Differential Effect of Allorecognition Loci on Phenotype in Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa) (original) (raw)
Abstract
The allorecognition complex of Hydractinia symbiolongicarpus is a chromosomal interval containing two loci, alr1 and alr2, that controls fusion between genetically distinct colonies. Recombination between these two loci has been associated with a heterogeneous class of phenotypes called transitory fusion. A large-scale backcross was performed to generate a population of colonies (N = 106) with recombination breakpoints within the allorecognition complex. Two distinct forms of transitory fusion were correlated with reciprocal recombination products, suggesting that alr1 and alr2 contributed differentially to the allorecognition response. Specifically, type I transitory fusion is associated with rapid and persistent separation of allogeneic tissues, whereas type II transitory fusion generates a patchwork of continuously fusing and separating tissues.
HYDRACTINIA symbiolongicarpus is a colonial cnidarian commonly found as an encrustation on hermit crab shells. Like many sessile, colonial invertebrates, Hydractinia can discriminate among allogeneic conspecific tissues. Many substrate-bound colonial marine invertebrates exhibit indeterminate growth through asexual propagation, which in turn can promote intense competition for space (Jackson 1977). When colonies sharing a substratum grow into contact, compatible colonies can fuse, giving rise to chimeras, while incompatible colonies reject, often leading to an antagonistic response (Hauenschild 1954, 1956; Müller 1964; Ivker 1972; Buss et al. 1984; Lange et al. 1989; Buss and Grosberg 1990; Shenk and Buss 1991; Grosberg et al. 1996; Cadavid et al. 2004). In the case in which two allogeneic colonies fuse, competition ensues between the nonidentical cell lines for contribution to the germline (Buss 1982; Stoner et al. 1999; Laird et al. 2005). Allorecognition mediates the nature of these competitive interactions, effectively controlling the switch between two different levels of natural selection—the colony and the cell line (Buss 1982).
While the topic has been studied for a century in a wide diversity of taxa, genetic techniques have been accessible in only two systems: the athecate hydroid Hydractinia and the compound ascidian Botryllus schlosseri, although other approaches have been pursued in a variety of other systems (e.g., Wiens et al. 2000, 2004). In both the hydroid and ascidian systems, allorecognition is controlled by a set of two linked loci, FuHc and fester in Botryllus and alr1 and alr2 in Hydractinia (Cadavid et al. 2004; De Tomaso et al. 2005; Nyholm et al. 2006), while allodeterminants in all other systems remain unknown. In Hydractinia, fusion, characterized by complete ectodermal and gastrovascular continuity, occurs when colonies share one or both alleles at both loci, whereas rejection, characterized by a failure to adhere and nematocyst discharge, occurs when colonies share no loci at either allele (Cadavid et al. 2004). The Botryllus system is similar in that fusion occurs if colonies match one or more alleles at FuHc and rejection occurs if colonies share no loci (De Tomaso et al. 2005).
An obvious question arises as to what phenotypes may occur in individuals recombinant across this interval. An earlier study demonstrated that transitory fusion phenotypes appear when colonies share alleles at one allorecognition locus and not at the other (Cadavid et al. 2004). Transitory fusion is characterized by an initial fusion of allogeneic tissues, followed by a variety of patterns of subsequent incompatibility (Hauenschild 1954, 1956; Grosberg et al. 1996; Cadavid et al. 2004). Transitory fusion is clearly a heterogeneous phenotypic category with known variation in the amount of time colonies remain fused, whether separation occurs, and whether the separation persists. Only 13 recombinants were isolated by Cadavid et al. (2004), motivating us to initiate a much larger backcross that yielded 106 recombinants. We report here that in this population two predominant classes of transitory fusion were observed and that these two categories covaried with the direction of recombination within the interval. This result clarifies considerably the diversity of Hydractinia allorecognition phenotypes and provides the first evidence that the two allorecognition loci contribute differentially to phenotype.
MATERIALS AND METHODS
Animals:
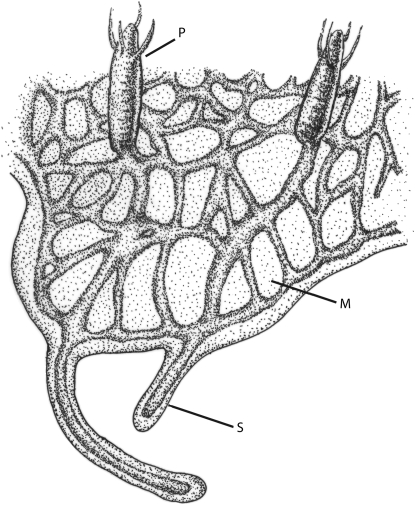
H. symbiolongicarpus grows as a colony by asexual budding. The colony is composed of various types of polyps (“P” in Figure 1), specialized for the different functions of feeding, sexual reproduction, and defense. Gastrovascular canals, called stolons (“S” in Figure 1), connect the polyps in a branching web-like network that over time becomes encased within the mat tissue (“M” in Figure 1).
