Uracil–DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms (original) (raw)
Abstract
DNA glycosylases UNG and SMUG1 excise uracil from DNA and belong to the same protein superfamily. Vertebrates contain both SMUG1 and UNG, but their distinct roles in base excision repair (BER) of deaminated cytosine (U:G) are still not fully defined. Here we have examined the ability of human SMUG1 and UNG2 (nuclear UNG) to initiate and coordinate repair of U:G mismatches. When expressed in Escherichia coli cells, human UNG2 initiates complete repair of deaminated cytosine, while SMUG1 inhibits cell proliferation. In vitro, we show that SMUG1 binds tightly to AP-sites and inhibits AP-site cleavage by AP-endonucleases. Furthermore, a specific motif important for the AP-site product binding has been identified. Mutations in this motif increase catalytic turnover due to reduced product binding. In contrast, the highly efficient UNG2 lacks product-binding capacity and stimulates AP-site cleavage by APE1, facilitating the two first steps in BER. In summary, this work reveals that SMUG1 and UNG2 coordinate the initial steps of BER by distinct mechanisms. UNG2 is apparently adapted to rapid and highly coordinated repair of uracil (U:G and U:A) in replicating DNA, while the less efficient SMUG1 may be more important in repair of deaminated cytosine (U:G) in non-replicating chromatin.
INTRODUCTION
Uracil is a common base lesion in DNA and is introduced into the genome by deamination of cytosine and misincorporation of dUMP instead of dTMP during replication. Spontaneous deamination of cytosine has been estimated to occur at a rate of 60–500 events per day in human cells (1–3). In addition, recent research has revealed that enzymatic deamination of cytosine at the Ig loci by activation-induced cytosine deaminase (AID) initiates the antigen-dependent antibody diversification processes (4). Uracil generated by deamination of cytosine is 100% miscoding, and result in C:G to T:A transition mutations if not repaired prior to replication. Misincorporated uracil is not directly miscoding, but it appears to be a critical source of spontaneously generated AP-sites (apurinic/apyrimidinic-sites) in the genome (5).
Uracil and some uracil analogs generated by oxidation of cytosine are excised from the genome by uracil-DNA glycosylases (UDGs). Mammalian cell nuclei contain at least four UDGs; UNG2, SMUG1, TDG and MBD4. Current evidence suggests that UNG2 (uracil-N-glycosylase 2) and SMUG1 (single-strand-selective monofunctional UDG) are the major enzymes responsible for repair of spontaneously deaminated cytosine (6–8), while post-replicative excision of misincorporated dUMP (U:A) and excision of AID-generated uracil (U:G) are performed mainly by UNG2 alone (9–11). Consistent with the role of UNG2 in replication associated repair, UNG2 binds PCNA and RPA, is localized to replication foci, and is cell cycle regulated with the highest levels in S-phase (9,12–15). Conversely, SMUG1, is not cell cycle regulated and is evenly distributed in the nucleoplasm (7). SMUG1 excises uracil from DNA with a much lower efficiency than UNG2, but has broader substrate specificity. Only SMUG1 excises thymine with an oxidized methyl group (7,16). UNG and SMUG1 belong to a superfamily that has apparently evolved from the same ancestral gene (17). Comparison of crystal structures of human UNG and Xenopus SMUG1 has revealed that these enzymes share a common fold and that the SMUG1 active site is a mosaic of features from UNG and MUG/TDG enzyme families (18).
UNG is widely distributed in bacteria, eukaryotes and even some large DNA viruses, while SMUG1 has previously been reported to be present in vertebrates and insects only (6,17). Here we report the existence of bacteria that contain SMUG1 as their only identified UDG. Interestingly, identification of these bacterial SMUG1 orthologs shed new light on the origin of SMUG1 and UNG. Vertebrates contain both SMUG1 and UNG, but their distinct roles in base excision repair (BER) of deaminated cytosine are still not fully defined. We have compared the repair mechanisms of human SMUG1 (hSMUG1) and human UNG2 (hUNG2) on deaminated cytosine, in vivo by using replicating ung− Escherichia coli cells containing AID-induced U:G lesions, and in vitro using purified enzymes (hSMUG1, hUNG2 and hAPE1) including a panel of hSMUG1 mutants. We find that only hUNG2 can complement E. coli Ung in repair of U:G mismatches, whereas hSMUG1 inhibits cell proliferation in the same system. In vitro analyses reveal that hSMUG1 and hUNG2 coordinate the initial steps of BER by distinct mechanisms. Furthermore, we characterize a specific motif in hSMUG1 that confers U:G-substrate preference and stabilizes the product AP-site binding. Finally, we propose a model for how SMUG1 and UNG2 initiate and coordinate repair of deaminated cytosine (U:G) by distinct mechanisms. This model is consistent with a role for the slow-acting SMUG1 in repair of deaminated cytosine in non-replicating chromatin, and efficient and highly coordinated repair by UNG2 of uracil (both U:G and U:A) in replicating DNA.
MATERIALS AND METHODS
DNA constructs and site-directed mutagenesis
To generate pAID an NcoI site flanking the ATG start codon of hAID cDNA (Image Clone: 4853069) was made by site-directed mutagenesis. The complete reading frame was then cloned into the NcoI–PstI sites of the expression vector pTrc99A (Amersham Biosciences). Cloning of pUNG2 (p658kan-UNG2) was published previously (7). pSMUG1 (p658kan-SMUG1) was constructed by cloning the complete reading frame of hSMUG1 cDNA (Image Clone: 726197) as an NdeI–BamHI fragment into the pJB658_cop_251kan vector (7,19). Cloning of 6 x His-tagged hSMUG1 (pET28a–SMUG1) and hUNG2 (pET28a–UNG2) was published previously (10,20). The hAPE1 expression vector (pET14b–APE1) (21) was a gift from Dr Ian Hickson (Cancer Research UK Laboratories, Oxford, UK). Site-directed mutagenesis was carried out using the Quick-Change™ kit (Stratagene), and the mutants were confirmed by sequencing.
In vivo U:G repair assay
Escherichia coli Ung-deficient strain NR8052 (Δpro-lac, thi-, ara, trp9777, ung1) or Ung-proficient strain NR8051 (Δpro-lac, thi-, ara, trp9777) (22) were transformed with the IPTG inducible constructs; pTrc99A-AID [encoding AID wild-type and ampicillin resistance (ampR)] or pTrc99A-AID-C87A (encoding catalytically inactive AID as control) followed by transformation with the toluic acid inducible p658kan-hSMUG1 or p658kan-hUNG2 constructs [encoding hSMUG1 and hUNG2, respectively and kanamycin resistance (kanR)]. Empty pJB658_cop_251kan vector (7) was used as control. Single ampR + kanR colonies were picked from plate and grown in 3 ml LB containing 100 µg/ml amp, 30 µg/ml kan, 1 mM IPTG and 1 mM toluic acid at 30°C over night. Aliquots were mixed with 3 ml soft agar and plated on LB-amp + kan plates and LB-amp + kan plates containing 100 µg/ml rifampicin (rif) 100 µg/ml. The numbers of (amp + kan)R colonies were counted after incubating the plates at 37°C for 24 h, while the numbers of (amp + kan + rif)R colonies were counted after 48 h. Mutation frequencies were calculated as the number of (amp + kan + rif)R colonies per 108 (amp + kan)R colonies.
Western analysis
Expression of hUNG2 and hSMUG1 in E. coli was confirmed by western analysis. Five microgram soluble cell lysate from cultures induced over night were separated by SDS–PAGE (NuPAGE®, Invitrogen) and electro-blotted onto Immobilon PVDF membranes (Millipore). hUNG2 and hSMUG1 were detected using the primary antibodies PU101 (23) and PSM1 (7), respectively, followed by HRP-conjugated swine anti-rabbit secondary antibody (DakoCytomation) and Super Signal West Femto substrate (Pierce Chemical Co.) The western blots were analysed on a Kodac Image station 2000R.
