Characterization of the Epidemic European Fusidic Acid-Resistant Impetigo Clone of Staphylococcus aureus (original) (raw)
Abstract
Resistance to the antibiotic fusidic acid in European strains of Staphylococcus aureus causing impetigo has increased in recent years. This increase appears to have resulted from clonal expansion of a strain we have designated the epidemic European fusidic acid-resistant impetigo clone (EEFIC), which carries the fusidic acid resistance determinant fusB on its chromosome. To understand better the properties of the EEFIC responsible for its success, we have performed detailed phenotypic and genotypic characterization of this clone. Molecular typing revealed the EEFIC to be ST123, spa type t171, and agr type IV and therefore unrelated to earlier prevalent fusB+ strains found in the United Kingdom. EEFIC strains exhibited resistance to fusidic acid, penicillin, and, in some cases, erythromycin, which are all used in the treatment of impetigo. PCR analysis of the EEFIC and complete DNA sequencing of the 39.3 Kb plasmid it harbors identified genes encoding several toxins previously implicated in impetigo (exfoliative toxins A and B and EDIN-C). The location of fusB was mapped on the chromosome and found to be associated with a novel 16.6-kb genomic island integrated downstream of groEL. Although this element is related to classical staphylococcal pathogenicity islands, it does not encode any known virulence factors and consequently has been designated SaRI_fusB_ (for “S. aureus resistance island carrying _fusB_”).
The antibiotic fusidic acid is used extensively for topical treatment of superficial skin infections caused by Staphylococcus aureus, including impetigo and atopic dermatitis (7, 13). Although fusidic acid has been used clinically throughout Europe since the early 1960s, the prevalence of resistance to this antibiotic in S. aureus remained low well into the 1990s (2, 7, 38). However, in the last decade there has been an increase in the prevalence of staphylococcal resistance to fusidic acid in a number of northern European countries; this resistance has been primarily associated with strains causing impetigo bullosa (9, 28, 30, 37, 38). In Sweden (30), Norway (38), and the United Kingdom (9), the fusidic acid-resistant S. aureus strains responsible for impetigo are highly clonal in each case and in fact constitute a single fusidic acid-resistant clone (28). Consequently, this strain has been designated the epidemic European fusidic acid-resistant impetigo clone (EEFIC) (29).
The EEFIC is resistant to fusidic acid as a consequence of recruitment of the fusB gene (28, 29), a determinant that protects the staphylococcal translation apparatus from inhibition by fusidic acid (27). Typically, this determinant is plasmid-borne (27), although in the EEFIC it is located on the chromosome (28). Since fusidic acid is a primary treatment for impetigo in many European countries (16), resistance to this antibiotic is likely to be one of the key properties of the EEFIC that has contributed to its success.
Although resistance to fusidic acid may help to explain the natural selection of this clone in locations in which the antibiotic is extensively used for treating impetigo, it does not provide an explanation for why this strain is so effective at causing impetigo in the first instance. Presumably, the clone possesses features that assist the establishment of impetigo. To better understand the EEFIC, we have subjected it to detailed phenotypic and genotypic characterization, with particular emphasis on establishing the properties of this strain that are responsible for its success. We have also examined the possibility that the EEFIC is related to the original fusB+ fusidic acid-resistant (FAR) S. aureus strains that were prevalent in the United Kingdom during the 1970s (18).
MATERIALS AND METHODS
Bacterial strains and routine culture.
Representatives of the EEFIC were identified by pulsed-field gel electrophoresis (PFGE) in an earlier study (28). EEFIC strain CS6 (28) was chosen for detailed molecular characterization. FAR strains, which were predominantly isolated in the United Kingdom in the early 1970s, have been previously described (18). Escherichia coli XL-10 Gold (Stratagene, Amsterdam, The Netherlands) and S. aureus RN4220 (11) were used as routine hosts for cloning. Unless otherwise stated, strains were cultured in Luria-Bertani broth or on Luria-Bertani agar at 37°C.
