Adaptive Radiation in Mediterranean Cistus (Cistaceae) (original) (raw)
Abstract
Background
Adaptive radiation in Mediterranean plants is poorly understood. The white-flowered Cistus lineage consists of 12 species primarily distributed in Mediterranean habitats and is herein subject to analysis.
Methodology/Principal Findings
We conducted a “total evidence” analysis combining nuclear (ncp_GS_, ITS) and plastid (trnL-trnF, trnK-matK,trnS-trnG, rbcL) DNA sequences and using MP and BI to test the hypothesis of radiation as suggested by previous phylogenetic results. One of the five well-supported lineages of the_Cistus-Halimium_ complex, the white-flowered_Cistus_ lineage, comprises the higher number of species (12) and is monophyletic. Molecular dating estimates a Mid Pleistocene (1.04±0.25 Ma) diversification of the white-flowered lineage into two groups (C. clusii and C. salviifolius lineages), which display asymmetric characteristics: number of species (2 vs. 10), leaf morphologies (linear vs. linear to ovate), floral characteristics (small, three-sepalled vs. small to large, three- or five-sepalled flowers) and ecological attributes (low-land vs. low-land to mountain environments). A positive phenotype-environment correlation has been detected by historical reconstructions of morphological traits (leaf shape, leaf labdanum content and leaf pubescence). Ecological evidence indicates that modifications of leaf shape and size, coupled with differences in labdanum secretion and pubescence density, appear to be related to success of new species in different Mediterranean habitats.
Conclusions/Significance
The observation that radiation in the Cistus salviifolius lineage has been accompanied by the emergence of divergent leaf traits (such as shape, pubescence and labdanum secretion) in different environments suggets that radiation in the group has been adaptive. Here we argued that the diverse ecological conditions of Mediterranean habitats played a key role in directing the evolution of alternative leaf strategies in this plant group. Key innovation of morphological characteristics is supported by our dated phylogeny, in which a Mediterranean climate establishment (2.8 Ma) predated the adaptive radiation of the white-flowered_Cistus_.
Introduction
The concept of adaptive radiation implies a rapid ecological diversification, which should be reflected in a greater morphological and/or physiological divergence among species in brief periods of rapid diversification from a single ancestor [1], [2]. Two mechanisms could generate adaptive radiations: (1) extrinsic causes due to new environmental circumstances [3], [4]; (2) intrinsic characters of organisms (key innovation) that allow a taxon to utilize existing niche space in a novel manner [5]. Remoteness and the rich diversity of habitats of island systems help ensure little competition and different environments to test the potential of plant radiations [6], [7]. In contrast to the wealth of studies documenting adaptive radiations in oceanic islands [see 3], [8],[9] and particular mainland habitats [see 10], [11], we have found in literature no study fully focused on the Mediterranean region.
The Mediterranean climatic type, characterized by a strong seasonality (hot dry summers, cool wet winters), occurs in California, South Africa, central Chile, southern Australia, and typically in the Mediterranean Basin [12], [13]. In all five of these areas the native vegetation is a dense scrub characterized by annuals, drought-tolerant deciduous and semi-deciduous malacophyllous species, and woody evergreen sclerophyllous species [14]. Sclerophyllous species are adapted to low water availability during summer by means of small, leathery and dark leaves covered with thick cuticles and small, thick-walled cells [15]. Small leaves and low specific leaf area have been viewed as adaptations to Mediterranean-type climates in many species of evergreen plants [16]. Indeed, sclerophylly is so successful that unrelated genera and families of woody plants converged into similar leaf traits. Two alternative origins have been proposed for the evolution of Mediterranean, woody plants: resprouters corresponding to older lineages (Tertiary with tropical to subtropical conditions) and seeders (such as Cistus) to younger lineages (Quaternary with Mediterranean conditions) [17]. Few studies have, however, addressed the origin of Mediterranean plant groups by means of phylogenetic approaches related to ecological preferences [but see 18].