Expression and purification of recombinant 6 x His-tagged enzymes
hSMUG1, hUNG2 and hAPE1 were expressed in E. coli BL21-CodonPlus (DE3)-RIL (Stratagene) and purified as described (7,10). Protein concentrations were measured by the Bradford protein assay (BioRad) using BSA as standard, and stored at –80°C in 50% glycerol and 1 mM DTT. The hSMUG1 mutants were confirmed by trypsin digestion followed by MALDI–TOF mass spectrometry.
UDG activity assays
Standard UDG assays were performed as previously described (7). Briefly, 10 nM SMUG1 and 1.8 µM [3H]dUMP-containing calf thymus DNA (U:A) with specific activity 0.5 mCi/μmol were incubated in a 20 μl assay mixture containing (final) 20 mM Tris–HCl pH 7.5, 10 mM NaCl, 7.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.5 mg/ml BSA (UDG assay buffer), for 10 min at 30°C. The amount of released uracil was measured as described (24).
Limited turnover oligonucleotide assays were performed by using equimolar amounts of enzyme and [33P]-5-end-labelled oligonucleotide (U141: CATAAAGTGUAAAGCCTGG). dsDNA substrates (U141A and U141G) were prepared and assays were performed as previously described (7). Enzyme 20 nM and substrate 20 nM were incubated in 10 µl UDG assay buffer at 30°C for 0, 5, 30 or 60 min. Substrate and product were quantified by phosphor imaging.
Multiple turnover oligonucleotide assays were performed as described (7) by using 0.4 nM enzyme and 20 nM oligonucleotide substrates in 10 µl UDG assay buffer at 30°C for 15 min. Substrate and product were quantified by phosphor imaging.
Kinetic assays for the determination of Km and kcat were performed under Michaelis–Menten conditions, using high molar excess of substrate: 20 nM [33P]-labelled and 1–20 µM non-labelled U141A, U141G and U141 oligonucleotide substrates in 10 µl samples were incubated at 30°C for 10 min. In this assay, enzymes (SMUG1-WT and SMUG1-P245A) were adjusted to give <30% uracil excision at each substrate concentration. 1 nM, 20 nM and 5 nM enzyme were used together with the U:G, U:A and Uss substrates, respectively. Kinetic parameters were calculated according to the method of Wilkinson using the Enzpack for Windows version 1.4 (Biosoft).
UDG activity in bacterial extracts were measured using 1 µg soluble cell lysate (from cultures induced with 1 mM toluic acid at 30°C over night) and 5 nM U141G oligonucleotide in 10 µl total volume containing 20 mM Tris–HCl pH 7.5, 35 mM NaCl, 3 mM EDTA and 1 mM DTT with or without 0.1 µg Ugi. The samples were incubated at 30°C for 15 min and analysed as described (7).
AP-site inhibition assays: AP-site inhibitors were generated from an uracil-containing oligonucleotide (U93:TGAAATTGUTATCCGCTCA). Fifteen nanomol U93 were incubated with 1 µg (40 pmol) hUNGΔ84 (23) in a total volume of 300 µl containing (final) 20 mM Tris–HCl pH 7.5, 10 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.5 mg/ml BSA at 37°C for 2 h. UNG was inactivated at 65°C for 5 min, followed by addition of the specific UNG protein inhibitor Ugi (160 pmol). To prepare dsAP-DNA the AP-oligonucleotide was annealed with 50% molar excess of complementary oligonucleotides containing either A (93A) or G (93G) opposite the AP-site, generating AP:A and AP:G, respectively. The inhibitory effects of AP-sites were analysed by standard UDG assays using 1.8 µM [3H]dUMP-containing calf thymus DNA substrate, 7.5 nM hSMUG1 (WT or mutants) and 62.5 nM AP:G or 500 nM AP:A inhibitor.
Electrophoretic mobility shift assay (EMSA)
SMUG1 (0.05–0.50 µM) or UNG2 (0.05–0.50 µM) was incubated with 4 nM [33P]-end-labeled oligonucleotide (U141G, U141A, T141A, or U141ss) in 10 μl UDG assay buffer containing 2.5% glycerol for 30 min at 30°C to generate AP-sites. Complete excision of U from the oligonucleotides was confirmed in parallel samples by piperidine cleavage and denaturing PAGE as described (7). After uracil-excision, binding of enzyme to end-products (AP-sites) was analysed by non-denaturing 8% PAGE (containing 2.5% glycerol) in 0.5 × TAE pH 8 buffer at room temperature for 15 min at 100 V followed by 30 min at 150 V. The gels were fixed, dried and analyzed by phosphorimaging. The amount of bound AP-site oligonucleotide was quantified and plotted using a sigmoid curve fit model in GraphPad Prism®. Kd values (concentration of enzyme giving 50% of maximum binding) were calculated for SMUG1 on AP:A and AP:G.
AP-endonuclease activity assay
Exonuclease III was purchased from New England Biolabs (#M0206S). AP-site substrate was prepared by incubating 3 pmol [33P]-end labelled U141-oligonucleotide with 37.5n g (1.2 pmol) hUNGΔ84 (23) in a total volume of 30 µl containing (final) 20 mM Tris–HCl pH 7.5, 10 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.5 mg/ml BSA at 37°C for 1 h. The UNG enzyme was inactivated by heating at 65°C for 10 min. To generate dsAP-DNA the AP-oligonucleotide was annealed with 50% molar excess of complementary oligonucleotide containing G (141G) opposite the AP-site.
AP-endonuclease assays were performed with 0.025 nM hAPE1 or E. coli ExoIII and 2 nM [33P]-labelled AP:G substrate in final volume of 10 µl UDG assay buffer and incubated at 30°C for 10 min. AP-endonuclease cleaved products and uncleaved AP-site substrates were separated by denaturing PAGE and quantified by phosphorimaging.
RESULTS
SMUG1 is present in both prokaryotes and eukaryotes
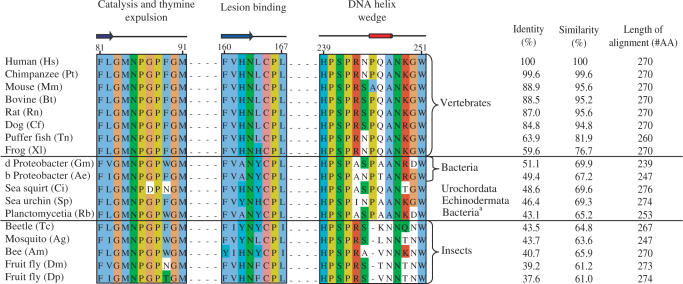
SMUG1 was suggested to be a relatively new evolutionary offspring in the UDG superfamily found only in vertebrates and insects (6,17). However, a BLAST search using the sequence of hSMUG1 protein as query revealed SMUG1 orthologs both in prokaryotes (proteobacteria and planctomycetes) and in marine non-vertebrates such as sea urchin and sea squirt (Figure 1). Remarkably, the vertebrate SMUG1 has highest similarity to sequences identified in bacteria, showing 51.1% identity and 69.9% similarity between human SMUG1 and SMUG1 from Geobacter metallireducens (Figure 1). We did not find UNG genes in the SMUG1-containing non-vertebrate organisms identified here except in sea urchin, neither is it present in insects. Moreover, the prokaryotes encoding SMUG1 also lack orthologs of other members of the UDG family [MUG, UDG 4 (25) and UDG 5 (26)], indicating that SMUG1 may be the only uracil-DNA glycosylase in these species.
Figure 1.