Antibiotic susceptibility determinations.
All susceptibility testing was performed by using Etest (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar according to the manufacturer's recommendations.
Strain typing.
Established methodologies were used for PFGE (25), phage typing (22), multilocus sequence typing (MLST) (10), and spa typing (14).
PCR-based detection of toxin genes.
PCR was used to detect genes encoding exfoliative toxins, Panton-Valentine leukocidin, and pyrogenic toxin superantigens (including tst) according to established methods (4, 5, 19, 23).
Plasmid purification.
Strain CS6 was grown without shaking at 37°C in brain heart infusion broth containing 0.5% (wt/vol) glycine for 16 h. Cells were pelleted by centrifugation, washed in sterile water, and resuspended in 50 mM Tris-HCl (pH 8), 10 mM EDTA, and 25% (wt/vol) sucrose. Lysis of cells and plasmid purification were carried out essentially as described previously (39). Purified plasmid preparations were treated with plasmid-safe ATP-dependent DNase (Epicenter, Madison, WI) to remove residual chromosomal DNA contamination.
Identification of the chromosomal location of fusB in the EEFIC.
Total DNA was prepared from strain CS6 by using a bacterial genomic DNA purification kit (Edge Biosystems, Gaithersburg, MD) from protoplasts generated by incubation with lysostaphin (100 μg/ml) for 30 min at 37°C. Southern hybridization for fusB (28) was performed on total DNA digested with a variety of restriction enzymes to identify suitably sized fragments for cloning; digestion with HindIII generated an ∼5-kb fragment carrying fusB (data not shown). Large-scale restriction digests of CS6 DNA were performed with HindIII, and fragments in the range 4 to 6 kb were excised from agarose gels, purified, and ligated into plasmid pCU1 (3). A DNA library was established in E. coli XL-10 Gold, which was recovered and electroporated (35) into S. aureus RN4220. FAR clones were identified by replica plating onto agar containing 1 μg of fusidic acid/ml, and the presence of fusB was confirmed by Southern hybridization. DNA inserts carrying fusB were sequenced by primer walking.
PCR analysis of the chromosomal location of fusB.
The procedure described above was successful in mapping one junction of the DNA element with which fusB was associated, and the EEFIC chromosome (see Results). A DNA fragment containing the other junction was generated by long PCR using the TripleMaster PCR system (Eppendorf, Cambridge, United Kingdom) and oligonucleotide primers specific for fusB (5′- TAAGCGGCCGCAAGATTCTTCAATATCGTCATCTA) and groEL (5′-GCAGCGGCCGCAGCTCAAGCAATGATTCAAGAAGG).
Shotgun DNA sequencing and sequence analysis.
DNA was shotgun sequenced according to standard methodology (34). Briefly, DNA was mechanically sheared into 1- to 3-kb fragments, end repaired, ligated to blunt pUC57, and electroporated into E. coli DH10β (Invitrogen, Paisley, United Kingdom) in the case of the EEFIC plasmid or ligated into blunt pcrSMART (Lucigen) and electroporated into E. coli 10G (Lucigen) in the case of the PCR amplicon carrying fusB. Plasmid clones were sequenced from forward and reverse vector priming sites, and regions of low coverage were filled by primer walking and/or the generation and DNA sequencing of appropriate PCR amplicons.
DNA sequences were assembled into contigs by using Sequencher 4.5 (GeneCodes), and open reading frames (ORFs) were identified and annotated by using Artemis 7 (32), Glimmer 2.13 (33), and BLAST (1).
Accession number.
The DNA sequence of the element carrying fusB (SaRI_fusB_) has been assigned GenBank accession number AM292600.
RESULTS
Strain typing.