Figure 1.—
Schematic showing the morphology of a Hydractinia colony. P, polyp; M, mat; S, stolon.
Animal care and breeding have been described in detail elsewhere (Müller 1973; Blackstone and Buss 1991; Cadavid et al. 2004). One modification of protocol was that newly metamorphosed polyps were not hand fed but instead immersed in seawater containing a high density of 2-day-old Artemia salina nauplii. Individuals unable to capture brine shrimp succumbed and were not tested further.
Genetic crosses and allorecognition nomenclature:
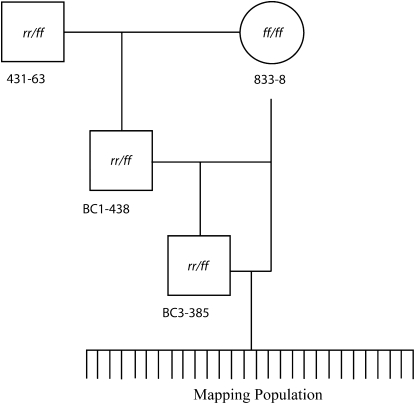
The mapping population was generated by backcrossing animals derived from inbred and congenic lines developed previously (Mokady and Buss 1996; Cadavid et al. 2004). In these laboratory lines, two haplotypes of the allorecognition complex, designated f and r, segregate. Parents of the crosses diagrammed in Figure 2 of this article can be found in the pedigree shown in Figure 2 of Cadavid et al. (2004). We adopt here a terminology slightly different from that used by Cadavid et al. (2004). Animals homozygous for the f haplotype are genetically designated as alr1-f alr2-f/alr1-f alr2-f, which is equivalent to the alr1-f, alr2-α/alr1-f, _alr2-_α designation used in Cadavid et al. (2004). Likewise, animals homozygous for the r haplotype are genetically designated as alr1-r alr2-r/alr1-r alr2-r, which is equivalent to the alr1-r, alr2-β/alr1-r, _alr2-_β designation used in Cadavid et al. (2004). As a shorthand, from here on we omit the locus designations and write simply ff/ff, rr/rr, or, in the case of heterozygotes, rr/ff. To describe the animals recombinant between alr1 and alr2, we use rf/ff (corresponding to alr1-r alr2-f/alr1-f alr2-f) or fr/ff (corresponding to alr1-f alr2-r/alr1-f alr2-f).
Figure 2.—
Pedigree used to generate mapping population. 431-63 is a heterozygote, and 833-8 is a homozygote. Squares represent males, and circles represent females. The lineage of 833-8 can be found in Figure 2 of Cadavid et al. (2004) and likewise the heterozygous male (431-63) was a member of the 431 population represented as the final product of the mating program in Cadavid et al. (2004).
Fusibility assays/determination of allorecognition phenotype:
All individuals identified as recombinant over the interval were tested for their fusibility phenotype in a colony assay unless they died between DNA sampling and genotyping. An inbred individual of genotype rr/rr was used as the tester colony in fusibility assays. Once the colony reached sexual maturity, a small excision from the colony mat bearing two to six feeding polyps was placed next to a similar excision of the tester individual on a glass slide. The excised tissues were held against the slide by a string tied around the slide and allowed to adhere over 2–3 days. The string was removed and the colonies were allowed to grow into contact. Their interaction was observed daily for at least the first 2 days of contact and 6 days/week thereafter for at least 2 weeks. Fusibility tests were noted for ectodermal and gastrovascular continuity. All tests were observed genotype blind and all tests were repeated at least twice, except in cases where this was not possible due to mortality.