Alignment of SMUG1 orthologs arranged in descending order of sequence similarity to hSMUG1. Sequences were obtained by TBLASTN 2.2.14 including GenBank, EMBL, DDBJ and PDB sequences (54). The alignment of the SMUG1 orthologs was generated using ClustalW (55); the final alignment was made with Jalview (56). Secondary structure of xSMUG1 is illustrated above the alignment. The alignment displays the important functional motifs characterizing the SMUG1 enzyme with its description above. Individual residues within each motif are coloured according to ClustalX colour coding. Species and accession number used are as follows: Hs (H. sapiens, AAL86910.1), Pt (Pan troglodytes, XP_509109), Mm (M. musculus, Q6P5C5), Bt (Bos Taurus, Q59I47), Rn (Rattus norvegicus, Q811Q1), Cf (Canis familiaris, XP_543623.2), Tn (Tetraodon nigroviridis, CAF95523.1), Xl (Xenopus laevis, Q9YGN6), Gm (Geobacter metallireducens GS-15, YP_282069.1), AE (Azoarcus sp. EbN1, YP_158606.1),Ci (Ciona intestinalis, AK115076.1), Sp (Strongylocentrotus purpuratus, XP_782746.1), Rb (Rhodopirellula baltica SH 1, NP_869403.1), Tc (Tribolium castaneum, XP_971699.1), Ag (Anopheles gambiae str. PEST, XP_312038.2), Am (Apis mellifera, XP_396883.2), Dm (Drosophila melanogaster, NP_650609.1), Dp (Drosophila pseudoobscura, EAL272349.1_)_.
hSMUG1 cannot complement E. coli Ung in repair of U:G mismatches
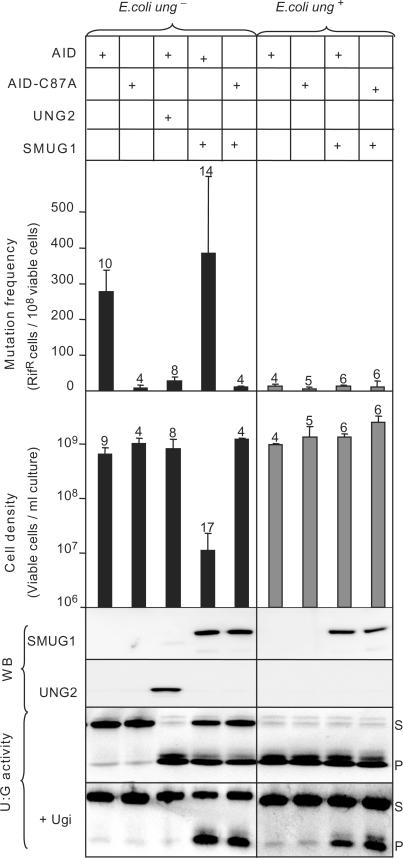
The observation that SMUG1 is apparently the sole UDG in some bacteria, prompted us to examine whether human SMUG1 can act as a functional homolog of Ung in E. coli that lacks SMUG1. Human SMUG1 has been reported to complement Ung1 in Saccharomyces cerevisiae (27). In the yeast study, antifolate agents were used to increase misincorporation of dUMP, generating U:A base pairs. However, mammalian SMUG1 is more likely involved in removal of deaminated cytosine rather than misincorporated uracil (6), thus U:A pairs may not represent the most relevant in vivo substrate for SMUG1. Here we used an in vivo system in which the cytosine deaminase AID was expressed in Ung-deficient E. coli to specifically generate promutagenic U:G mispairs (28). In this background, we expressed hSMUG1 or hUNG2. Mutation frequencies were monitored by the rifampicin resistance (rifR) assay (29), and cell growth was analysed by counting viable cells. Expression of SMUG1 and UNG2 in the cells was verified by western blot analysis and uracil-excision activity (U:G substrate) in clarified lysate from induced cultures. The results are summarized in Figure 2.
Figure 2.
Mutation frequency and cell density in cultures of Ung- deficient and Ung-proficient E. coli ung- cells co-expressing AID and SMUG1 or UNG2. U:G mispairs were introduced into chromosomal DNA by expressing AID in E. coli ung- cells (black bars) and E. coli ung+ cells (grey bars). A catalytically inactive AID mutant (AID-C87A) and empty vector were included as controls. Mutation frequencies were measured by the rifampicin resistance assay (# rifR/108 cells). The error bars represent standard deviations calculated from the number of experiments given on top of each bar. Expression of SMUG1 and UNG2 was confirmed by western blot (WB) analysis and UDG activity were assayed on a U:G oligonucleotide substrate (U:G activity). Ugi was added (+Ugi) to the extracts to verify SMUG1 activity. Uncleaved substrate (S) and cleaved product (P) are indicated.
Expression of AID in Ung-deficient E. coli yields a mutator phenotype, which is reversed by co-expression of UNG2. SMUG1 did not suppress the mutator phenotype, but markedly inhibited cell growth in AID-expressing Ung-deficient cells. Notably, hSMUG1 was neither growth inhibitory nor mutagenic in ung+ cells. The level of genomic uracil is enhanced more than 30-fold (to 31 uracil per 106 nucleotides) in Ung-deficient E. coli cells, probably mostly U:A base pairs caused by replicative incorporation of dUMP (30). Importantly, SMUG1 did not influence growth or mutation frequency in _ung_− cells expressing the inactive AID-C87A mutant (Figure 2), neither did induction of only SMUG1 in Ung-deficient cells inhibit cell growth (data not shown). This demonstrates that the growth inhibitory effect of SMUG1 is dependent on U:G-lesions or U:G-repair intermediates. Taken together these results reveal that SMUG1 does not act as a functional homolog of Ung in U:G repair in proliferating E. coli cells.
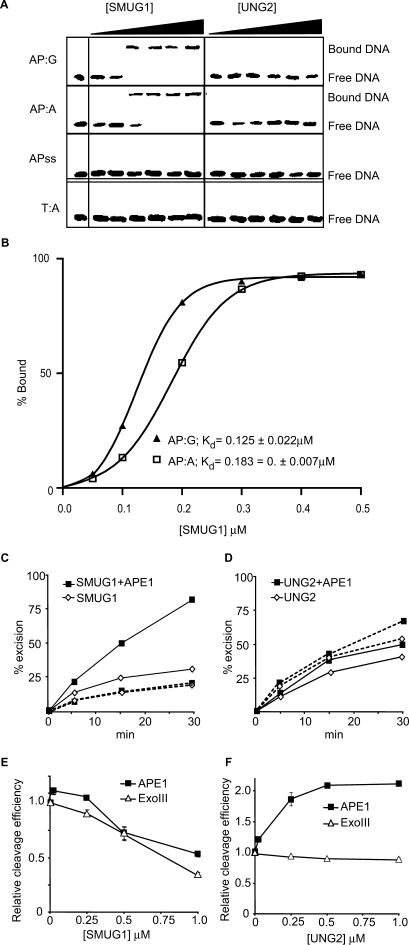
SMUG1 binds to AP-sites and inhibits APE1, while UNG2 does not bind AP-sites and stimulates APE1
To elucidate the molecular mechanisms underlying the observed in vivo effects of SMUG1 and UNG2, we analysed the product (AP-site) binding subsequent to uracil-excision by purified human SMUG1 and UNG2 using electrophoretic mobility shift assays (EMSA). The results, illustrated in Figure 3A, demonstrate that SMUG1 readily binds to AP-sites in dsDNA (AP:G and AP:A), while no binding to AP-sites in single-stranded DNA (APss) or dsDNA without AP-site (T:A) was detected. In contrast, we did not observe binding of UNG2 to the same set of oligonucleotides. SMUG1 binds AP:G with slightly higher affinity than AP:A with Kd values (concentration of enzyme yielding 50% of maximum binding) calculated to 0.125 ± 0.022 µM and 0.183 ± 0.007 µM, respectively (Figure 3B). The sigmoid curve plotted in Figure 3B represents the EMSA data in Figure 3A. The binding experiments were, however, repeated several times and consistently revealed higher affinity for AP:G than for AP:A.
Figure 3.