The EEFIC was originally delineated through use of PFGE (28). To characterize this clone more extensively, phage typing and molecular typing methods (spa, MLST) were used. The basic phage type of the EEFIC was 3C, 55, or 71 (Table 1). Typing according to spa indicated that the EEFIC corresponds to t171, or single-locus variants (SLVs) thereof (Table 1). Based on a combination of direct determination of MLST types, and sequence type prediction based on spa type, the EEFIC is ST123 (or, in the case of strain CS21, a novel SLV of ST123) (Table 1). The EEFIC is agr type IV (data not shown).
TABLE 1.
Typing and exfoliative toxin status of representative EEFIC and FAR strains
| Strain | Phage type | MLSTa | spa repeat | spa type | Presence (+) or absence (−) of: | |
|---|---|---|---|---|---|---|
| eta | etb | |||||
| CS11 | 3C/55/71 | ST123 | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| CS6 | 3C/55/71 | ST123 | 14-44-13-12-17-17-22-18 | t874 | + | + |
| CS604 | 3C/55/71 | ST123 | 14-44-13-12-17-17-17-23-18 | t659 | + | - |
| CS607 | 3C/55/71 | ST123 | 14-44-13-12-17-17-17-22-18 | t659 | + | - |
| CS1152 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-23-18 | t659 | + | + |
| CS1160 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-22-18 | t875 | + | + |
| CS1163 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| CS1213 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-17-23-18 | t408 | + | - |
| CS1215 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-17-23-18 | t408 | + | + |
| CS18 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| CS4 | 3A/3C/55 | ST123* | 14-44-13-12-17-17-23-18-17 | t159 | + | + |
| H17 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | - |
| H18 | 3C/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | - |
| H19 | 3C/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| H44 | 3C/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| H45 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| H47 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | - |
| H49 | 3C/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | - |
| H78 | 3C/55/71 | ST123* | 14-44-13-12-17-17-17-17-23-18 | t171 | + | + |
| H83 | 3C/71 | ST123* | 14-44-13-12-17-17-17-23-18 | t659 | + | + |
| CS21 | 3C/55/71 | ST123slv | 14-44-13-12-17-17-17-17-02-18 | t171 | + | - |
| FAR10 | NTc | ST45* | 09-02-16-34-13-17-34-16-34 | t065 | - | - |
| FAR11 | 95 | ST45* | 09-02-16-34-13-17-34-16-34 | t065 | - | - |
| FAR19 | 47/54/75/77 | ST45* | 09-02-16-34-13-17-34-16-34 | t065 | - | - |
| FAR4 | 95 | ST45* | 09-02-16-34-34 | t880 | - | - |
| FAR8 | 95 | ST45* | 09-02-16-34-34 | t880 | - | - |
| FAR6 | 95 | NDb | NT | NT | - | - |
Detection of genes encoding virulence factors.
All representative EEFIC strains tested negative by PCR for genes encoding toxic shock syndrome toxin and Panton-Valentine leukocidin (tst and lukS-lukF, respectively) but positive for enterotoxin genes seg and sei, and for the exfoliative toxin A gene, eta. Approximately two-thirds (13 of 21) of the EEFIC strains examined tested positive for the gene encoding exfoliative toxin B, etb (Table 1). Characterization of the EEFIC plasmid (see below) revealed that these strains also carry the gene encoding epidermal differentiation inhibitor C (EDIN-C), an ADP-ribosylating exotoxin that had previously been implicated in impetigo and related superficial staphylococcal skin infections (40).
Relationship to earlier FAR isolates.
In the early 1970s, fusidic acid resistance in clinical isolates of S. aureus was reported to be predominantly plasmid mediated (18, 31). A single plasmid (pUB101) was thought to be responsible (18), a suggestion that we have corroborated in the present study by restriction analysis of the plasmids from a cross-section of these original FAR strains (Table 1) and Southern hybridization for fusB (data not shown). In contrast to the earlier study (18), we found that strains carrying pUB101 appear to be clonal (Table 1). Since the simplest possibility for the evolution of the EEFIC would be recruitment of fusB from pUB101 to the chromosome in these original strains, we examined whether these earlier strains might represent the progenitor of the EEFIC. However, classical and molecular typing methods showed that these strains are not related (Table 1). Furthermore, the earlier strains possess a different agr type (I versus IV for the EEFIC [data not shown]) and do not carry the exfoliative toxins associated with the EEFIC (Table 1).
