which cannot be found in the oxygenated zone. It is known that extant microbial communities are a result of either geochemical conditions that result in selection of a community or founding populations that may be endemic rather than cosmopolitan or both. Previous studies of the Washington margin and Puget Sound suggested that the denitrifying bacterial populations in these two sites were very different even though the geochemical conditions were similar and that the difference was greater than the difference associated with overlying water depth (8). This means that water depth also affected selection of the denitrifier community in the study of Braker et al. Since oxygen is a key factor in controlling denitrifying community structure in the Mexico samples, the difference between the denitrifier communities in the Pacific Northwest marine sediments and the denitrifier communities in the sediments used in this study could also be associated with differences in the oxygen concentration at the sediment surface between the two sites based on the results of this study. (iv) Interestingly, there were a few clones (three nirS clones and one nirK clone) in this study that clustered with the clones from the Pacific Northwest sediments, indicating that some denitrifying bacteria might be cosmopolitan. These clones were all isolated from sediments at station 318, which was within the oxygen-deficient zone and was also the shallowest of the four stations studied (Fig. 2B and Table 1). However, these clones represented only small fractions of the total clone population (1.3 and 1.7% of the nirS and nirK clone populations).
The results presented here provide baseline data about denitrifier communities in marine sediments within the oxygen-deficient zone. Besides contributing information on the genetic diversity of denitrifying bacteria in the marine environment, in this study we also present a view of the biogeochemical factors that influence this important group of bacteria. Even though oxygen affected selection of the denitrifier communities in our study, it should be considered that our results reflect only a snapshot of the succession of denitrifier communities in the marine sediments. The effects of different environmental factors may underlie temporal changes, and so the denitrifier communities associated with temporal changes need to be investigated in order to increase our understanding of the linkage between the functional processes and microbial community structure involved in nitrogen cycling.
Acknowledgments
X. Liu and S. M. Tiquia contributed equally to this work.
This research was supported by the Biotechnology Investigations-Ocean Margins and Natural and Accelerated Bioremediation Research Programs, Office of Biological and Environmental Research, Office of Science, U.S. Department of Energy. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DOE-AC05-00OR22725.
REFERENCES
- 1.Altabet, M. A., R. Francois, D. W. Murray, and W. L. Prell. 1995. Climate-related variations in denitrification in the Arabian Sea from sediment 15N/14N ratios. Nature 373**:**506-509. [Google Scholar]
- 2.Anderson, L. 1979. Simultaneous spectrophotometric determination of nitrate and nitrite by flow injection analysis. Anal. Chim. Acta 110**:**123-128. [Google Scholar]
- 3.Bender, M. L., W. Martin, J. Hess, F. Sayles, L. Ball, and C. Lambert. 1987. A whole core squeezer for interfacial pore water sampling. Limnol. Oceanogr. 32**:**1214-1225. [Google Scholar]
- 4.Berg, P., N. Risgaard-Petersen, and S. Rysgaard. 1998. Interpretation of measured concentration profiles in the sediment porewater. Limnol. Oceanogr 43**:**1500-1510. [Google Scholar]
- 5.Berner, R. A. 1989. Biogeochemical cycles of carbon and sulfur and their effect on atmospheric oxygen over Phanerozoic time. Palaeogeogr. Palaeoclimatol. Palaeoecol. 73**:**97-122. [Google Scholar]
- 6.Braker, G., H. A. Ayala-del Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67**:**1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braker, G., A. Fesefeldt, and K. P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64**:**3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66**:**2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandes, J. A., and A. H. Devol. 1995. Simultaneous nitrate and oxygen respiration in coastal sediments: evidence for discrete diagenesis. J. Mar. Res. 53**:**771-797. [Google Scholar]
- 10.Brandes, J. A., A. H. Devol, T. Yoshinari, D. A. Jayakumar, and S. W. A. Naqvi. 1998. Isotopic composition of nitrate in the central Arabian Sea and eastern tropical North Pacific: a tracer for mixing and nitrogen cycles. Limnol. Oceanogr. 43**:**1680-1689. [Google Scholar]
- 11.Capone, D. 2001. Marine nitrogen fixation: what's the fuss? Curr. Opin. Microbiol. 4**:**341-348. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, J. P., J. W. Murray, A. H. Devol, and L. A. Codispoti. 1987. Denitrification in continental shelf sediments has major impact on the oceanic nitrogen budget. Global Biogeochem. Cycles 1**:**97-116. [Google Scholar]
- 13.Codispoti, L. A. 1989. Phosphorus vs nitrogen limitation of new and export production, p. 377-394. In W. H. Berger, V. S. Smetacek, and G. Wefer (ed.), Production of the oceans: present and past. Wiley and Sons, New York, N.Y.