Product binding, effects of the AP-endonuclease APE1 and effects on AP-endonucleases APE1 and Exo III. (A) EMSA analysis of SMUG1 and UNG2 on AP-sites; 50, 100, 200, 300, 400 and 500 nM hSMUG1 and hUNG2 were incubated with 4 nM ds oligonucleotide (with complementary bases as indicated) or ss oligonucleotide, following uracil-excision binding to the product AP-sites were analysed by non-denaturating PAGE. (B) Percent bound AP:G and AP:A oligo plotted against the concentration of SMUG1 (µM) using a sigmoid curve fit model in GraphPad Prism®. The Kd values were calculated and represent the concentration of SMUG1 giving 50% of maximal binding to the AP-oligo. (C) Multiple turnover oligonucleotide assay with 0.85 nM SMUG1 and 20 nM U:G substrate (solid lines) or 20 nM Uss substrate (dotted lines) in the absence or presence of 8.5 nM APE1. (D) Multiple turnover oligonucleotide assay with 0.085 nM UNG2 and 20 nM U:G substrate (solid lines) or 20 nM Uss substrate (dotted lines) in the absence or presence of 8.5 nM APE1. Uracil excision was quantified after piperidine cleavage of the AP-site and separation by PAGE. (E) AP-cleavage activity of APE1 and ExoIII in the presence of SMUG1. (F) AP-cleavage activity of APE1 and ExoIII in the presence of UNG2. In (E) and (F), the AP-endonucleases (0.025 nM) were incubated for 10 min with 2 nM oligonucleotide substrate containing a central AP-site opposite guanine (AP:G) together with increasing amounts (0–1 µM) of SMUG1 or UNG2. AP-endonucleolytic cleavage was quantified after separation by PAGE.
To gain more insight into the coordination of the first and the second step of BER of deaminated cytosine, we investigated the effect of the major human AP endonuclease, APE1, on uracil-excision from U:G substrate by SMUG1 and UNG2. The excision rate from U:G substrate by SMUG1 was 2–3-fold stimulated by APE1, while no stimulatory effect was observed with Uss substrates (Figure 3C). In contrast, uracil-excision by UNG2 was only weakly stimulated with both substrates (Figure 3D). This confirms our previous quantitation of APE1 stimulation of SMUG1 and UNG2 measured with U:A-containing substrate (7), and is in accordance with previously published data on SMUG1 (6). Together with the EMSA results (Figure 3A and B), this indicates that uracil-turnover by SMUG1 is increased by alleviation of product-binding after cleavage of the AP-site. In support of this, we observed that bacterial AP endonuclease Endo IV also stimulates SMUG1 activity and that SMUG1 does not bind to nicked AP-sites (data not shown).
Based on the different AP-site binding properties of SMUG1 and UNG2 we analysed their effects on the activity of human APE1 and bacterial ExoIII. A molar excess of hSMUG1 inhibited both hAPE1 and ExoIII activity (Figure 3E), indicating that SMUG1 and AP endonucleases compete for binding to AP-sites. In contrast, hUNG2 markedly stimulated the activity of hAPE1 but had no effect on ExoIII (Figure 3F). Thus, SMUG1 probably binds strongly to the product until it is displaced by an AP endonuclease that cleaves the AP-site, whereas UNG2 may physically interact with APE1 to coordinate and facilitate the first and the second step in BER.
Single amino acid substitutions in the active site pocket of SMUG1 have moderate effect on catalytic activity
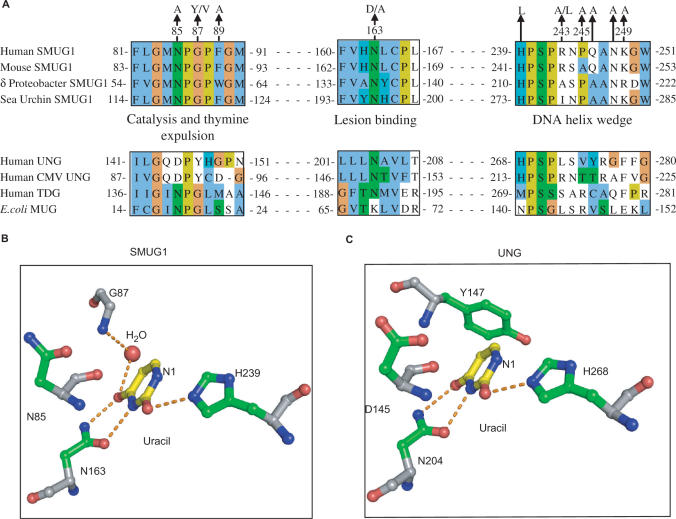
We have previously characterized substrate specificities and kinetic constants of human SMUG1 and UNG2 (7). In addition, mechanisms of uracil recognition, substrate binding and catalysis by human UNG have been extensively studied (23,31–39). The active site of SMUG1 is a mosaic of features from UNG and MUG/TDG enzyme families (18) (Figure 4A). To further explore the molecular mechanisms of SMUG1, active-site mutants were generated (Figure 4A), purified and activities were measured by standard UDG assays (see ‘Materials and Methods’ section) using 1.8-µM [3H]dUMP-containing DNA substrate (U:A). Residual activities of corresponding hSMUG1 and hUNG mutants, analysed by identical activity assays, are compared and listed in Table 1.
Figure 4.
Alignment of important motifs and structural organization of active-site residues. (A) SMUG1 orthologs from human (AAL86910.1), mouse (XP_509109.1), δ proteo-bacteria (YP_383069.1) and sea urchin (XP_782746.1) aligned in ClustalW (55) with human UNG (NP_550433.1), human Cytomegalovirus (CMV) UNG (P16769), human TDG (NP_003202.3) and E. coli MUG (P0A9H1). Final alignment was made in Jalview (56) and individual residues coloured according to ClustalX colour codes. SMUG1 residues mutated in this work are indicated above. (B) Structural organization of the active sites of SMUG1 (PDB entry 1OE5) and human UNG (PDB entry 1SSP), showing key residues (numbered according the hSMUG1 orthologs) in catalysis, lesion binding and specificity.
Table 1.
Activity of SMUG1 active-site mutants compared with activity of corresponding UNG mutants
| Residue function | hSMUG1 mutant | Activity (% of WT) | hUNG mutant | Activity (% of WT) | |
|---|---|---|---|---|---|
| U:A | U:G | U:A | |||
| H2O coordination | N85A | 3.0 ± 0.2 | 7.0 ± 0.4 | D145N | 0.04a |
| Thymine expulsion | G87V | 21.8 ± 1.5 | 58 ± 2 | Y147A | 0.05b |
| G87Y | Not detectable | Not detectable | |||
| Substrate binding | N163D | 11.2 ± 1.5 | 32 ± 0.2 | N204D | 0.04b |
| Stabilisation of transition state | H239L | 28.6 ± 0.9 | 26 ± 5 | H268L | 0.32a |
In the structures of xenopus SMUG1 (xSMUG1) and hUNG an asparagine at the bottom of the catalytic pocket binds to O4 and N3 of uracil (Figure 4B). Mutating this residue to aspartic acid in hSMUG1 (N163D) resulted in 11% residual activity, the residual activity of the corresponding UNG mutant (N204D) was only 0.04% (33). H268 in hUNG is believed to have a critical function in stabilization of the transition state intermediate (40), and accordingly the hUNG-H268L mutant displayed only 0.32% activity compared with WT. Interestingly, the equivalent mutation in hSMUG1 (H239L) retained 28.6% residual activity. To rule out a possible contribution to activity from contaminating UNG, we analysed the H268L mutant in the presence of the specific UNG inhibitor Ugi. Ugi did not have any inhibitory effect on activity, demonstrating that the hSMUG1-H268L mutant was not contaminated by UNG (data not shown). Mutation of the asparagine (hSMUG1-N85A) proposed to coordinate the active water molecule in SMUG1 (18) resulted in 3% residual activity, whereas mutation of the corresponding residue in hUNG (D145N) reduced the activity to 0.04% compared with WT.