SNP discovery/generation of markers:
Five amplified fragment length polymorphisms (AFLPs) were previously identified as linked to the allorecognition trait (Cadavid et al. 2004). To increase the throughput of this study, a strategy was employed that enabled the simultaneous, codominant genotyping of all five markers spanning the interval. The technology used, Sequenom MassARRAY Homogenous MassEXTEND (hME) assay, required a large collection of SNPs to identify a subset of markers that were compatible in a multiplex reaction. Therefore, the five markers identified by Cadavid et al. (2004) were used as starting markers from which to identify physically linked SNPs by a variety of approaches (see supplemental materials at http://www.genetics.org/supplemental/). A collection of 85 SNPs was identified over the chromosomal interval. Sequenom SpectroDESIGNER 2.0 software was used to analyze the SNP collection for potential multiplex assays that included at least one SNP linked to each of the original five AFLPs. The multiplex assay was tested and validated on 95 samples of known alr genotypes.
DNA extraction:
To minimize the time required to maintain the testcross progeny, we developed a nondestructive technique to extract DNA from young colonies at the earliest possible developmental time. A single polyp was removed from the colony with forceps (sterilized in 10% bleach and rinsed with dH2O) and dipped into 100 μl rinse buffer (50 mm Tris–HCl, pH 8, 20 mm NaCl, 1 mm EDTA, pH 8) to remove excess seawater. The polyp was placed into a well of a 96-well microtiter plate containing 2.5 μl digestion buffer (50 mm Tris–HCl, pH 8, 20 mm NaCl, 1 mm EDTA, pH 8, 0.2% SDS, 0.1% proteinase K) and incubated at 55° for 30 min, followed by 5 min at 99.9° to destroy residual proteinase activity. The heat-treated extract was then diluted in 20 μl sterile dH2O and centrifuged at 4000 × g for 30 min. Supernatant (15 μl) was transferred to a new plate well. Samples were then diluted further to 1:200 in dH2O to a concentration of ∼2 ng/μl, 1 μl of which was used in the genotyping assay.
Genotyping:
Sequenom hME genotyping was performed by either the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT) or at the Harvard Partners Center for Genetics and Genomics (HPCGG, Cambridge, MA). PCR conditions prior to the Keck Foundation processing were the following: 2 ng template DNA, 1 pm primers (for primer sequences see Table 1), QIAGEN (Valencia, CA) HotStar Taq buffer 1×, 1.625 mm supplemental magnesium chloride, 0.5 mm dNTP, 0.03 unit QIAGEN HotStarTaq in a 5-μl total volume reaction. Thermal cycling conditions were 95° for 15 min, 95° for 20 sec, 56° for 30 sec, and 72° for 1 min repeated 45 times, followed by 72° for 3 min. For samples genotyped by HPCGG, genomic template was provided to their facilities where PCR and downstream manipulations were performed according to internal protocols. Animals identified as recombinant were genotyped a second time from an independent DNA extraction for verification. Animals identified as nonrecombinant were discarded.
TABLE 1.
MassARRay primers (5′ → 3′)
| Marker | First PCR primera | Second PCR primera | Amplicon length | Extension primer |
|---|---|---|---|---|
| 194m6 | GACATACATAATTTACGTAAC | CCATTGATTTTAGGACGTGAC | 120 | GGACGTGACTTAACAGA |
| 18m1 | TGCAGCAAATGGTGATGTAC | ACAGACGAAATGGGAAATCC | 114 | CCATGTTGTAAAATACGCC |
| 28m6 | AGCGACGGGCTTAAGGTTTT | CGAAGTTCTTGATACACATGC | 112 | TTGTTAATTTTCTCACTCGTAA |
| 174m4 | AAATATTCAAACTGATGCTC | AGCCGAAACAGTTATCAGTC | 115 | AATTTTTTATTGTGCGGAAC |
| 29m9 | GAATTCATTTTTCTAAACAG | TGCATTGGATTGAAGCAAAG | 107 | GATTGAAGCAAAGAAGTTTAG |
A small percentage of the population (182 individuals or 5%) was screened with only the two flanking markers of the allorecognition complex (194 and 29). Marker 194 genotyping was performed as in Poudyal et al. (2007), with PCR primers that assay a size fragment polymorphism physically linked to 194m6. Marker 29 genotyping was performed using a cleaved amplified polymorphic sequence (CAPS) marker (Konieczny and Ausubel 1993) by amplifying a 736-bp product (forward primer 5′-CACAAACGGTGAAATATCAAAGCA-3′ and reverse primer 5′-TGACAGGATGTTGCGTTAAGGT-3′), followed by digestion with SspI, which does not cut the r allele, but cuts the f allele into two fragments (441 and 295 bp). This CAPS marker assays a SNP physically linked to 29m9. Any animals identified as recombinants between markers 194 and 29 were subsequently genotyped with all other markers across the interval to further localize the breakpoint.