Complete DNA sequence of the EEFIC plasmid.
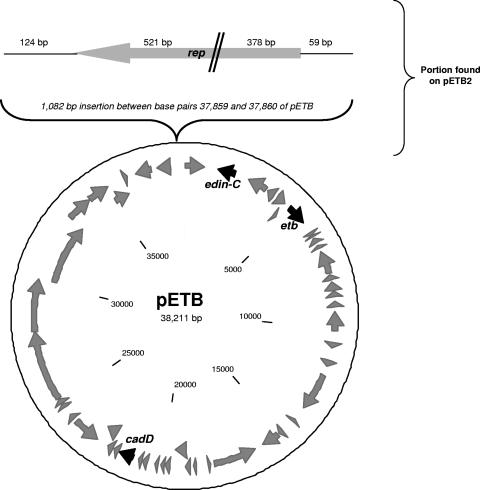
The EEFIC harbored a single, large plasmid of approximately 40 kb (data not shown). To investigate whether this plasmid carried genes that might be linked to the success of the EEFIC, the entire plasmid from strain CS6 was subjected to shotgun DNA sequencing to >5-fold coverage. The final 39,307-bp sequence was essentially identical to that of the previously sequenced pETB plasmid from S. aureus (40), recovered from an impetigo-causing strain (Fig. 1). Consequently, we designated this new plasmid pETB2 (Fig. 1). Aside from a limited number of nucleotide polymorphisms, the only difference between pETB and pETB2 is the presence in the latter of an additional 1,082-bp tract (Fig. 1), which accounts almost entirely for the difference in their sizes (39.3 kb for pETB2 versus 38.2 kB for pETB). This region carries a plasmid replication (rep) gene that has become disrupted following a substantial deletion (Fig. 1). (The pETB2 plasmid is so similar to pETB that we have not submitted it to GenBank, although the sequence is available upon request.) The pETB2 plasmid is the source of the etb gene in the EEFIC and also carries the gene encoding EDIN-C. This plasmid encodes resistance to cadmium (cadD), although no other antimicrobial resistance genes are present (Fig. 1). In addition, this plasmid encodes one or more bacteriocins capable of killing other strains of S. aureus (40).
FIG. 1.
Genetic architecture of the pETB plasmid and the closely related plasmid (pETB2) found in the EEFIC. ORFs are indicated as gray arrows, and genes of interest are indicated as black arrows. The top of the figure shows the 1,082-bp portion carrying a disrupted rep gene that is present in pETB2 but not in pETB.
Antibiotic resistance.
MIC determinations established that representative EEFIC strains were relatively susceptible to antibiotics. Based on BSAC susceptibility breakpoints (21), all of the strains were sensitive to tetracycline, mupirocin, rifampin, gentamicin, ciprofloxacin, oxacillin, and vancomycin (Table 2). In addition to resistance to fusidic acid, all strains examined were resistant to penicillin, and three of five strains were resistant to erythromycin (Table 2). Of the erythromycin-resistant strains, two (CS604 and CS607) exhibited inducible resistance to clindamycin (data not shown).
TABLE 2.