In UNG, Tyr147 blocks the entrance of thymine to the active site pocket (29,31,33). SMUG1 has glycine in the corresponding position, explaining its ability to accept uracil with hydrophilic substitutions at C5 position (Figure 4A and B). Substitution of this glycine with the much larger tyrosine (SMUG1-G87Y) resulted in a catalytically dead enzyme, suggesting a side-chain orientation of this residue that completely blocks entrance of all substrates to the binding pocket. However, except from this latter substitution, corresponding active-site mutations have generally less effect on SMUG1 activity than on UNG activity, when measured on a U:A substrate.
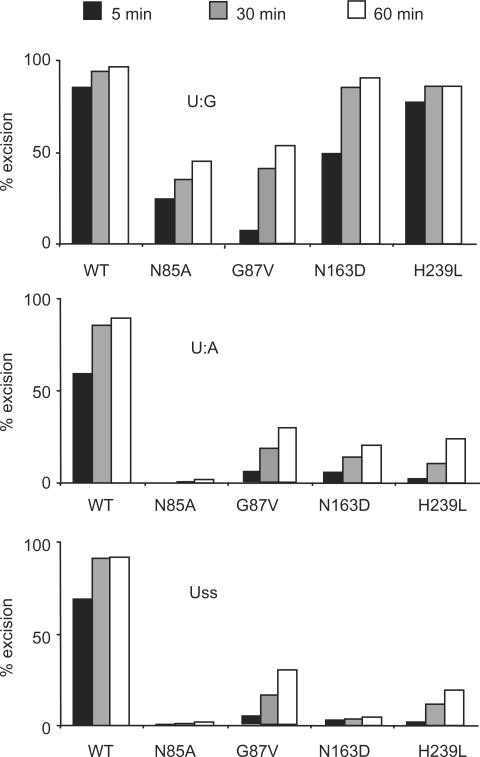
To quantify residual activity of the hSMUG1 mutants towards U:G, we used a multiple turnover oligonucletide assay (see “Materials and Methods” section) with a 50-fold molar excess of U:G substrate. Interestingly, the residual activities of the mutants were even higher when analyzed with U:G substrate as compared with U:A substrate (Table 1). To minimize the effect of product binding, the SMUG1 mutants were also analysed using equimolar amounts of enzyme and oligonucleotides with uracil in either U:G, U:A or Uss contexts. Under these conditions, the U:G preference of the SMUG1 mutants was even more pronounced (Figure 5). A stable enzyme-substrate complex with a long residence time will increase the possibility for catalysis to occur, thus these results indicate that SMUG1 binds U:G substrate (not only AP:G product) with higher affinity than U:A and Uss substrates.
Figure 5.
Uracil-substrate preference of SMUG1 active-site mutants. Limited turnover oligonucleotide assay of SMUG1 mutants. Equimolar amounts (20 nM) of enzyme and [33P]-labelled uracil oligonucleotide substrate (U:G, U:A and Uss) were incubated for 5, 30 and 60 min. Uracil excision was quantified after piperidine cleavage of the AP-site and separation by denaturing PAGE.
A wedge motif in SMUG1 confers sequence specificity, U:G-substrate preference and stabilizes AP:G product binding
The crystal structure of the xSMUG1–DNA complex reveals a DNA-helix penetrating wedge formed by a loop followed by a short α-helix (18). This structural architecture suggests a more invasive interaction with dsDNA than observed for UNG, and the wedge motif apparently also contacts base pairs adjacent to the flipped out lesion (Figure 6A and B). Thus, we examined to what extent the nature of these bases influences the catalytic activity SMUG1 and UNG2. As demonstrated in Figure 6C, SMUG1 activity was markedly influenced by the bases flanking uracil, with a preference for A:T base pairs. Conversely, no such preference was observed for UNG2 using the same dsDNA substrates (Figure 6D). These results support the hypothesis that SMUG1 interacts with the base pairs flanking the lesion in the DNA helix, and possibly disrupts the Watson–Crick hydrogen bonds.
Figure 6.
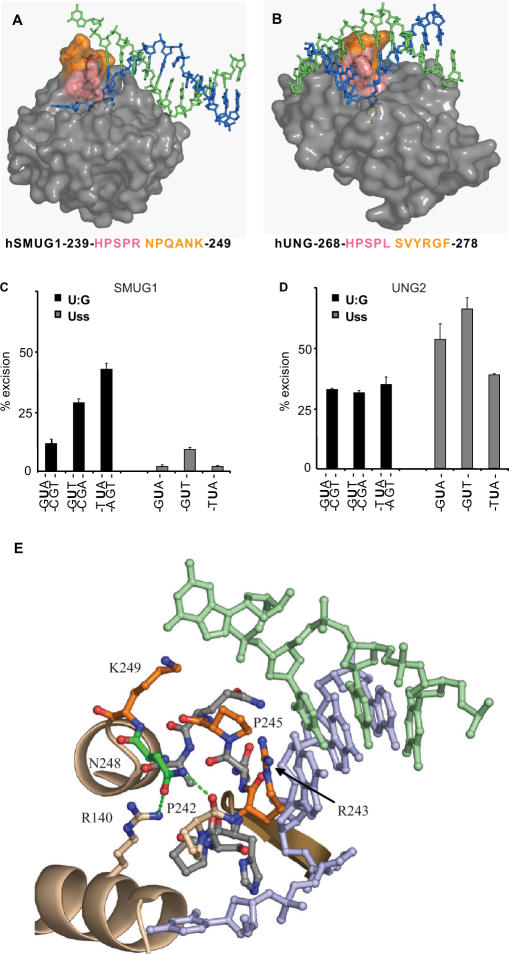
DNA interactions and uracil-context dependence of SMUG1 and UNG. (A, B) Surface models of the DNA complex structure of xSMUG1 (PDB entry 1OE5) and hUNG (PDB entry 1EMH), respectively. The hSMUG1-specific motif 244-NPQANK-249 (xSMUG1 255-NPQANK-260) (orange) points towards the distal strand (green), while the conserved uracil-flipping motif—HPSPR/L—(pink) points towards the uracil-containing proximal strand (blue). (C) Sequence context dependence of hSMUG1. Multiple turnover oligonucleotide assays (0.4 nM SMUG1 and 20 nM substrate) on oligonucleotides with uracil in different sequence contexts. (D) Sequence context dependence of UNG2. Multiple turnover oligonucleotide assays (0.04 nM UNG2 and 20 nM substrate) on substrates with uracil in different sequence contexts. (E) Close-up presentation of the SMUG1 wedge motif (PDB entry 1OE5). Residues with side-chains facing the distal strand (hSMUG1-R243, -P245, -K249) are colored orange. hSMUG1-N248 (colored green) makes two hydrogen bonds (green dots) to hSMUG1-P242 and –R140, respectively. The residues are labeled according to the human SMUG1 sequence.
The N-terminal part of the wedge (239-HPSPR-243) faces the uracil-containing strand, and resembles the intercalating leucine loop in UNG (268-HPSPL-272) (34), except that SMUG1 has arginine in the position corresponding to hUNG-Leu272 (Figure 6A and B). This residue aids expulsion of the uracil residue from the DNA-helix and subsequently fills the gap left by the flipped out nucleotide (34). Similar to SMUG1, UNG encoded by human cytomegalo- and vaccinia virus has arginine at this position (Figure 4A). To analyse this uracil-flipping residue in SMUG1 in more detail we mutated the Arg243 to leucine and alanine. The R243L mutation reduced the activity to about 3% on U:A substrate and 7% on U:G substrate, while the R243A mutation had little effect on enzyme activity (Table 2). Notably, there is alanine at this position in SMUG1 from bacteria and sea squirt and isoleucine in sea urchin (Figures 1 and 4A). Thus, different UNG and SMUG1 family members may either have a large hydrophilic (Arg) or a large hydrophobic side chain (Leu or Ile) or surprisingly even a small side chain (Ala) as the residue expelling uracil.