Linkage analysis:
Map distances between the five SNPs, alr1, and alr2 were estimated using MapMaker (Lander et al. 1987). This analysis excluded the individuals genotyped with only the PCR markers 194 and 29. Because the alr1 and alr2 genes have not yet been identified, the genotype of alr1 and alr2 in recombinant animals had to be inferred from phenotype analysis. Note that in this study animals identified as nonrecombinant were not fusibility tested because alr1 and alr2 were assumed to match the genotype of flanking markers. This assumption relied on the small genetic intervals in question over which double recombination is highly unlikely and also on the Cadavid et al. (2004) study, which phenotyped all 490 animals of the mapping population and found no violation of the principal fusibility phenotype predicted by flanking markers.
RESULTS
Allorecognition phenotypes:
Recombination between the five SNPs assayed was detected in 106 animals. Of these, 73 were viable for a period long enough to be tested in colony fusibility assays to determine their allorecognition phenotype. In total, four distinct phenotypes were observed: fusion, rejection, and two types of transitory fusion. Additionally, a low frequency of cases (6) exhibited ambiguous phenotypes that were not easily classifiable. These are referred to as “exceptional phenotypes” (see below).
As was previously reported (Cadavid et al. 2004), recombination between alr1 and alr2 resulted in animals that underwent transitory fusion against an rr/rr tester. Recombination outside of this interval resulted in animals that underwent simple fusion or rejection. In this study, careful observation of the transitory fusions revealed that these fusions could be further subdivided into two distinct categories, which we refer to here as type I and type II transitory fusion.
In type I transitory fusions, colonies initially fused (Figure 3) and remained fused for ∼1–3 days before initial signs of rejection began. The first indication of this change was a gray or whitish opaqueness occurring in a demarcation along the interaction zone. Generally within a day, loss of gastrovascular transport across this line was observed and was followed by separation of the gastrovascular canals accompanied by separation of the mat. As the gastrovascular canals ceased to connect across the interaction zone, they began to branch laterally (parallel to the contact zone) and to make connections with canals on the same side of the fusion boundary (Figure 4). Frequently, a border of white fibrous material began to accumulate between the two sides until they no longer appeared to be in physical contact (arrow, Figure 4). The separation between colonies was stable unless both colonies began to grow vertically over the border in such a way as to recontact their neighbor or if the periphery was externally disrupted, e.g., by manually causing tissue trauma or by removing the fibrous border. In these cases, the transitory fusion process began again from fusion to separation.
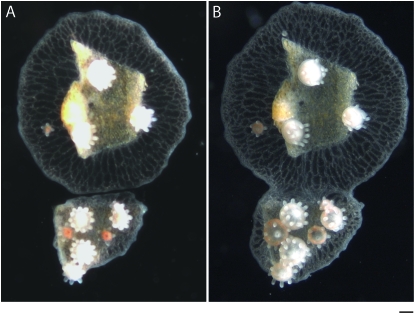
Figure 3.—
Initial stages of fusion and transitory fusion. (A) Colony assay prior to contact where the two colonies grow side by side. (B) Fusion 48 hr later showing continuity between mat tissue and gastrovascular canals. Bar, 200 μm.
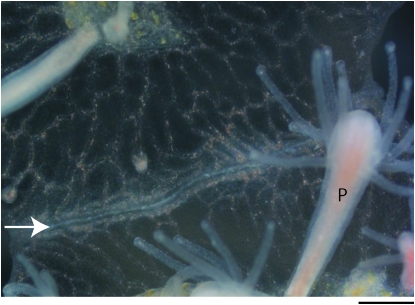
Figure 4.—
Outcome of type I transitory fusion. This close-up of interacting tissue between colonies depicts the outcome of type I transitory fusion. The two colonies have already undergone fusion and initial stages of rejection and have separated, indicated by the white fibrous material between the colonies that creates a border (arrow). Note also the gastrovascular canals that branch laterally along the line of separation and rejoin one another. P, polyps. Bar, 200 μm.
