Chimpanzee Reservoirs of Pandemic and Nonpandemic HIV-1 (original) (raw)
. Author manuscript; available in PMC: 2008 Jul 2.
Published in final edited form as: Science. 2006 May 25;313(5786):523–526. doi: 10.1126/science.1126531
Abstract
Human immunodeficiency virus type 1 (HIV-1), the cause of human acquired immunodeficiency syndrome (AIDS), is a zoonotic infection of staggering proportions and social impact. Yet uncertainty persists regarding its natural reservoir. The virus most closely related to HIV-1 is a simian immunodeficiency virus (SIV) thus far identified only in captive members of the chimpanzee subspecies Pan troglodytes troglodytes. Here we report the detection of SIVcpz antibodies and nucleic acids in fecal samples from wild-living P. t. troglodytes apes in southern Cameroon, where prevalence rates in some communities reached 29 to 35%. By sequence analysis of endemic SIVcpz strains, we could trace the origins of pandemic (group M) and nonpandemic (group N) HIV-1 to distinct, geographically isolated chimpanzee communities. These findings establish P. t. troglodytes as a natural reservoir of HIV-1.
Since the first detection of an HIV-1–related lentivirus in chimpanzees (1, 2), this species has been suspected as the source of the human AIDS pandemic. However, a crucial missing link in the chain of evidence implicating SIVcpz in the origin of HIV-1 and AIDS has been the absence of a recognizable virus reservoir in wild-living apes. Chimpanzees (Pan troglodytes) are classified into four subspecies on the basis of differences in mitochondrial DNA sequence (3): P. t. verus in west Africa; P. t. vellerosus in Nigeria and northern Cameroon; P. t. troglodytes in southern Cameroon, Gabon, and the Republic of Congo; and P. t. schweinfurthii in the Democratic Republic of Congo and countries to the east (Fig. 1). Two of these subspecies, P. t. troglodytes and P. t. schweinfurthii, are known to harbor SIVcpz, and their viruses form divergent subspecies-specific phylogenetic lineages (SIVcpz_Ptt_ and SIVcpz_Pts_) (4). HIV-1 is most closely related to SIVcpz_Ptt_ (5), but this virus has been detected only rarely and then only in captive apes (1, 5–7). There is no counterpart of SIVcpz_Pts_ that is known to infect humans (4, 8–10).
Fig. 1.
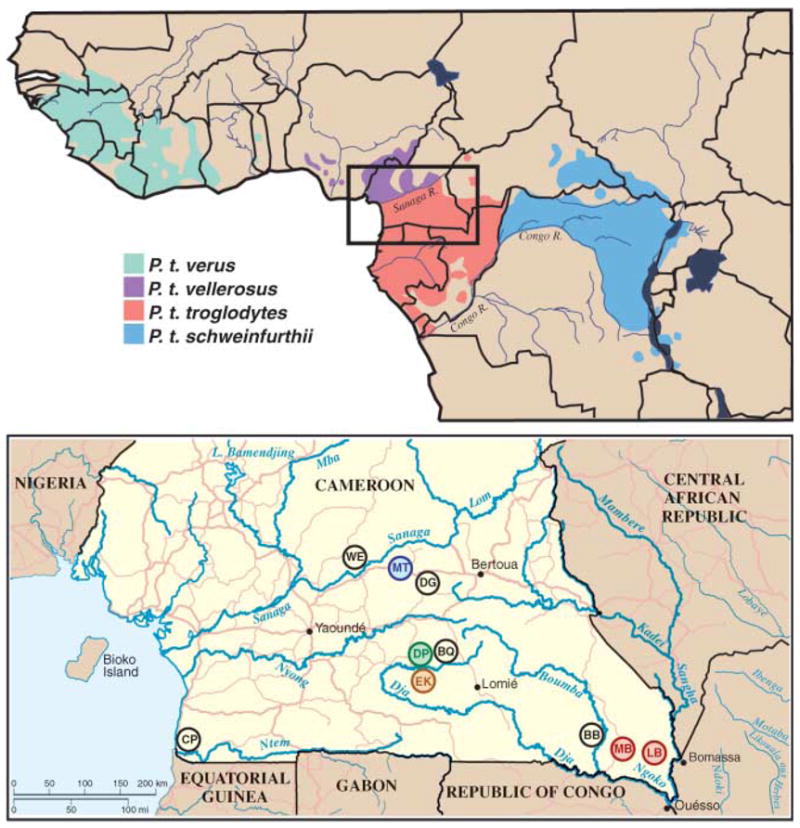
Natural ranges of the four chimpanzee subspecies (top) and locations of wild chimpanzee study sites WE, MT, DG, DP, BQ, EK, CP, BB, MB, and LB in southern Cameroon (inset and bottom). Field sites with endemic SIVcpz_Ptt_ infection are color-coded to correspond with the SIVcpz_Ptt_ lineages shown in Figs. 3 and 4.