Antibiotic susceptibility profiles of representative EEFIC strains
| Strain | MIC (μg/ml) ofa: | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tet | Ery | Mup | Pen | Rif | Gen | Cip | Oxa | Van | Fus | |
| CS6 | 0.25 | 0.75 | 0.125 | 0.75 | 0.008 | 0.25 | 0.38 | 0.38 | 2 | 4 |
| CS11 | 0.38 | 0.38 | 0.125 | 0.75 | 0.008 | 0.25 | 0.38 | 0.38 | 2 | 4 |
| CS21 | 0.38 | 0.5 | 0.125 | 1.5 | 0.006 | 0.75 | 0.5 | 0.75 | 2 | 4 |
| CS604 | 0.38 | >256 | 0.125 | 0.75 | 0.008 | 0.25 | 0.38 | 0.38 | 2 | 4 |
| CS607 | 0.38 | >256 | 0.19 | 0.75 | 0.008 | 0.25 | 0.38 | 0.38 | 2 | 4 |
Recruitment of the fusidic acid resistance determinant, fusB, to the chromosome.
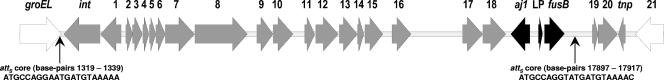
We previously established that the EEFIC is resistant to fusidic acid as a consequence of carrying the fusB gene on its chromosome (28). Shotgun cloning of genomic DNA mapped fusB downstream of SA1815 (S. aureus N315 gene designation), the locus occupied in some S. aureus strains by a pathogenicity island (PI) (17). The other junction of the DNA region encoding fusB and the chromosome was recovered by long PCR between fusB and the next conserved chromosomal gene in the other direction (groEL). The resulting ∼15-kb amplicon was subjected to DNA sequencing by using a combination of shotgun sequencing and primer walking.
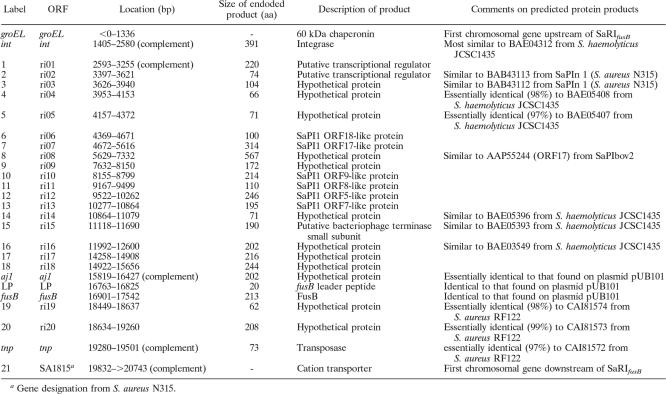
The fusB determinant was located on what initially appeared to be an ∼18-kb genomic island (GI) comprising 24 ORFs encoding predicted proteins ≥50 amino acids in length, flanked at one end by an integrase gene (Fig. 2). However, identification of the core attachment (attS) sites of this GI (ATGCCAGGAATGATGTAAAAA and ATGCCAGGTATGATGTAAAAC; Fig. 2) indicated that the element carrying fusB is actually only 16,599 bp in length, with fusB representing the right-hand terminal gene (Fig. 2). The region of the GI responsible for conferring resistance to fusidic acid exhibited almost complete DNA sequence identity to the equivalent region found on plasmid pUB101 and comprised fusB and two upstream genes (aj1 and the gene encoding the fusB leader peptide (27) (Fig. 2). The remainder of the ORFs encode hypothetical proteins, a number of which are homologous to those encoded by characterized staphylococcal PIs (Fig. 2). No virulence genes were apparent on this GI. Consequently, we have designated this element “S. aureus resistance island carrying fusB_” (SaRI_fusB).
FIG. 2.
Genetic architecture and chromosomal context of SaRI_fusB_. The predicted termini of SaRI_fusB_ are indicated by the vertical arrows. Chromosomal genes up- and downstream of SaRI_fusB_ are indicated in white, genes apparently acquired from plasmid pUB101 are indicated in black, and other predicted genes are indicated in gray. The genes and the products they encode are described in the table below the diagram.