Table 2.
Activity and AP-site inhibition of SMUG1 mutated in the distal strand interacting motif
| hSMUG1 mutant | Activity (% of WT) | AP-site inhibition (%) | ||
|---|---|---|---|---|
| U:A | U:G | AP:A | AP:G | |
| WT | 100 | 100 | 67 ± 3 | 70 ± 1 |
| R243Aa | 94 ± 2 | 94 ± 10 | 59 ± 2 | 47 ± 2 |
| R243L | 3 ± 1 | 7 ± 2 | Not determined | Not determined |
| N244A | 77 ± 5 | 139 ± 8 | 72 ± 2 | 76 ± 1 |
| P245Aa | 82 ± 6 | 227 ± 7 | 65 ± 2 | 47 ± 1 |
| Q246A | 98 ± 7 | 168 ± 22 | 67 ± 1 | 68 ± 1 |
| N248A | 127 ± 3 | 268 ± 13 | 60 ± 1 | 62 ± 3 |
| K249Aa | 122 ± 6 | 169 ± 11 | 57 ± 3 | 48 ± 1 |
The C-terminal part of the wedge is completely different in SMUG1 and UNG (Figure 6A and B). Whereas no direct interaction between enzyme and the bases in the distal strand was observed in the hUNG-DNA crystal structure (32), the xSMUG-DNA structure indicates that the C-terminal part of the wedge in SMUG1 interacts with the distal strand. To investigate this in more detail, a series of site-specific alanine mutants were generated in the C-terminal part of the wedge motif (243-RNPQANK-249) (Figures 4A and 6E).
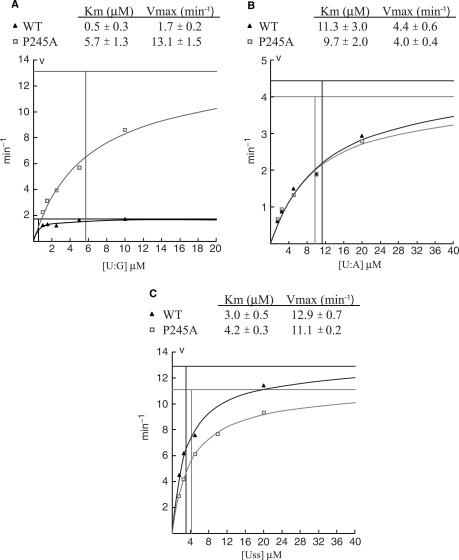
Strikingly, mutations in the 244-NPQANK-249 region of hSMUG1 significantly increased the U:G activity (Table 2), an effect that was most pronounced for the SMUG1-P245A mutant. To investigate this phenomenon in more detail we performed kinetic analysis of SMUG-WT and SMUG1-P245A on U:G, U:A and Uss oligonucleotide substrates. The effect of mutating Pro245 to Ala turned out to be specific (7-fold increase in kcat) to U:G substrates, since no significant changes were observed using U:A and Uss substrates (Figure 7). This strongly suggests that SMUG1-P245 is involved in making specific interactions with guanines opposite uracil-lesions probably by pushing into the base-stack opposite the lesion as suggested by Wibley and colleagues (18) (Figure 6E). Thus, mutation of Pro245 most likely increases turnover by destabilizing binding to the AP:G product. Supporting this view we have previously reported that AP-sites in dsDNA, but not in ssDNA, are inhibitors of hSMUG1, and that AP-sites opposite guanine are much more potent inhibitors than AP-sites opposite adenosine (7). The activities of the SMUG1 wedge mutants were therefore analysed in the presence of oligonucleotides containing AP-sites opposite adenine (AP:A) or guanine (AP:G) (Table 2). SMUG1 mutated in residues pointing towards the distal strand, SMUG1-R243A, SMUG1-P245A and SMUG1-K249A (Figure 6E), were less inhibited (∼47%) by AP:G than WT (70%), indicating a weaker binding to AP:G. As expected from the U:A activity, the wedge mutations had a less pronounced effect on the inhibition by AP:A (Table 2).
Figure 7.
Michaelis–Menten plots of SMUG1-WT and SMUG1-P245A. Each point is the mean value calculated from three independent experiments. Uracil excision at each point is below 30%. Kinetic parameters (Km, kcat) were calculated by the Wilkinson method using the Enzpack 1.4 (Biosoft) software package. (A) 1 nM SMUG1-WT and SMUG1-P245A measured on U:G oligonucleotide substrate (1–10 µM). (B) 20 nM SMUG1-WT and SMUG1-P245A measured on U:A oligonucleotide substrate (2–20 µM). (C) 5 nM SMUG1-WT and SMUG1-P245A measured on Uss oligonucleotide substrate (2–20 µM). Note the different values on the axis.
The side chain of SMUG1-Asn248 is not oriented towards the distal strand, but stabilizes the wedge by making hydrogen bonds to hSMUG1 residues Pro242 and Arg141 (Figure 6E). Mutating this residue (SMUG1-N248A) increased both U:G and U:A activity. Inhibition by AP-sites was, however, only marginally reduced compared with wild-type. This indicates that the increased enzyme activity of the SMUG1-N248A mutant probably is the result of a more flexible wedge, and not due to reduced AP-site binding.
Taken together, these results support our proposed function for the C-terminal part of the wedge motif in binding the distal strand base with a preference for guanine opposite the product AP-site.
DISCUSSION
The existence of SMUG1 orthologs in bacteria suggests that SMUG1 is of older origin than previously assumed. Notably, non-vertebrates (except from sea urchin) appear to have UDGs of either the SMUG1- or the UNG-type, while vertebrates contain both. In vertebrates, SMUG1 and UNG2 have probably evolved to carry out different and specialized functions in processing of genomic uracil (and some uracil analogous) in the most appropriate way depending on uracil context, gene locus (e.g. Ig-genes), cell type, proliferative status, cell cycle phase, sub-nuclear localization and mutagenic potential of the lesion. In the present work, we have compared and characterized some of these specialized molecular functions of hSMUG1 and hUNG2 by both in vivo and in vitro experiments, and demonstrated that they coordinate the initial steps in BER by different molecular mechanisms.
We show that hUNG2, but not hSMUG1, can repair U:G lesions in proliferating E. coli cells in vivo. In contrast, hSMUG1 expression inhibits cell growth in this system. Interestingly, it was reported that hSMUG1 can functionally compensate for Ung1 in yeast cells treated with antifolate agents to increase misincorporation of uracil in the genome (27). In WT cells, antifolate treatment results in S-phase arrest and cellular toxicity due to uracil excision and single-strand breaks. Antifolate-treated ▵ung1 cells are, however, able to complete DNA replication, but when hSMUG1 is expressed the cells are arrested in S-phase like the WT. This indicates that SMUG1 can target misincorporated uracil in the yeast genome and generate cytotoxic AP-sites. However, complete repair of the lesion was not monitored in the yeast study. Since the authors use S-phase arrest and cellular toxicity as a measure of complementation, their results are in agreement with our results, although their conclusion is different. In conclusion, SMUG1 and UNG2 can both target uracil residues, but only UNG2 can initiate complete BER in rapidly growing cells. It cannot be excluded, however, that SMUG1 can compensate for Ung in U:A repair both in yeast and in bacteria. Furthermore, it would be interesting to find out whether SMUG1 from a prokaryote can complement Ung-deficient bacteria.