Wild-living chimpanzees are reclusive and highly endangered and live in remote jungle areas. To study chimpanzees in their natural habitat, we developed methods to detect SIVcpz-specific antibodies and nucleic acids in fecal samples collected from the forest floor (9–11). In addition, we developed genotyping approaches to amplify host mitochondrial and genomic markers (polymorphic microsatellite loci) from these same specimens for species, gender, and individual identification (11, 12). These methods were validated in captive and habituated apes of known infection status (13). We used these noninvasive approaches to conduct the first molecular epidemiological field study of SIVcpz in wild-living nonhabituated chimpanzees in west central Africa.
Cameroon is home to two chimpanzee subspecies, P. t. vellerosus in the north and P. t. troglodytes in the south, with the Sanaga River forming the boundary between their ranges (Fig. 1). In the present study, we collected 599 fecal specimens at 10 forest sites throughout the southern part of Cameroon (Fig. 1). All field sites, except one (WE), were in the range of the P. t. troglodytes subspecies. To establish the species and subspecies origin of each sample, a 498–base pair (bp) mitochondrial DNA (mtDNA) (D-loop) fragment was amplified from fecal DNA and subjected to phylogenetic analysis (13). Eighty-six specimens were degraded, and 67 samples contained gorilla mtDNA sequences (table S1). The remaining 446 samples were of chimpanzee origin: 423 from P. t. troglodytes and 23 from P. t. vellerosus. These comprised 82 unique mtDNA haplotypes (fig. S1 and table S2). Consistent with the recognized ranges of the two subspecies, all 23 P. t. vellerosus specimens were collected north of the Sanaga River, whereas 421 of 423 P. t. troglodytes samples were collected south of the river (table S1).
All mtDNA-positive fecal samples were tested for virus-specific antibodies with a sensitive immunoblot assay specifically developed for surveys at remote field sites (13). This analysis identified 34 specimens, all from P. t. troglodytes apes, that contained antibodies reactive with HIV-1 antigens (Fig. 2). Twelve samples exhibited a strong and broadly cross-reactive Western blot profile that was virtually indistinguishable from the HIV-1–positive human plasma control. Eighteen additional samples reacted with both the HIV-1 envelope (gp160) and major core (p24) proteins, thus also meeting formal criteria for HIV-1/SIVcpz antibody positivity. Four samples (EK502, EK506, MB245, and MB248) reacted only faintly with a single HIV-1 protein (p24) and were classified as indeterminant. None of 23 P. t. vellerosus or 67 gorilla specimens exhibited detectable Western blot reactivity to any HIV-1 protein (table S1).
Fig. 2.