DISCUSSION
This study confirms our earlier PFGE analysis which indicated that the EEFIC is a clone (28). The EEFIC corresponds to phage type 3C, 55, and/or 71, spa type t171 (or SLVs), and ST123. Previously reported ST123 strains are all United Kingdom isolates (http://www.mlst.net/), which suggests that the EEFIC may have originated in the United Kingdom. This ST has been described as a nasal carriage strain (8), although it also caused soft tissue infection in an intravenous drug user (24).
The EEFIC carries a number of determinants that are either known to be important, or implicated, in the establishment of impetigo by S. aureus. For example, exfoliative toxins are almost always found in S. aureus strains causing both bullous and nonbullous impetigo (12, 15), and the EEFIC invariably carried eta, with the majority of strains also carrying etb. In addition, the presence of the ADP-ribosylating exotoxin, EDIN-C, likely contributes to the EEFIC's ability to cause superficial disease (40). EDIN proteins cause inhibition of keratinocyte differentiation in vitro, hyperplasia of the epidermis in vivo, and loss of barrier function of the endothelium (6, 36). The presence on pETB/pETB2 of an operon encoding bacteriocin(s) with antistaphylococcal activity (40) presumably affords the EEFIC a competitive advantage over other S. aureus strains on the skin.
The EEFIC is generally susceptible to antibiotics, but representatives of this clone are resistant to several agents that are of particular importance in the treatment of impetigo, including macrolides and penicillin (16) (Table 1). Most importantly, the EEFIC is resistant to fusidic acid, often the treatment of choice for impetigo in many European countries (16).
To investigate how recruitment of the fusB gene has occurred in the EEFIC, the location of this determinant on the chromosome was mapped. We found that fusB was associated with a GI of 16.6 kb (Fig. 2) that has become integrated into the chromosome at the 3′ end of groEL, a recognized site for insertion of PIs (e.g., SaPIn1) (17).
Factors apart from the integration site indicate that the GI we identified is closely related to staphylococcal PIs. For example, it is very similar in size to classical PIs (∼15 to 20 kb) (26), is bordered at one end by a gene encoding an integrase, and encodes homologues of a number of the hypothetical proteins encoded by other SaPIs (Fig. 2).
In contrast to SaPIs, the GI we identified does not harbor any known virulence factors (Fig. 2) and does not therefore appear to be a PI. However, it does confer resistance to fusidic acid by virtue of harboring the fusB gene. We have therefore chosen to designate this GI SaRI_fusB_ (for “S. aureus resistance island carrying fusB_”). On the basis of almost complete DNA sequence identity, it is clear that the region of this element responsible for conferring resistance to fusidic acid has been recruited from the pUB101 plasmid (or from a shared genetic source). This recruitment has occurred in a very precise fashion, i.e., essentially the minimal pUB101 fragment required for full functional expression of fusidic acid resistance (27) has been recruited to SaRI_fusB (Fig. 2).
Several of the ORFs identified on SaRI_fusB_ encode homologues of proteins from S. haemolyticus (Fig. 2), a finding that may point to the source of this SaRI. Furthermore, we note that the fusB determinant has been detected in S. haemolyticus (GenBank accession no. AJ302698).
We identified an architecturally similar GI, resident in the same chromosomal location as SaRI_fusB_, in the genome sequence of S. aureus RF122 (GenBank accession no. AJ938182). However, in place of fusB, this SaRI carries a gene (SAB1892c) encoding a multidrug resistance protein, and presumably additional SaRI variants exist which carry other antibiotic resistance genes. Based on the fact that SaPIs are mobile (20), it seems likely that the closely related SaRI elements are also mobile.
Identification of SaRI_fusB_ indicates that GIs other than classical transposons, plasmids, and staphylococcal chromosomal cassette elements participate in the recruitment of antibiotic resistance genes to the chromosome in S. aureus.
Acknowledgments
We are grateful to Richard Novick (Skirball Institute, New York University Medical Center, New York) for advice and helpful discussion regarding SaRI_fusB_.