Significant shrub components in the European-African Mediterranean ecosystems (e.g., “maquis”, “garrigue”) belong to Cistaceae (Tuberaria, Halimium,Cistus). Cistus is a genus of 21 frutescent and suffrutescent shrub species with a predominantly Mediterranean distribution [19], except for five species endemic to the Canary Islands (Table 1). Previous phylogenetic studies revealed the separation of the_Cistus_-Halimium lineage and identification of two major natural groups: one of purple-flowered Cistus_species (hereafter the purple-flowered lineage) and other containing the white-flowered species of Cistus, plus the pinkish-flowered_C. parviflorus (hereafter the white-flowered lineage)[20], [21], [22]. Moreover, the white-flowered lineage is divided in two groups: one containing C. clusii and_C. munbyi_ species (hereafter the C. clusii group) and other containing the rest of the white-flowered_Cistus_ species (9), plus C. parviflorus (hereafter the C. salviifolius group) (Fig. 1). Despite the two lineages (the C. clussi and the C. salviifolius groups) are inhabiting the Mediterranean basin, the C. salviifolius group has undergone higher differentiation and displays greater variation in leaf trichome density, size, shape and tissue thickness than do the _C. clusii_group. These properties influence the resistance to drought stress and solar irradiance [14]. Indeed, ecological analyses of leaf morphological and physiological characters in dry environments [23], [24] appear to be related to speciation of Mediterranean plants.
Table 1. List of species used in the phylogenetic analysis.
| Taxon | Distribution | Locality/source | Voucher |
|---|---|---|---|
| Cistus L. | |||
| Cistus albanicus E.F. Warb. ex Heywood | Albania, Greece | Cultivated | R. G. Page 8cBGA04 (MA) |
| Cistus albidus L | Iberia, S France, N Italy, N Africa, Corsica, Sardigna | Spain, Madrid, Aldea del Fresno | P. Vargas 25PV03 (MA) |
| _Cistus chinamadensis_Bañares et Romero | La Gomera, Tenerife (Canary Islands) | Canary Islands, La Gomera | Á. Fernández & J. Leralta 44BGA04 (MA) |
| R. G. Page 8bBGA04 (MA) | |||
| Cistus clusii Dunal subsp. clusii | Spain, Italia, N Africa, Sicily | Spain, Málaga, Mijas | C. Navarro et al.(MA618671) |
| Cistus clusii Dunal subsp. multiflorus Demoly | Balear Islands, SE Iberia Peninsula | Spain, Balear Islands, Mallorca, Sa Rápita | P. Vargas 209PV04 (MA) |
| Cistus creticus L. | Mediterranean Basin | Greece, Olympus | B. Guzmán 58BGA04 (MA) |
| Cistus crispus L. | Iberia, S France, N Italy, N Africa, Corsica, Sicily | Spain, Córdoba, Posadas | B. Guzmán 99BGA04 (MA) |
| _Cistus heterophyllus_Desf. | SE Spain, N Africa | Morocco, Beni-Hadifa | B.Guzmán 2BGA05 (MA) |
| Cistus horrens Demoly | Gran Canaria (Canary Islands) | Canary Islands, Gran Canaria, Ayacata | B. Guzmán 109BGA04 (MA) |
| Cistus ladanifer L. subsp. africanus | S Spain, N Africa | Morocco, Targuist | B. Guzmán 7BGA03 (MA) |
| Cistus ladanifer L. subsp. ladanifer | S France, Iberia, N Africa, Cyprus | Spain, Madrid, Boadilla del Monte | B. Guzmán 29BGA04 (MA) |
| Cistus ladanifer L. subsp. sulcatus | S Portugal | Portugal, Sagres | B. Guzmán 13BGA03 (MA) |