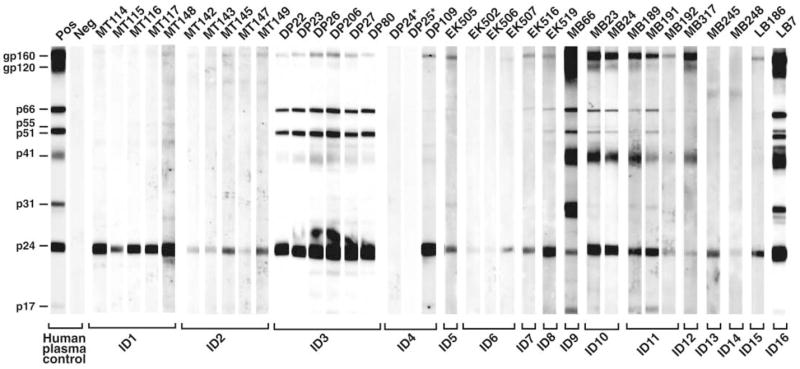
Detection of SIVcpz antibodies in chimpanzee fecal samples. Fecal samples from wild-living chimpanzees were tested by enhanced chemiluminescent Western blot using HIV-1 antigen–containing strips. Samples are numbered, with letters indicating their collection site as shown in Fig. 1. Samples from the same individual (ID) are grouped. Asterisks indicate two antibody-negative but virion RNA–positive samples (also see table S3). Molecular weights of HIV-1 proteins are indicated. The banding patterns of plasma from HIV-1–infected (Pos) and –uninfected (Neg) humans are shown as controls.
To corroborate the fecal antibody results, RNA was extracted from all immunoblot-reactive samples and subjected to reverse transcription polymerase chain reaction amplification using consensus env and pol primers. In addition, fecal DNA was used to amplify polymorphic microsatellite loci to identify and distinguish individual apes and to amplify a portion of the amelogenin gene for gender determination (13). These analyses revealed that the 34 immunoblot-reactive samples represented 16 different P. t. troglodytes apes (7 males and 9 females). Each of these apes had detectable virion RNA in one or more fecal samples (table S3). SIVcpz env (~390 bp) and/or pol (~890 bp) sequences were amplified from 31 of 34 (91%) immunoblot-reactive samples, including all four specimens with indeterminant Western blot reactivity (Fig. 3 and table S3). These data, together with previous findings for SIVcpz_Pts_-infected apes (10), indicate that fecal antibody reactivity to a single HIV-1 Gag protein is indicative of SIVcpz_Ptt_ infection (14).
Fig. 3.
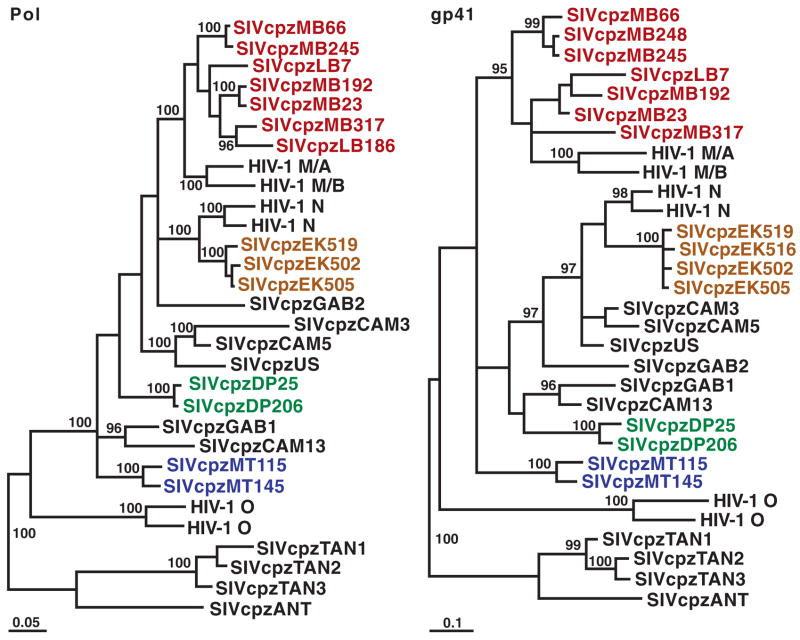
Phylogenetic analysis of SIVcpz_Ptt_ strains from wild P. t. troglodytes apes. Newly identified SIVcpz_Ptt_ strains are highlighted by colors reflecting their collection sites (Fig. 1). Representative strains of HIV-1 groups M, N, and O and SIVcpz_Pts_ (TAN1, TAN2, TAN3, and ANT) are shown. Trees were inferred by the Bayesian method; numbers on nodes are percentage posterior probabilities (only values above 95% are shown). The scale bars represent 0.05 and 0.1 substitutions per site. Pol, polymerase; gp41, envelope transmembrane protein.