This study was funded by grants from LEO Pharma to the University of Leeds (I.C.) and the Statens Serum Institut (R.S.).
Footnotes
▿
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, J., J. Ashby, G. Jevons, N. Lines, and R. Wise. 1999. Antimicrobial resistance in gram-positive pathogens isolated in the UK between October 1996 and January 1997. J. Antimicrob. Chemother. 43**:**689-698. [DOI] [PubMed] [Google Scholar]
- 3.Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K. D. Entian, and F. Gotz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204**:**1149-1154. [DOI] [PubMed] [Google Scholar]
- 4.Becker, K., A. W. Friedrich, G. Lubritz, M. Weilert, G. Peters, and C. Von Eiff. 2003. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 41**:**1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, K., R. Roth, and G. Peters. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36**:**2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, L., A. Doye, M. Rolando, G. Flatau, P. Munro, P. Gounon, R. Clement, C. Pulcini, M. R. Popoff, A. Mettouchi, L. Landraud, O. Dussurget, and E. Lemichez. 2006. Induction of transient macroapertures in endothelial cells through RhoA inhibition by Staphylococcus aureus factors. J. Cell Biol. 173**:**809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, E. M., and P. Thomas. 2002. Fusidic acid resistance in Staphylococcus aureus isolates. Lancet 359**:**803. [DOI] [PubMed] [Google Scholar]
- 8.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292**:**114-116. (Retraction, **295:**971, 2002.) [DOI] [PubMed] [Google Scholar]
- 9.El-Zimaity, D., A. M. Kearns, S. J. Dawson, S. Price, and G. A. Harrison. 2004. Survey, characterization and susceptibility to fusidic acid of Staphylococcus aureus in the Carmarthen area. J. Antimicrob. Chemother. 54**:**441-446. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38**:**1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairweather, N., S. Kennedy, T. J. Foster, M. Kehoe, and G. Dougan. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect. Immun. 41**:**1112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravet, A., P. Couppie, O. Meunier, E. Clyti, B. Moreau, R. Pradinaud, H. Monteil, and G. Prevost. 2001. Staphylococcus aureus isolated in cases of impetigo produces both epidermolysin A or B and LukE-LukD in 78% of 131 retrospective and prospective cases. J. Clin. Microbiol. 39**:**4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenwood, D. 2003. Fusidanes, p. 297-299. In R. G. Finch (ed.), Antibiotic and chemotherapy, 8th ed. Churchill Livingstone, London, England.
- 14.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41**:**5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koning, S., A. van Belkum, S. Snijders, W. van Leeuwen, H. Verbrugh, J. Nouwen, M. Op't Veld, L. W. van Suijlekom-Smit, J. C. van der Wouden, and C. Verduin. 2003. Severity of nonbullous Staphylococcus aureus impetigo in children is associated with strains harboring genetic markers for exfoliative toxin B, Panton-Valentine leukocidin, and the multidrug resistance plasmid pSK41. J. Clin. Microbiol. 41**:**3017-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koning, S., A. P. Verhagen, L. W. A. van Suijlekom-Smit, A. Morris, C. C. Butler, and J. C. van der Wouden. 2004. Interventions for impetigo (Cochrane Review). The Cochrane Library. John Wiley & Sons, Ltd., Chichester, United Kingdom. [DOI] [PubMed]
- 17.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357**:**1225-1240. [DOI] [PubMed] [Google Scholar]
- 18.Lacey, R. W., and V. T. Rosdahl. 1974. An unusual “penicillinase plasmid” in Staphylococcus aureus; evidence for its transfer under natural conditions. J. Med. Microbiol. 7**:**1-9. [DOI] [PubMed] [Google Scholar]