| Cistus laurifolius L. | N Africa, Iberia, France, Italy, Corsica, Turkey | Spain, Jaén, Sierra de Segura | R. G. Page 149BGA04 (MA) |
| Cistus libanotis L. | Portugal, S Spain, Argelia | Spain, Córdoba | B. Guzmán 35BGA04 (MA) |
| Cistus monspeliensis L. | Mediterranean Basin, Canary Islands | Portugal, Sagres | O. Filippi 4BGA04 (MA) |
| Cistus munbyi Pomel | Algeria, Morocco | Morocco | R. G. Page 8BGA04 (MA) |
| Cistus ochreatus C. Sm. ex Buch | Gran Canaria (Canary Islands) | Canary Islands, Gran Canaria | P. Escobar 48/05 (MA) |
| _Cistus osbeckiifolius_Webb ex Christ | Tenerife (Canary Islands) | Canary Islands, Tenerife | O. Filippi 6BGA04 (MA) |
| Cistus parviflorus Lam. | Greece, Turkey, Italy, Cyprus, N Libia, Lampedusa | Greece, Crete | B. Guzmán 20BGA04 (MA) |
| Cistus populifolius L. subsp. major (Dunal) Heywood | Iberia, N Morocco | Portugal, Ourique | P. Vargas 5PV03 (MA) |
| Cistus populifolius L. subsp. populifolius | Iberia, S France | Spain, Ávila, Arenas de San Pedro | R. G. Page 8tBGA04 (MA) |
| Cistus pouzolzii Delile | Algeria, N Morocco, France | France | P. Vargas 7PV03 (MA) |
| _Cistus psilosepalus_Sweet | Iberia, France | Spain, Ávila, Arenas de San Pedro | P. Vargas 6PV03 (MA) |
| Cistus salviifolius L. | Mediterranean Basin | Spain, Ávila, Arenas de San Pedro | B. Guzmán 143BGA04 (MA) |
| _Cistus symphytifolius_Lam. | El Hierro,La Palma,La Gomera,Tenerife,Gran Canaria | Canary Islands, La Palma, La Cumbrecita | |
| Fumana (Dunal) Spach | |||
| Fumana thymifolia (L.) Spach ex Webb | Mediterranean Basin | Portugal, Ferrerías | B. Guzmán 53BGA04 (MA) |
| Halimium (Dunal) Spach | |||
| _Halimium atlanticum_Humbert & Maire | N Africa | Morocco, Tazzeka | RDG14/2006/5 |
| Halimium atriplicifolium(Lam.) Spach | Spain, N Morocco | Spain, Granada, Sierra Nevada | P. Vargas 120PV04 (MA) |
| Halimium calycinum (L.) K. Koch | Iberia, NW Morocco | Portugal, Cabo Sardao | B. Guzmán 49BGA04 (MA) |
| Halimium halimifolium(L.) Willk. halimifolium | Iberia, Morocco | Spain, Málaga, Marbella | A. Segura (MA580185) |
| Halimium lasianthum(Lam.) Spach lasianthum | SW Iberia, N Morocco | Spain, Málaga | P. Vargas 3PV06 |
| Halimium lasiocalicynum(Boiss. & Reut.) Gross ex Engl. subsp.riphaeum (Pau & Font Quer) Maire | N Africa | Morocco, Bab-Berred | P. Escobar 665/04 (MA) |
| Halmium ocymoides (Lam.) Willk. | Iberia Peninsula, N Morocco | Portugal, Coimbra | R. G. Page 158BGA04 (MA) |
| Halimium umbellatum (L.) Spach | Mediterranean Basin | Spain, Madrid, Tres Cantos | P. Vargas 71BGA04 (MA) |
| Helianthemum Mill. | |||
| Helianthemum squamatum(L.) Dum. Cours. | Iberia, N Africa | Cultivated | B. Guzmán 70BGA04 (MA) |
| Tuberaria Dunal | |||
| Tuberaria guttata (L.) Fourr. | W Europe, Mediterranean Basin, Canary Islands | Portugal, Vila do Vispo | B. Guzmán 44BGA04 (MA) |
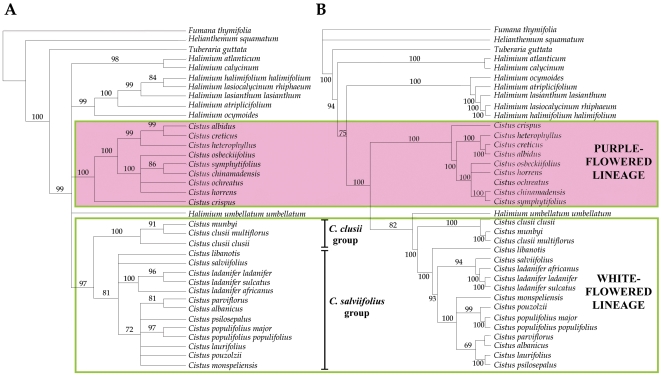
Figure 1. Phylogenetic hypothesis based on plastid (trnL-F, trnK-matK,trnS-trnG, rbcL) and nuclear (ITS, ncp_GS)_ sequences.
(A) Strict consensus of 104 equally parsimonious trees of 1317 steps (CI = 0.82, RI = 0.80), showing bootstrap support for clades above branches; (B) Bayesian inference tree (50% majority rule consensus tree) showing posterior probabilities above branches.