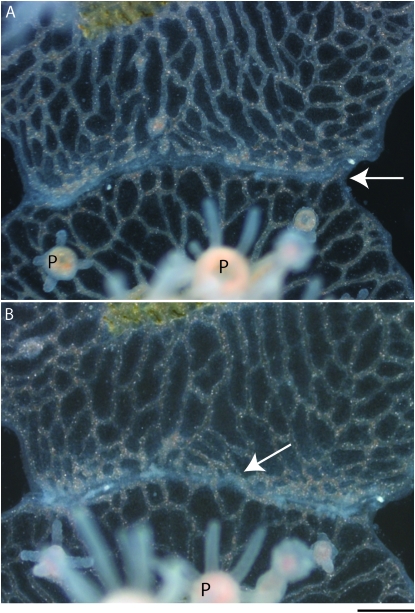
The second category of transitory fusion, type II, initiated in a manner identical to type I, but differed in later stages of the phenotype's progression. Colonies fused upon contact but within 1–3 days showed signs of separation. Tissue at the interaction zone took on a gray or white opaque quality that was followed within 24–48 hr by the onset of loss of gastrovascular continuity accompanied by ectodermal separation and some lateral branching (Figure 5A). Up to this point in the interaction, the phenotype was indistinguishable from type I transitory fusion. Subsequently, however, the interaction became distinct. A ring canal was almost never fully reformed and, instead of lasting separation, the colonies continued to show contact with gastrovascular canals refusing throughout the separated zone (Figure 5B). This gastrovascular discontinuity and refusion along the interaction zone continued for the duration of observation (weeks or even months), forming a patchwork of serially fusing and separating areas.
Figure 5.—
Two stages of type II transitory fusion. This region of interacting tissue between two colonies depicts the separation and refusion phases that occur in type II transitory fusion. (A) The separation phase after the colony has already undergone contact and fusion. The zone of interaction/separation is indicated by an arrow. This is an early stage of separation common to both type I and II transitory fusion where the mat and gastrovascular canals are separate, but unlike Figure 4, there is no border accumulation or extensive lateral branching of canals. (B) Refusion 24 hr later. The arrow indicates one of several areas where colonies have rejoined their gastrovascular canals. P, polyps. Bar, 200 μm.
Type I and type II transitory fusion phenotypes correlated with specific genotypes. Consider that, in our testcross, recombination between alr1 and alr2 would be expected to generate one of two reciprocal allotypes: rf/ff or fr/ff. Given that all recombinants were fusion tested against a homozygous rr/rr colony, the two interacting colonies were expected to share either one allele at alr1 and no alleles at alr2 (i.e., rf/ff) or to share one allele at alr2 and no alleles at alr1 (i.e., fr/ff). The correlation of the type of transitory phenotype and recombinant genotype was evident and is illustrated in Table 2. The sharing of one allele at alr1 and no shared alleles at alr2 correlated with type II transitory fusion and, reciprocally, no shared alleles at alr1 but one shared allele at alr2 correlated with the observation of type I transitory fusion. These results suggested a differential contribution of alr1 and alr2 loci to the allorecognition process.
TABLE 2.
Distribution of phenotype with respect to alr genotype
| Genotype | Type I transitory fusion | Type II transitory fusion |
|---|---|---|
| fr/ff | 27 | 1 |
| rf/ff | 0 | 24 |
Exceptional phenotypes occurred at a low frequency:
In addition to type I and type II transitory fusion phenotypes described above, six colonies exhibited exceptional phenotypes that could not be easily categorized. For each animal exhibiting an exceptional phenotype, the colony assay was repeated three separate times. In all cases, the colonies initially fused to the rr/rr tester, but within ∼3–4 days a gray line appeared across the interaction zone, followed by occasional, transient separations of mat and gastrovascular canals and accumulation of debris over the interaction zone. In contrast to the other two transitory phenotypic classes described here, the colonies failed to achieve complete separation. The most prominent feature of exceptional phenotypes was a necrotic character in the interaction zone, meaning that the tissue there appeared gray, inflamed, granular, and easily torn, as opposed to the smooth and elastic character of healthy mat tissue. The necrotic appearance occurred in a diffuse swath between the two colonies as opposed to the precise line of separation that appeared between colonies undergoing transitory fusion, as shown in Figure 4. Individual colonies varied in visual characteristics such as transparency, width of gastrovascular canals, frequency of branching, and thickness of mat, which made it possible to observe that in some cases the interaction zone shifted as though one colony's tissue was overtaking the other. This “conversion” of tissue was variable, appearing in only some colony assays and in only a subset of replicates.
The occurrence of exceptional phenotypes did not show a clear relationship to breakpoint location. Three cases occurred when the breakpoint was between 194m6 and 18m1 (colonies AP100-1279, AP100-1369, AP100-2801). Two cases occurred when the breakpoint was between 18m1 and 28m6 (colonies AP100-51 and AP100-2388). One case occurred when the breakpoint was between 28m6 and 174m4 (colony AP100-1546). In all these cases, marker 194m6 was in a heterozygous state. For the purpose of linkage analysis, no value was assigned to alr1 or alr2 for these colonies.