The prevalence of SIVcpz_Ptt_ infection in wild chimpanzee communities was estimated for each of the 10 field sites (table S1). For the DP, EK, MB, BB, and LB communities, this was done based on the proportion of infected individuals as determined by microsatellite analyses, taking into consideration assay sensitivities and specimen degradation (tables S1 and S4). For the remaining sites, prevalence rates were estimated based on the proportion of antibody- and/or SIVcpz virion RNA–positive fecal samples, while also adjusting for repeat sampling (13). The results indicated widespread but notably uneven SIVcpz_Ptt_ infection of wild-living P. t. troglodytes apes, with prevalence rates ranging from 23 to 35% in the LB, EK, and MB communities; 4 to 5% in the DP and MT communities; and the absence of infection in the WE, DG, BQ, BB, and CP communities.
To determine the evolutionary relationships of the 16 new SIVcpz_Ptt_ viruses to each other and to previously characterized SIVcpz and HIV-1 strains, pol and env sequences were subjected to phylogenetic analyses. All of the newly identified SIVcpz strains were found to fall within the radiation of SIVcpz_Ptt_ strains from captive P. t. troglodytes apes, which also includes HIV-1 groups M (pandemic) and N (nonpandemic) but not group O or SIVcpz_Pts_ (Fig. 3). The new P. t. troglodytes viruses exhibited significant phylogeographic clustering: SIVcpz sequences from the EK, DP, MT, and MB/LB collection sites formed well-separated clades corresponding to their field site of origin. One of these clades included closely related SIVcpz strains (EK519, EK516, EK502, and EK505), probably reflecting recent virus transmission within that community. The remaining clades were each composed of more divergent but still monophyletic SIVcpz strains (Fig. 3). Thus, chimpanzee populations separated by long distances or major geographical barriers such as rivers (Fig. 1) harbored distinct SIVcpz lineages (such as populations EK, DP, and MT), whereas neighboring communities not separated by such barriers harbored viruses that were phylogenetically interspersed (such as populations MB and LB).
The phylogeographic clustering of the newly identified SIVcpz_Ptt_ strains allowed us to trace the origins of present-day human AIDS viruses to distinct chimpanzee communities. In sub-genomic pol and env regions, SIVcpz_Ptt_ strains from the MB/LB and EK sites were much more closely related to HIV-1 groups M and N, respectively, than were any previously identified SIVcpz strains (Fig. 3). Full-length genome analysis of 4 of the 16 new viruses confirmed and extended these findings, revealing strong statistical support for the clustering of HIV-1 groups M and N with the MB/LB and EK lineages of SIVcpz_Ptt_, respectively (Fig. 4). Moreover, inclusion of the new viruses reduced the lengths of the branches marking the cross-species transmission events for all genomic regions by almost half (arrows in Fig. 4). Given these short branch lengths, it is highly unlikely that other SIVcpz_Ptt_ strains exist that are significantly more closely related to HIV-1 groups M and N than are the viruses from the MB/LB and EK communities. Indeed, expanded field studies in southern Cameroon by our group have identified additional SIVcpz_Ptt_ strains, including nine from the MB/LB area, whose sequences support this conclusion and corroborate the phylogenetic relationships shown in Figs. 3 and 4 (15). Thus, an extensive set of molecular epidemiological data all points to chimpanzees in southeastern and south central Cameroon as the sources of HIV-1 groups M and N, respectively.
Fig. 4.