- 19.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29**:**1128-1132. [DOI] [PubMed] [Google Scholar]
- 20.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29**:**527-543. [DOI] [PubMed] [Google Scholar]
- 21.MacGowan, A. P., and R. Wise. 2001. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J. Antimicrob. Chemother. 48(Suppl. 1)**:**17-28. [DOI] [PubMed] [Google Scholar]
- 22.Marples, R. R., and V. T. Rosdahl. 1997. International quality control of phage typing of Staphylococcus aureus. J. Med. Microbiol. 46**:**511-516. [DOI] [PubMed] [Google Scholar]
- 23.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38**:**1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monk, A. B., S. Curtis, J. Paul, and M. C. Enright. 2004. Genetic analysis of Staphylococcus aureus from intravenous drug user lesions. J. Med. Microbiol. 53**:**223-227. [DOI] [PubMed] [Google Scholar]
- 25.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41**:**1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick, R. P. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49**:**93-105. [DOI] [PubMed] [Google Scholar]
- 27.O'Neill, A., J., and I. Chopra. 2006. Molecular basis of _fusB_-mediated resistance to fusidic acid in Staphylococcus aureus. Mol. Microbiol. 59**:**664-676. [DOI] [PubMed] [Google Scholar]
- 28.O'Neill, A. J., A. R. Larsen, A. S. Henriksen, and I. Chopra. 2004. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob. Agents Chemother. 48**:**3594-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill, A. J., A. R. Larsen, R. Skov, A. Henriksen, and I. Chopra. 2005. Characterization of the epidemic European fusidic acid-resistant impetigo clone of Staphylococcus aureus, abstr. O212. 15th Eur. Cong. Clin. Microbiol. Infect. Dis. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [DOI] [PMC free article] [PubMed]
- 30.Osterlund, A., T. Eden, B. Olsson-Liljequist, S. Haeggman, and G. Kahlmeter. 2002. Clonal spread among Swedish children of a Staphylococcus aureus strain resistant to fusidic acid. Scand. J. Infect. Dis. 34**:**729-734. [DOI] [PubMed] [Google Scholar]
- 31.Rosdahl, V. T. 1976. Fusidic acid-resistant Staphylococcus aureus. Zentbl. Bakteriol. 1976(Suppl. 5)**:**1021-1026. [Google Scholar]
- 32.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16**:**944-945. [DOI] [PubMed] [Google Scholar]
- 33.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26**:**544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 35.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73**:**133-138. [DOI] [PubMed] [Google Scholar]
- 36.Sugai, M., K. Hashimoto, A. Kikuchi, S. Inoue, H. Okumura, K. Matsumoto, Y. Goto, H. Ohgai, K. Moriishi, B. Syuto, et al. 1992. Epidermal cell differentiation inhibitor ADP-ribosylates small GTP-binding proteins and induces hyperplasia of epidermis. J. Biol. Chem. 267**:**2600-2604. [PubMed] [Google Scholar]
- 37.Sule, O., N. Brown, D. F. Brown, and N. Burrows. 2002. Fusidic acid cream for impetigo: judicious use is advisable. BMJ 324**:**1394. [PubMed] [Google Scholar]
- 38.Tveten, Y., A. Jenkins, and B. E. Kristiansen. 2002. A fusidic acid-resistant clone of Staphylococcus aureus associated with impetigo bullosa is spreading in Norway. J. Antimicrob. Chemother. 50**:**873-876. [DOI] [PubMed] [Google Scholar]
- 39.Vriesema, A. J., S. A. Zaat, and J. Dankert. 1996. A simple procedure for isolation of cloning vectors and endogenous plasmids from viridans group streptococci and Staphylococcus aureus. Appl. Environ. Microbiol. 62**:**3527-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi, T., T. Hayashi, H. Takami, M. Ohnishi, T. Murata, K. Nakayama, K. Asakawa, M. Ohara, H. Komatsuzawa, and M. Sugai. 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect. Immun. 69**:**7760-7771. [DOI] [PMC free article] [PubMed] [Google Scholar]