We have previously characterized the catalytic domain of hUNG in detail (23,32,33). Here we generated active site mutants of hSMUG1 and compared their activities with those of the corresponding mutants of hUNG. Interestingly, using the same standard UDG assay protocol, single active-site mutations in SMUG1 have less effect on catalytic activity than the corresponding mutations in UNG. Analysis of hSMUG1 active-site mutants has also been published by another group (41), and they report a more dramatic reduction in activity of several of the mutants. This is probably because they measured SMUG1 activity using a very high molar excess of enzyme (up to 670-fold) and substrate concentrations about three orders of magnitude below the Km value of hSMUG1-WT (7) (Figure 7A). Such assay conditions will, however, mainly reflect substrate affinity and not catalytic turnover, because the substrate is the limiting factor. By using a high substrate concentration (1.8 µM), we here focused on catalytic turnover and less on substrate affinity. The relatively high residual uracil-excision activity (measured with molar excess of substrate) in the SMUG1 active-site mutants, especially against U:G substrates, may in part be explained by the specific helix-inserting motif that probably is important for binding to dsDNA substrate and product. When SMUG1 is bound to the substrate, the DNA substrate itself may be important to drive the reaction forward by so-called ‘substrate autocatalysis’. For glycosylases in the UDG superfamily, it has been reported that the substrate itself is a major contributor to lowering of the activation energy, thus explaining residual activity in mutants lacking catalytic key residues (38). This ‘substrate autocatalysis’ phenomena may also explain the discrepancy between residual activity of SMUG1 active-site mutants measured at very low substrate concentrations (41) and those presented here measured at high substrate concentrations.
Interestingly, one of the active-site SMUG1 mutants was, however, catalytically dead. Introducing a UNG like thymine expulsion residue in SMUG1 (SMUG1-G87Y) abolished the activity completely. The superimposed structures of xSMUG1 and hUNG reveal that the thymine expulsion loops do not follow the same path, bringing the side-chains of SMUG1-G87Y and UNG-Y147 in different orientation in the substrate binding pockets of the enzymes. Additionally, in SMUG1 this residue is sandwiched between two prolines that restrict conformational flexibility in this loop segment. Thus, a large-residue in this position will most likely block the entrance of the substrate in the active site pocket of SMUG1.
We find that hSMUG1 binds to product AP-sites in dsDNA in vitro, with the strongest binding to AP:G, while no product binding was observed for UNG2. The growth-inhibitory effect of SMUG1 in E. coli cells containing U:G lesions is most likely explained by this product binding. SMUG1 attached to AP-sites may probably interfere with replication, and thereby prevent cell division, a situation that is especially prominent when SMUG1 is over-expressed. Under these circumstances, the endogenous level of AP-endonuclease activity is likely insufficient to alleviate the product binding and replication is blocked. In support of this, we find that SMUG1 inhibits activity of both the human APE1 and the bacterial ExoIII AP-endonucleases (Figure 3D). Thus, a high level of SMUG1 likely interferes with the downstream processing of the AP-sites and prevents complete repair. This is in agreement with the observation that expression of hSMUG1 in Δ_ung1_ yeast cells did not suppress the spontaneous mutator phenotype, but rather caused an increased mutation frequency (27).
It is well known that APE1 stimulates the excision activity of many DNA glycosylases (6,7,42–44). However, stimulation of APE1 by a DNA-glycosylase has to our knowledge not previously been reported. Here we find that UNG2 stimulates the cleavage activity of APE1, indicating that UNG2 may physically interact with APE1. In support of this, we have previously isolated UNG2-associated complexes containing all factors required for complete BER of uracil, including APE1, from human cell extracts (45).
We have examined whether sequence context of the substrate influenced on uracil-excision activity of SMUG1 and UNG2. Surprisingly, the nature of the bases flanking the uracil had only impact on uracil-excision activity of SMUG1 and not of UNG2, measured on the same double-stranded oligonucleotide substrates. The latter was rather unexpected since sequence preference of UNG from several sources [a truncated form of UNG purified from calf thymus, E. coli Ung and the catalytic domain of human UNG (UNGΔ84)] has previously been demonstrated in our laboratory (23,46,47) and by others (herpes simplex virus UDG) (48). However, all these enzymes lack the regulatory N-terminal sequence. In the present study we have analysed the full-length human UNG2 enzyme in presence of Mg2+, which has a strong stimulatory effect particularly on UNG2 (7,20,49). It is possible that the N-terminal domain of UNG2 diminishes the sequence specificity observed for the truncated forms of UNG in order to obtain the most efficient repair of uracil in all contexts at the replication fork.
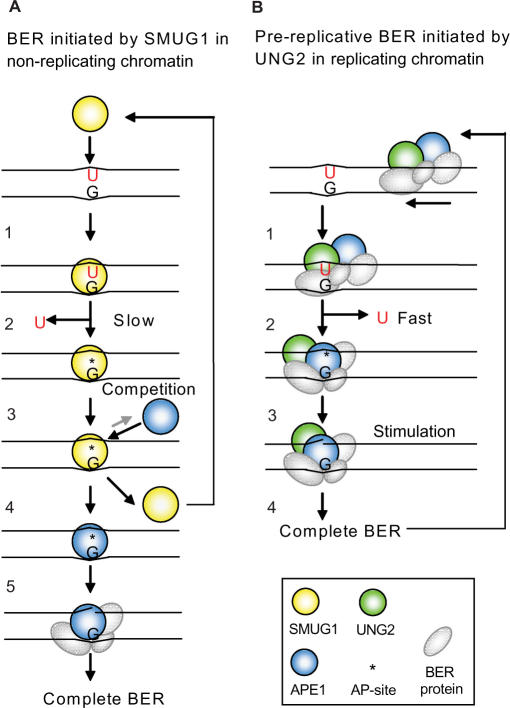
Taken together, it is clear that SMUG1 and UNG2 have evolved distinct mechanisms for the coordination of the second step in BER. Based on previous results and the new data presented here, we propose a model for how SMUG1 and UNG2 initiates and coordinates repair of deaminated cytosine (U:G) by distinct ‘hand-over’ mechanisms (Figure 8). This model is consistent with a role for SMUG1 in repair of deaminated cytosine in non-replicating chromatin and repair of uracil (U:G and U:A) by UNG2 in replication foci. The catalytically highly efficient and context-independent UNG2 enzyme is probably important in rapidly dividing cells to remove deaminated cytosine in front of the moving replication fork (pre-replicative repair), in addition to post-replicative repair of misincorporated uracil (13). This pre-replicative repair of U:G by UNG2 is supported by the observed 5.2-fold increased mutation frequency in Ung-deficient mouse embryonic fibroblasts (MEFs), mostly G:C to A:T transitions (8). SMUG1, on the other hand, is not designed to rapidly repair uracil during replication, and is probably more important in non-replicating chromatin, outside S-phase and in resting cells where the level of UNG2 is low (15). However, SMUG1 counteracts mutations also in cycling mouse cells (MEFs). Knocking down Smug1 by siRNA in MEFs resulted in 2.4-fold increased mutation frequency at the HPRT locus (8). Thus, the slow-acting, product-binding SMUG1 may efficiently recognize deaminated and some oxidized cytosine derivatives in non-replicating dsDNA (especially in A-T rich regions where the cytosine deamination rate is expected to be higher due to increased DNA breathing), excise the lesion and remain attached to the cytotoxic AP-site product until APE1 arrives and initiates further repair.
Figure 8.
Repair of deaminated cytosine; model illustrating distinct coordination of BER initiated by SMUG1 and UNG2 in non-replicating chromatin and in replicating chromatin (foci), respectively. (A) 1. SMUG1 binds to the lesion and interacts with both strands in the DNA-helix. Uracil is probably flipped out of the helix and into the active site. 2. The catalysis is not very efficient because the active site is relaxed to be able to bind several other lesions. SMUG1 stays bound to the AP-site after excision. 3. APE1 competes with SMUG1 for AP-site binding. 4. SMUG1 is released form the product and is free to bind new lesions. 5. APE1 cuts the DNA strand, and Polβ/XRCC1/LigIIIα is recruited and completes BER. (B) 1. UNG2 is likely part of a highly coordinated and efficient repair complex scanning for lesions (U:G) in front of the replication fork (UNG2 is localized in replication foci). 2. Encountering the lesion uracil is flipped out of the DNA-helix and into the highly specific catalytic pocket of UNG2. Uracil is released by efficient hydrolysis of the _N_-glycosidic bond. Note, UNG2 interacts only with the uracil-containing DNA strand. UNG2 is immediately released from the AP-site and APE1 binds. 3. UNG2 stimulates APE1 cleavage of the AP-site and BER is completed.