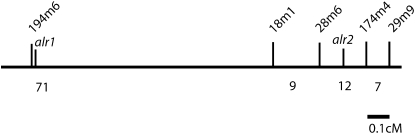
High-resolution genetic mapping of the Hydractinia allorecognition complex:
The backcross used to generate recombinants permitted us to refine the genetic map of the allorecognition interval. Linkage analysis of the five SNPs distributed across the allorecognition complex (supplemental data at http://www.genetics.org/supplemental/) produced a refined map (Figure 6). Of the 3565 animals genotyped, 7 (numbered AP100-644, AP100-993, AP100-2146, AP100-2224, AP100-2635, AP100-2728, AP100-3562) appeared to be recombinant but were genotyped three or more times from independent DNA extractions and failed to give unambiguous results and so were not included in linkage analysis.
Figure 6.—
Genetic map of the allorecognition complex. Markers are named as in Table 1. Loci are alr1 and alr2. Bar, 0.1 cM as shown. The total mapped interval represents 1.7 cM. Numbers below the map represent how many recombinants were recovered between each of the markers.
Mendelian segregation in the backcross population used here predicts that half of the individuals should be homozygous for the genetic markers tested and half should be heterozygous. In contrast to this prediction, a deficit was observed in the number of homozygous offspring recovered (46% of the total population) and a surplus in the number of heterozygous offspring (54% of the total population) for each of the five markers tested. These results differed significantly (P < 0.0001 for each marker) from a 1:1 ratio in a χ2 goodness-of-fit test.
DISCUSSION
Transitory fusions are a class of phenomena defined by fusion upon initial contact followed by a heterogeneous set of subsequent events. The archetypal response is that described by Hauenschild (1954, 1956) and later by Cadavid et al. (2004) and here identified as type I. The type II response has also been observed. Grosberg et al. (1996, p. 2224) reported an “ambiguous” phenotype occurring at a low frequency, which they described as having “different areas of interclonal contact simultaneously and persistently involved fusion and rejection.” Other forms of variation are also known, most notably colonies that remain fused for prolonged intervals only to separate at a large size (Shenk and Buss 1991) and colonies in which the separation process begins but is not completed (Cadavid et al. 2004). Clarification of this phenomenological zoo would be desirable.
Two factors affect allorecognition phenotypes: allorecognition genetics and colony ontogeny. Allorecognition genetics determine the compatibility of two colonies, while the ontogeny of the allorecognition phenotype in large part depends on developmental and environmental variation. One example of such variation is “passive” vs. “aggressive” rejection as described by Buss and Grosberg (1990). Rejection between two incompatible colonies can be either passive or aggressive, depending on whether tissue contact is between mat or stolon, respectively. In passive rejection, nematocysts are deployed and cause tissue damage, but eventually a stable border forms between the colonies and they cease growth on the front of contact. In aggressive rejection, the stoloniferous tissue proliferates and becomes hyperplastic, swollen with nematocytes. Tissue damage and cell lysis can be observed wherever hyperplastic stolons contact other tissues. Typically, this aggressive interaction escalates until only one colony survives (Ivker 1972; Buss et al. 1984; Buss and Grosberg 1990). In both cases, the genetics of allorecognition is the basis for rejection, but factors unrelated to allorecognition genetics cause a dramatic phenotypic difference.
Similarly, the occurrence of transitory fusion here had a clear genetic basis, but the phenotype depended on several sources of variation. The rejection following an initial fusion depended on (1) an as-yet-unknown process that caused tissue death in the interaction zone when colonies were still fused and that allowed the colonies to separate, (2) the lateral branching between isogeneic gastrovascular canals that accompanied separation, (3) the accumulation of a fibrous material that formed a border between colonies, and (4) whether and how quickly colonies grew back into contact after having separated. The relative rates at which these events occurred in large part determined the phenotypic outcome.
These biological processes occurred at different rates in different colonies and led to subtle variations between tests. For example, Table 2 notes that 27 individuals of the fr/ff recombinant genotype underwent type I transitory fusion, but 1 individual (number AP100-815) exhibited a phenotype more characteristic of type II transitory fusion. In two independent tests, AP100-815 succeeded in separating from the rr/rr tester but gastrovascular canals failed to laterally branch following separation. Instead, the unfused ends of the canals grew immediately forward again to reconnect with the tester colony before any significant border had accumulated, leading to repeated cycles of fusion and rejection. This could have been due to the strength of allorecognition signaling, to epistatic effects, or to an idiosyncratic effect of AP100-815's growth pattern.