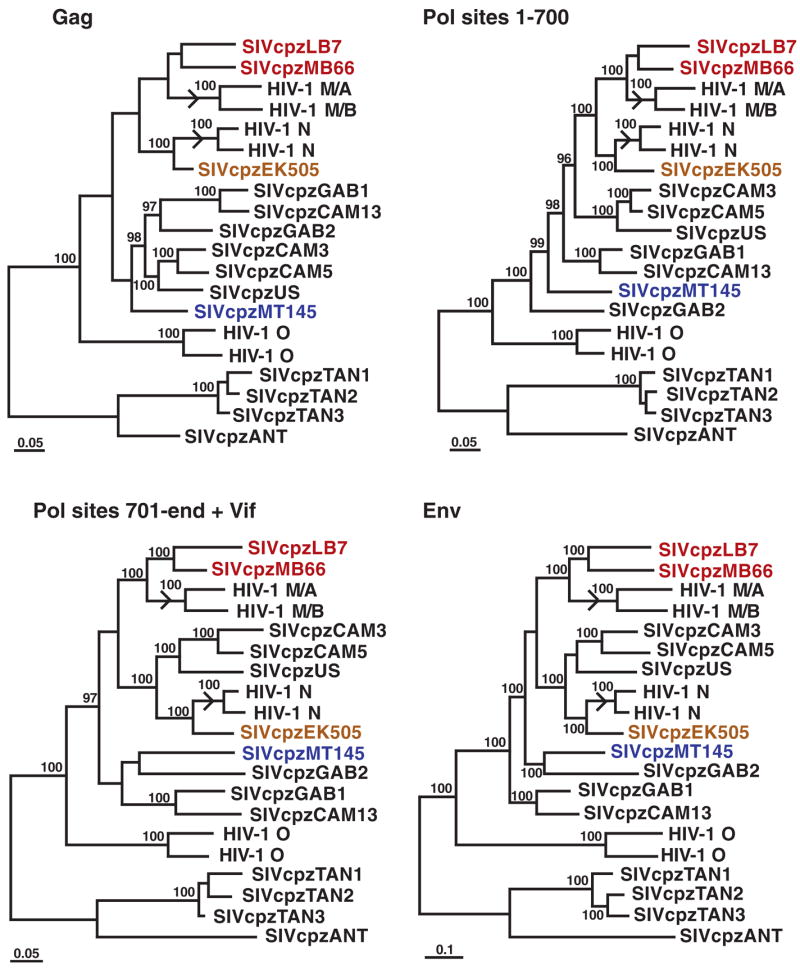
Evolutionary relationships of SIVcpz and HIV-1 strains based on full-length sequences. Trees were inferred by the Bayesian method for Gag, Pol, and Env; the Pol protein was separated into two fragments at a recombination breakpoint previously identified in HIV-1 group N (5, 21); the C terminus of Pol was concatenated with downstream Vif sequences. Sequences are color-coded as in Fig. 3. Numbers on internal branches indicate estimated posterior probabilities (only values above 95% are shown). The scale bars represent 0.05 and 0.1 substitutions per site. Arrows indicate branches where cross-species transmissions gave rise to HIV-1 groups M and N.
The findings presented here, together with prior studies, provide for the first time a clear picture of the origin of HIV-1 and the seeds of the AIDS pandemic. SIVcpz, the progenitor of HIV-1, arose as a recombinant of ancestors of SIV lineages presently infecting red-capped mangabeys and Cercopithecus monkeys in west-central Africa (16). Chimpanzees acquired this recombinant virus, or its progenitors, by cross-species transmission some time after the split of P. t. verus and P. t. vellerosus from the other subspecies (fig. S1) but possibly before the divergence of P. t. schweinfurthii from P. t. troglodytes (4). This explains the absence of SIVcpz infection in present-day P. t. verus and P. t. vellerosus apes, the presence of SIVcpz infection in P. t. troglodytes and P. t. schweinfurthii apes, and the phylogenetic separation of SIVcpz_Ptt_ from SIVcpz_Pts_ viruses (4, 7, 9, 15). HIV-1 groups M, N, and O each resulted from independent cross-species transmissions of SIVcpz_Ptt_ from P. t. troglodytes to humans early in the 20th century (17–19). We show here that the SIVcpz_Ptt_ strain that gave rise to HIV-1 group M belonged to a viral lineage that persists today in P. t. troglodytes apes in southeastern Cameroon. That virus was probably transmitted locally. From there it appears to have made its way via the Sangha River (or other tributaries) south to the Congo River and on to Kinshasa where the group M pandemic was probably spawned (20). HIV-1 group N, which has been identified in only a small number of AIDS patients from Cameroon (21, 22), derived from a second SIVcpz_Ptt_ lineage in south central Cameroon and remained geographically more restricted. The source of HIV-1 group O remains unknown but will probably yield to further study of wild ape populations not yet sampled. Given the extensive genetic diversity and phylogeographic clustering of SIVcpz now recognized, and the vast areas of west central Africa not yet sampled (Fig. 1), it is quite possible that still other SIVcpz lineages exist that could pose risks of human infection and prove problematic for HIV diagnostics and vaccines. The present report describes molecular tools and noninvasive strategies that can be used to explore these possibilities as well as the molecular ecology of pathogens in endangered species more generally.