- 14.Codispoti, L. A. 1995. Is the ocean losing nitrate? Nature 376**:**724. [Google Scholar]
- 15.Codispoti, L. A. J. W. Brandes, J. P. Christensen, A. H. Devol, S. W. A. Naqvi, H. W. Paerl, and T. Yoshinari. 2001. The oceanic fixed nitrogen and nitrous oxide budgets: moving targets as we enter the anthropocene? Sci. Mar. 65(Suppl. 2)**:**85-105. [Google Scholar]
- 16.Conrad, R. 1996. Soil organisms as controllers of atmospheric trace gases (H2, CO2, CH4, OCS, N2O, and NO). Microbiol. Rev. 60**:**609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devol, A. H. 1991. Direct measurements of nitrogen gas fluxes from continental sediments. Nature 349**:**319-321. [Google Scholar]
- 18.Devol, A. H., and J. P. Christensen. 1993. Benthic fluxes and nitrogen cycling in sediments of the continental margin of the eastern North Pacific. J. Mar. Res. 51**:**345-372. [Google Scholar]
- 19.Devol, A. H., L. A. Codispoti, and J. P. Christensen. 1997. Summer and winter denitrification rates in western Arctic shelf sediments. Cont. Shelf Res. 9**:**1029-1050. [Google Scholar]
- 20.Ganeshram, R. S., T. F. Pedersen, S. E. Calvert, and J. W. Murray. 1995. Large changes n oceanic nutrient inventories from glacial to interglacial periods. Nature 376**:**755-758. [Google Scholar]
- 21.Genetics Computer Group. 1994. GCG program manual. Genetics Computer Group, Madison, Wis.
- 22.Gruber, N., and J. L. Sarmiento. 1997. Global patterns of marine nitrogen fixation and denitrification. Global Biogeochem. Cycles 11**:**235-266, [Google Scholar]
- 23.Hartnett, H. E., and A. H. Devol. 2003. The role of a strong oxygen deficient zone in the preservation and degradation of organic matter: a carbon budget for the continental margins of NW Mexico and Washington State. Geochim. Cosmochim. Acta 67**:**247-264. [Google Scholar]
- 24.Hedges, J. I., and R. G. Keil. 1995. Sedimentary organic matter preservation: an assessment and speculative synthesis. Mar. Chem. 49**:**81-115. [Google Scholar]
- 25.Hedges, J. I., and J. H. Stern. 1984. Carbon and nitrogen determinations of carbonate-containing solids. Limnol. Oceanogr. 29**:**657-663. [Google Scholar]
- 26.Hurt, R. A., X. Qui, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67**:**4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowles, R. 1982. Denitrification. Microbiol. Rev. 46**:**43-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen, E., A. H. Devol, and H. E. Hartnett. 1999. Organic matter diagenesis in sediments on the continental shelf and slope of the eastern tropical and temperate North Pacific. Cont. Shelf Res. 19**:**1331-1351. [Google Scholar]
- 29.Longhurst, A., S. Sathyendranath, T. Platt, and C. Caverhill. 1995. An estimate of global primary production in the ocean from satellite radiometer data. J. Plankton Res. 17**:**1245-1271. [Google Scholar]
- 30.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28**:**173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Middleburg, J. J., K. Soetaert, P. M. J. Herman, and C. H. R. Heip. 1996. Denitrification in marine sediments: a model study. Global Biogeochem. Cycles 10**:**661-674. [Google Scholar]
- 32.Qui, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67**:**880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reimers, C. E., R. A. Jahnke, and D. M. McCorkle. 1992. Carbon fluxes and burial rates over the continental slope and rise off central California with implications for global carbon cycle. Global Biogeochem. Cycles 6**:**199-224. [Google Scholar]
- 34.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (_nos_Z) gene-specific PCR primers for detection of denitrifiers and three _nos_Z genes from marine sediments. FEMS Microbiol. Lett. 162**:**61-68. [DOI] [PubMed] [Google Scholar]
- 35.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65**:**1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66**:**1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma, S. 1996. Applied multivariate techniques. John Wiley, New York, N.Y.
- 38.Smith, S. W., R. Overbeek, C. R. Woese, W. Gilbert, and P. M. Gillevet. 1994. The genetic data environment: an expandable GUI for multiple sequence-analysis. CompUT. Applic. Biosci. 10**:**671-675. [DOI] [PubMed] [Google Scholar]
- 39.Stookey, L. L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42**:**779-781. [Google Scholar]
- 40.Strickland, J. D. H., and T. R. Parsons. 1972. A practical handbook of seawater analysis. Fisheries Research Board of Canada, Ottawa, Ontario.
- 41.Strunk, O., and W. Ludwig. 1996. ARB. Technical University of Munich, Munich, Germany.
- 42.Van Mooy, B. A. S., R. G. Keil, and A. H. Devol. 2002. Impact of suboxia on sinking particulate organic carbon: enhanced carbon flux and preferential degradation of amino acids via denitrification. Geochim. Cosmochim. Acta 66**:**457-465. [Google Scholar]
- 43.Walsh, J. J. 1991. Importance of the continental margins in the marine biogeochemical cycling of carbon and nitrogen. Nature 350**:**53-55. [Google Scholar]
- 44.Ward, B. B. 1995. Diversity of culturable denitrifying bacteria—limits of rDNA RFLP analysis and probes for the functional gene, nitrite reductase. Arch. Microbiol. 163**:**167-175. [Google Scholar]
- 45.Zhou, J., M. R. Fries, J. C. Chee-Sanford., and J. M. Tiedje. 1995. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus toluliticus sp. nov. Int. J. Sys. Bacteriol. 45**:**500-506. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62**:**316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, J., A. V. Palumbo, and J. M. Tiedje. 1997. Selective detection of a novel class of toluene-degrading denitrifiers, Azoarcus tolulyticus, with small-subunit rRNA primers and probes. Appl. Environ. Microbiol. 63**:**2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, J., B. Xia, D. S. Treves, L. Y. Wu, T. L. Marsh, R. V. O'Neil, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68**:**326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]