In this study, we used a molecular phylogenetic approach of DNA sequence data, sampled from both the nuclear (ITS, ncp_GS_) and the plastid (trnL-trnF, trnK-matK,trnS-trnG, rbcL) genomes, to test the explicit hypothesis of adaptive radiation. We first explored single ancestry in the _Cistus_-Halimium complex and differentiation in short periods of time by means of phylogenetic and molecular clock analyses [1], [25]. To test evolution in Mediterranean conditions, we chose a lineage exclusive to the Mediterranean basin (C. salviifolius lineage). Phenotype-environment correlation was further conducted to infer the role of ecological and vegetative characteristics [26], [27], [28] involved in speciation of this group.
Results
Phylogenetic analyses
The characteristics of the six sequence data sets are summarized in Table 2. MP analysis using Fitch parsimony resulted in 104 shortest trees of length 1317 steps (Fig. 1) for the combined sequence matrix. The consistency index (CI) for these trees was 0.82 and the retention index (RI) was 0.80. The BI tree displayed similar topology (except for the Halimium umbellatum position) and support values. Plastid and nuclear datasets yielded a similar phylogenetic pattern, although plastid sequences provided a more resolved tree (results not shown). In addition to strong (99% BS, 94 PP) support for the monophyly of the Cistus-Halimium complex, parsimony and Bayesian consensus trees were consistent at different places: (1)Cistus species were not monophyletic; (2)Cistus species were divided in two lineages, one of purple-flowered species (except C. parviflorus) (100% BS, 100 PP) and other of white-flowered species plus C. parviflorus(97% BS, 100 PP); (3) Cistus crispus was the sister-group of the rest of purple-flowered species (100% BS, 100 PP); and (4) a sister-group relationship existed between the C. clusii group (100% BS, 100 PP) and the rest of the white-flowered species plus _C. parvifloru_s (81% BS, 100 PP).Halimium umbellatum appears to be related to the white-flowered lineage in the Bayesian analysis, but not in the MP analysis (Fig. 1).
Table 2. Characteristics of each of the DNA sequence regions used in the phylogenetic analysis of Cistaceae and the white-flowered Cistus.
| trnS-trnG | trnL-trnF | trnK-matK | rbcL | ITS | ncp_GS_ | |
|---|---|---|---|---|---|---|
| Cistaceae | ||||||
| Length (bp) | ||||||
| Total aligned length | 1084 | 516 | 1403 | 1404 | 697 | 402 |
| Length range - ingroup | 617–824 | 399–461 | 1302–1357 | 1403–1404 | 644–650 | 340–452 |
| Length range - outgroup | 158–684 | 377–422 | 1301–1316 | 1404 | 585–654 | 318 |
| Number of characters | ||||||
| Total included | 713 | 516 | 1403 | 1379 | 697 | 402 |
| Variable/parsimony-informative | 148/54 | 128/52 | 280/108 | 103/44 | 203–69 | 86/17 |
| Mean G+C content | 21% | 33% | 33% | 43% | 65% | 40% |
| Maximum sequence divergence (GTR) | 17.92% | 14.1% | 14.08% | 4.11% | 20.37% | 35.4% |
| Sequence evolution model (Akaike Test) | GTR+G | GTR+G | GTR+G | GTR+I | GTR+I+G | HKY+G |
| White-flowered Cistus plus C. parviflorus | ||||||
| No. of variable/parsimony-informative characters | 45/25 | 28/11 | 33/12 | 20/10 | 75/33 | 25/8 |
| Maximum sequence divergence (GTR) | 1.90% | 3.15% | 0.85% | 0.74% | 4.21% | 3.11% |
| Sequence evolution model (Akaike Test) | GTR+I | F81+I | GTR | HKY | HKY+I+G | HKY+G |
Evaluating patterns of trait evolution
The range of interspecific variation in leaf morphology and ecological requirements is shown in Table 3. Character reconstruction of three morphological and three ecological characters mapped on the Bayesian consensus tree (Fig. 2) using MacClade optimization and Bayesian inference to investigate patterns of evolution. The most relevant results from the historical reconstructions are following described:
Table 3. Morphological and environmental characteristics of the white-flowered Cistus lineage. Data were taken from Grosser [75], Martín & Guinea [76], Dansereau [77] *, Warburg [78], Demoly and Montserrat [79], Greuter [80], Gülz_et al_. [49] **and own observations.