In related effects, certain areas of some assays exhibiting type II transitory fusion remained either stably separated or stably continuous in ways that seemed to depend on local colony architecture. Some patches remained stably separated in a particular location for the duration of observation if they succeeded in reforming a ring canal at that spot, such that two ring canals lay parallel, sandwiched against each other, but on either side of this spot the colonies fused and separated serially. Some gastrovascular canals of large diameter and strong gastrovascular flow were observed to maintain continuity throughout the duration of the test, perhaps because the strong flow allowed by the large diameter visibly carried away blockages that would otherwise accumulate and eventually block gastrovascular exchange via the canal. In a similar sense, given that the exceptional phenotypes did not map to a particular location, an alternative interpretation of the phenotype could be that they were type II transitory fusions (since all were heterozygous at the marker 194, which is most closely linked to alr1) that had only a weak ability to break continuity with a non-isogeneic partner.
Nonetheless, type I and type II transitory fusion showed a clear relationship to genotype: the phenotypic difference mapped onto the reciprocal recombination products fr/ff and rf/ff. The first of these two possible products correlated with type I transitory fusion and the latter with type II transitory fusion. Effectively, the difference between the recombinant and the rr/rr tester in this case was whether alleles were shared only at alr1 or only at alr2. Therefore, a system of only two loci accounted for these phenotypic differences.
The fact that reciprocal recombinant haplotypes did not lead to the same fusibility outcome shows that alr1 and alr2 components contribute unequally to the allorecognition process. The nonequivalence of alr1 and alr2 could be due to an epistatic interaction between these loci or it could depend on allelic interactions. In other words, it is not yet clear whether the same phenotype would be observed whenever, e.g., alr1 matches one allele and alr2 none, or whether a different phenotype would be observed if alleles other than f and r were present. Our finding that two major categories of transitory fusion are explained by differential locus or allele-specific contributions does not explain all known forms of variation in transitory fusion. For example, we do not yet know what controls the timing of separation, which may be rapid as in type I and type II transitory fusion, but can also be delayed, as observed by Shenk and Buss (1991). Similarly, the fact that some colonies begin to separate but thereafter return to a permanently fused state (Cadavid et al. 2004) remains unexplained. Conceivably, other alr alleles or as-yet-undiscovered modifying loci may be at play. In addition to discriminating between two major categories of transitory fusion, our work yielded a refined genetic map for the interval, identified a deficit of homozygotes, and found no compelling evidence for additional loci in the interval. Specifically, an increase in sample size from past populations (N = 490) to the current population (N = 3565) has shrunk the interval from 3.4 to 1.7 cM. The slight deficit of homozygotes recalls similar observations noted in genetic studies of the allorecognition system of Botryllus (De Tomaso and Weissman 2004). Finally, the failure to observe any novel phenotype cosegregating with marker genotype is consistent with the suggestion that no further loci impacting allorecognition segregate within our defined genetic lines.
Acknowledgments
We thank Christina Glastris and Krista Joy Altland for providing animal care, James Signorovitch for helpful discussions, and Karl Hager for aid in designing the Sequenom hME assay. This work was supported by National Institutes of Health (NIH) grant 1R21-AI066242 (to F.G.L., L.W.B., and S.L.D.) and by a New Directions Grant for Established Investigators from the American Society of Nephrology (to F.G.L.). A.E.P. was supported by NIH Training Grant in Genetics T32-GM07499.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. EU020112–EU020116.