Supplementary Material
References and Notes
- 1.Peeters M, et al. AIDS. 1989;3:625. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Huet T, et al. Nature. 1990;345:356. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 3.Groves CP. In: Mammalian Species of the World: A Taxonomic and Geographic Reference. 2. Wilson DE, Reader DM, editors. Smithsonian Institution Press; Washington, DC: 1993. pp. 243–277. [Google Scholar]
- 4.Sharp PM, Shaw GM, Hahn BH. J Virol. 2005;79:3891. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao F, et al. Nature. 1999;397:436. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 6.Corbet S, et al. J Virol. 2000;74:529. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nerrienet E, et al. J Virol. 2005;79:1312. doi: 10.1128/JVI.79.2.1312-1319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanden Haesevelde MM, et al. Virology. 1996;221:346. doi: 10.1006/viro.1996.0384. [DOI] [PubMed] [Google Scholar]
- 9.Santiago ML, et al. Science. 2002;295:465. doi: 10.1126/science.295.5554.465. [DOI] [PubMed] [Google Scholar]
- 10.Worobey M, et al. Nature. 2004;428:820. doi: 10.1038/428820a. [DOI] [PubMed] [Google Scholar]
- 11.Santiago ML, et al. J Virol. 2003;77:7545. doi: 10.1128/JVI.77.13.7545-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santiago ML, et al. J Virol. 2005;79:12515. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Materials and methods are available as supporting material on Science Online.
- 14.In contrast to plasma samples from uninfected humans, which exhibit false positive Western blot reactivity to HIV-1 p24 in as many as 10 to 15% of individuals (www.fda.gov/cber/products/hiv1cam052898.htm), we have found no such nonspecific cross-reactivity of chimpanzee immunoglobulin extracted by the RNA_later_ method from over 2000 fecal specimens.
- 15.Van Heuverswyn F, et al. 13th Conference on Retroviruses and Opportunistic Infections; 2006. Abstract 132, available at www.retroconference.org/2006/Abstracts/26513.htm. [Google Scholar]
- 16.Bailes E, et al. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 17.Hahn BH, Shaw GM, De Cock KM, Sharp PM. Science. 2000;287:607. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 18.Korber BTK, et al. Science. 2000;288:1789. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 19.Sharp PM, et al. Philos Trans R Soc London Ser B. 2001;356:867. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidal N, et al. J Virol. 2000;74:10498. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon F, et al. Nat Med. 1998;4:1032. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi J, et al. AIDS Res Hum Retroviruses. 2006;22:83. doi: 10.1089/aid.2006.22.83. [DOI] [PubMed] [Google Scholar]
- 23.We thank the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to perform this study; J. Dupain, C. Neel, and M. Epanda (Projet Grands Singes); L. Usongo, D. Dontego, F. Espedi, and B. Tshikangwa (World Wildlife Fund); and G. Etoga and D. M’bohand (Ministry of Environment and Forestry) for assistance in the field. This work was supported in part by the NIH (grants R01 AI50529, R01 AI58715, and P30 AI 27767), the Bristol Myers Freedom to Discover Program, the Institut de Recherche pour le Développement, and the Howard Hughes Medical Institute. New SIVcpz_Ptt_ sequences are available at GenBank under accession numbers DQ370366-DQ370419 and DQ373063-DQ373066; chimpanzee mtDNA sequences are available under DQ367532-DQ367613 and DQ370307-DQ370365.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.