Notably, in mouse SMUG1, the residue corresponding to the conserved Pro245 in the hSMUG1 wedge motif is alanine (Figure 4A). Kinetic analysis of the hSMUG1-P245A mutant, mimicking mouse SMUG1, revealed that this mutant has more than a 7-fold increased turnover number (kcat) on U:G substrate compared with WT (Figure 7A). The increased U:G activity of this mouse SMUG1 mimicking mutant could thus provide a mechanistic explanation for the apparently higher SMUG1 activity in extracts from mouse cells than from human cells (6,7). This observation should be kept in mind when using mice as model organisms for uracil repair in mammals.
The presence of at least one family member of the uracil-removing glycosylases in all known organisms points to the importance of this repair mechanism. The present article demonstrates new distinct properties of SMUG1 and UNG2 that point to different mechanisms for coordination of the initial steps in BER. Considering functional differences, SMUG1 still seems to be able to compensate for UNG-deficiency in most somatic tissues (6), and is apparently sufficient to maintain genomic stability in some organisms. However, from Ung−/− mice and human UNG-deficient patients it is evident that SMUG1 is not able to compensate for UNG2 in Ig diversification in B-cells (10,11,50,51). Furthermore, old Ung−/− mice develop B-cell lymphomas (52,53). Whether human individuals lacking UNG will develop malignancies remain unknown since they are yet too few identified and too young for conclusions to be made (51). A more comprehensive knowledge of the short-term and long-term consequences of deficient uracil removal require further studies of the Ung−/− mice and generation and characterization of Smug1−/− mice and Ung/Smug1 double knockout mice.
Supplementary Material
[Supplementary Material]
ACKNOWLEDGEMENTS
We wish to thank Petter Aslaksen for verification of the SMUG1 mutants by mass spectrometry, Nina Beate Liabakk for purification of UNG2 and Samuel Bennett for the Ugi expression construct. This work was sponsored by the National Programme for Research in Functional Genomics in Norway (FUGE) in the Research Council of Norway, the Norwegian Cancer Association, the Cancer Fund at St Olav's Hospital Trondheim, the Svanhild and Arne Must Fund for Medical Research and the European Union Integrated Project on DNA Repair. Funding to pay the Open Access publication charges for this article was provided by the Cancer Fund at St Olav's Hospital, Trondheim.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 3.Lari SU, Chen CY, Vertessy BG, Morre J, Bennett SE. Quantitative determination of uracil residues in Escherichia coli DNA: Contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst.) 2006;5:1407–1420. doi: 10.1016/j.dnarep.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem. Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- 5.Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 8.An Q, Robins P, Lindahl T, Barnes DE. C –> T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 10.Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, Durandy A, Krokan HE, Slupphaug G. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 2005;201:2011–2021. doi: 10.1084/jem.20050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Noia JM, Rada C, Neuberger MS. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, et al. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 13.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slupphaug G, Olsen LC, Helland D, Aasland R, Krokan HE. Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res. 1991;19:5131–5137. doi: 10.1093/nar/19.19.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haug T, Skorpen F, Aas PA, Malm V, Skjelbred C, Krokan HE. Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res. 1998;26:1449–1457. doi: 10.1093/nar/26.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L, Koonin EV. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-4-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 19.Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 20.Scaramozzino N, Sanz G, Crance JM, Saparbaev M, Drillien R, Laval J, Kavli B, Garin D. Characterisation of the substrate specificity of homogeneous vaccinia virus uracil-DNA glycosylase. Nucleic Acids Res. 2003;31:4950–4957. doi: 10.1093/nar/gkg672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothwell DG, Hang B, Gorman MA, Freemont PS, Singer B, Hickson ID. Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilises substrate binding. Nucleic Acids Res. 2000;28:2207–2213. doi: 10.1093/nar/28.11.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 24.Krokan H, Wittwer CU. Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981;9:2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 26.Sartori AA, Fitz-Gibbon S, Yang H, Miller JH, Jiricny J. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 2002;21:3182–3191. doi: 10.1093/emboj/cdf309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elateri I, Tinkelenberg BA, Hansbury M, Caradonna S, Muller-Weeks S, Ladner RD. hSMUG1 can functionally compensate for Ung1 in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 2003;2:315–323. doi: 10.1016/s1568-7864(02)00221-5. [DOI] [PubMed] [Google Scholar]
- 28.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 29.Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 2000;19:5542–5551. doi: 10.1093/emboj/19.20.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lari SU, Chen CY, Vertessy BG, Morre J, Bennett SE. Quantitative determination of uracil residues in Escherichia coli DNA: Contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst.) 2006 doi: 10.1016/j.dnarep.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 32.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 33.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA-general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianchet MA, Seiple LA, Jiang YL, Ichikawa Y, Amzel LM, Stivers JT. Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase. Biochemistry. 2003;42:12455–12460. doi: 10.1021/bi035372+. [DOI] [PubMed] [Google Scholar]
- 37.Krosky DJ, Bianchet MA, Seiple L, Chung S, Amzel LM, Stivers JT. Mimicking damaged DNA with a small molecule inhibitor of human UNG2. Nucleic Acids Res. 2006;34:5872–5879. doi: 10.1093/nar/gkl747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 39.Ma A, Hu J, Karplus M, Dinner AR. Implications of alternative substrate binding modes for catalysis by uracil-DNA glycosylase: an apparent discrepancy resolved. Biochemistry. 2006;45:13687–13696. doi: 10.1021/bi061061y. [DOI] [PubMed] [Google Scholar]
- 40.Drohat AC, Xiao G, Tordova M, Jagadeesh J, Pankiewicz KW, Watanabe KA, Gilliland GL, Stivers JT. Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187. Biochemistry. 1999;38:11876–11886. doi: 10.1021/bi9910880. [DOI] [PubMed] [Google Scholar]
- 41.Matsubara M, Tanaka T, Terato H, Ohmae E, Izumi S, Katayanagi K, Ide H. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res. 2004;32:5291–5302. doi: 10.1093/nar/gkh859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia L, Zheng L, Lee HW, Bates SE, Federico L, Shen B, O'Connor TR. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J. Mol. Biol. 2005;346:1259–1274. doi: 10.1016/j.jmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29:743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akbari M, Otterlei M, Pena-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eftedal I, Guddal PH, Slupphaug G, Volden G, Krokan HE. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21:2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsen H, Yazdankhah SP, Eftedal I, Krokan HE. Sequence specificity for removal of uracil from U.A pairs and U.G mismatches by uracil-DNA glycosylase from Escherichia coli, and correlation with mutational hotspots. FEBS Lett. 1995;362:205–209. doi: 10.1016/0014-5793(95)00244-4. [DOI] [PubMed] [Google Scholar]
- 48.Bellamy SR, Baldwin GS. A kinetic analysis of substrate recognition by uracil-DNA glycosylase from herpes simplex virus type 1. Nucleic Acids Res. 2001;29:3857–3863. doi: 10.1093/nar/29.18.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko R, Bennett SE. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair (Amst.) 2005;4:1421–1431. doi: 10.1016/j.dnarep.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 51.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 52.Nilsen H, Stamp G, Andersen S, Hrivnak G, Krokan HE, Lindahl T, Barnes DE. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 53.Andersen S, Ericsson M, Dai HY, Pena-Diaz J, Slupphaug G, Nilsen H, Aarset H, Krokan HE. Monoclonal B-cell hyperplasia and leukocyte imbalance precede development of B-cell malignancies in uracil-DNA glycosylase deficient mice. DNA Repair (Amst.) 2005;4:1432–1441. doi: 10.1016/j.dnarep.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]