References
- Blackstone, N. W., and L. W. Buss, 1991. Shape variation in hydractiniid hydroids. Biol. Bull. 180**:** 394–405. [DOI] [PubMed] [Google Scholar]
- Buss, L. W., 1982. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. USA 79**:** 5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss, L. W., and R. K. Grosberg, 1990. Morphogenetic basis for phenotypic differences in hydroid competitive behavior. Nature 343**:** 63–66. [Google Scholar]
- Buss, L. W., C. S. McFadden and D. R. Keene, 1984. Biology of hydractiniid hydroids. 2. Histocompatibility effector system/competitive mechanism mediated by nematocyst discharge. Biol. Bull. 167**:** 139–158. [Google Scholar]
- Cadavid, L. F., A. E. Powell, M. L. Nicotra, M. Moreno and L. W. Buss, 2004. An invertebrate histocompatibility complex. Genetics 167**:** 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tomaso, A. W., and I. L. Weissman, 2004. Evolution of a protochordate allorecognition locus. Science 303**:** 977. [DOI] [PubMed] [Google Scholar]
- De Tomaso, A. W., S. V. Nyholm, K. J. Palmeri, K. J. Ishizuka, W. B. Ludington et al., 2005. Isolation and characterization of a protochordate histocompatibility locus. Nature 438(7067): 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg, R. K., D. R. Levitan and B. B. Cameron, 1996. The evolutionary genetics of allorecognition in the colonial hydrozoan Hydractinia symbiolongicarpus. Evolution 50**:** 2221–2240. [DOI] [PubMed] [Google Scholar]
- Hauenschild, C., 1954. Genetische und Entwicklungsphysiologie Untersuchungen über Intersexualität und Gewebeverträglichkeit bei Hydractinia echinata. Rouxs Arch. Dev. Biol. 147**:** 1–41. [DOI] [PubMed] [Google Scholar]
- Hauenschild, C., 1956. Über die Vererbung einer Gewebeverträglichkeits-Eigenschaft bei dem Hydroidpolypen Hydractinia echinata. Z. Naturforsch. 11b**:** 132–138. [Google Scholar]
- Ivker, F. B., 1972. A hierarchy of histo-compatibility in Hydractinia echinata. Biol. Bull. 143**:** 162–174. [Google Scholar]
- Jackson, J. B. C., 1977. Competition on marine hard substrata: the adaptive significance of solitary and colonial strategies. Am. Nat. 111**:** 743–767. [Google Scholar]
- Konieczny, A., and F. M. Ausubel, 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4(2): 403–410. [DOI] [PubMed] [Google Scholar]
- Laird, D. J., A. W. De Tomaso and I. L. Weissman, 2005. Stem cells are units of natural selection in a colonial ascidian. Cell 123**:** 1351–1360. [DOI] [PubMed] [Google Scholar]
- Lander, E., P. Green, J. Abrahamson, A. Barlow, M. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1**:** 174–181. [DOI] [PubMed] [Google Scholar]
- Lange, R., G. Plickert and W. Müller, 1989. Histocompatibility in a low invertebrate, Hydractinia echinata: analysis of the mechanism of rejection. J. Exp. Zool. 249**:** 284–292. [Google Scholar]
- Mokady, O., and L. W. Buss, 1996. Transmission genetics of allorecognition in Hydractinia symbiolongicarpus (Cnidaria:Hydrozoa). Genetics 143**:** 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, W., 1964. Experimentelle Untersuchungen über Stockentwicklung, Polypdifferenzierung und Sexualchimären bei Hydractinia echinata. Rouxs Arch. Dev. Biol. 155**:** 181–268. [DOI] [PubMed] [Google Scholar]
- Müller, W., 1973. Induction of metamorphosis by bacteria and ions in the planulae of Hydractinia echinata: an approach to mode of action. Publ. Seto Marine Biol. Lab. 20**:** 195–208. [Google Scholar]
- Nyholm, S. V., E. Passegue, W. B. Ludington, A. Voskoboynik, K. Mitchel et al., 2006. Fester, a candidate allorecognition receptor from a primitive chordate. Immunity 25(1): 163–173. [DOI] [PubMed] [Google Scholar]
- Poudyal, M., S. Rosa, A. E. Powell, M. Moreno, S. L. Dellaporta et al., 2007. Embryonic chimerism does not induce tolerance in an invertebrate model organism. Proc. Natl. Acad. Sci. USA 104**:** 4559–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk, M. A., and L. W. Buss, 1991. Ontogenic changes in fusibility in the colonial hydroid Hydractinia symbiolongicarpus. J. Exp. Zool. 257**:** 80–86. [Google Scholar]
- Stoner, D. S., B. Rinkevich and I. L. Weissman, 1999. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc. Natl. Acad. Sci. USA 96**:** 9148–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens, M., A. Krasko, I. M. Müller and W. E. G. Müller, 2000. Increased expression of the potential proapoptotic molecule DD2 and increased synthesis of leukotriene B4 during allograft rejection in a marine sponge. Cell Death Differ. 7**:** 461–469. [DOI] [PubMed] [Google Scholar]
- Wiens, M., S. Perovic-Ottstadt, I. M. Müller and W. E. G. Müller, 2004. Allograft rejection in the mixed cell reaction system of the demosponge Suberites domuncula is controlled by differential expression of apoptotic genes. Immunogenetics 56**:** 579–610. [DOI] [PubMed] [Google Scholar]