| Soil | Climate conditions | Altitude (m) | Insolation conditions*, environment | Leaf shape (length×width in mm) | |
|---|---|---|---|---|---|
| C. albanicus | serpentines | mesic, Mediterranean mountain | 1000–1500 | submesophyllous, Abies cephalonica woodlands | elliptic (3–5×0.8–1.5)1 |
| C. clusii | calcicolous | dry to semi-arid, Mediterranean coast | 0–1500 | helioxerophyllous, bushy vegetation | linear (10–26×1–2) |
| C. ladanifer | silicolous | dry, Mediterranean | (0) 300–1000 (1500) | subheliophyllous, degradated_Quercus suber/ilex_woodlands | linear-lanceolate (40–80×6–21) |
| C. laurifolius | silicolous | mesic, Mediterranean mountain | 1900 (400–2800) | submesophyllous, degradated_Q. pyrenaica/faginea_and Pinus woodland | ovate-lanceolate (40–90×17–30) |
| C. libanotis | silicolous, sandy | dry, Mediterranean coast | 0–500 (1200) | subsciophyllous, degradated_Pinus halepensis/pinea_and Quercus suber woodlands | linear (22–40×2–5) |
| C. monspeliensis | silicolous | dry, Mediterranean | 0–800 (1200) | subheliophyllous, degradated_Quercus suber/ilex_ and_Pinus_ woodlands | linear-lanceolate (15–45×2–7) |
| C. munbyi | calcicolous | Mediterranean coast | 0–100 | helioxerophyllous, bushy vegetation | linear (6–30×1–4) |
| C. parviflorus | calcicolous | dry, Mediterranean coast | 0–600 | helioxerophyllous, scrub vegetation | ovate (15–30×7–27) |
| C. populifolius | silicolous | dry, Mediterranean | 200–1500 | submesophyllous, degradated_Quercus_ and_Pinus_ woodlands | ovate-lanceolate (50–95×25–55) |
| C. pouzolzii | silicolous | dry, mountain Mediterranean | 800–1800 | subheliophyllous, degradated_Quercus suber_/ilex and_Pinus_ woodlands | lanceolate-elliptic (20–31×4–11) |
| C. psilosepalus | silicolous | humid, woodlands of Atlantic influence | 0–800 (1100) | submesophyllous, scrub vegetation | lanceolate-elliptic (30–65×10–23) |
| C. salviifolius | silicolous/calcicolous | humid to dry, Mediterranean and Eurosiberian regions | 0–1800 | subheliophyllous/submesophyllous, degradated woodlands of many types | ovate (8–18×7–12) |
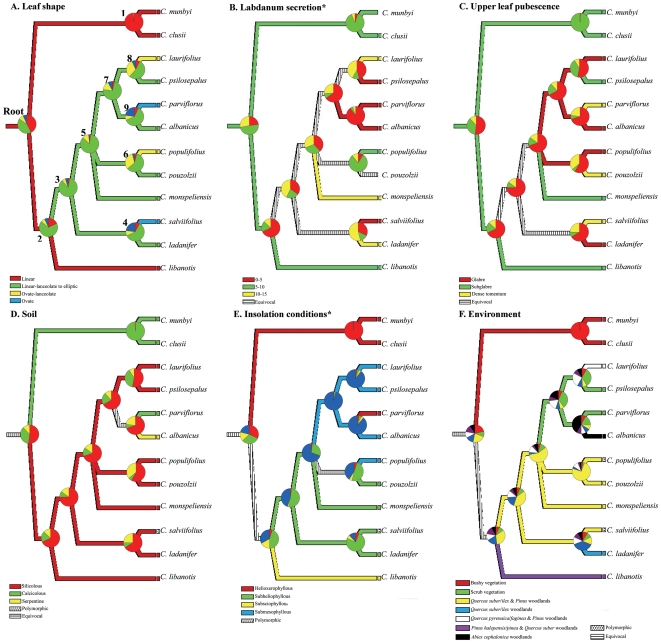
Figure 2. Historical patterns of leaf characters and ecological attributes.
Key characteristics are mapped onto the MacClade optimization tree as inferred by the Bayesian analysis: (A) Leaf shape, (B) Leaf labdanum secretion [49], (C) Pubescence of upper leaf surface, (D) Soil requirements, (E) Insolation conditions [77], (F) Environment. Pie charts at nodes represent the posterior probabilities of Bayesian inference character state evolution (Table 2). Node coding above branches in Fig. 2A.
- Leaf shape ( Fig. 2A ). The character state reconstruction showed linear or linear-lanceolate to elliptic leaves as a plesiomorphic state. Ovate-lanceolate and ovate shapes evolved twice in the _C. salviifolius_lineage.
- Labdanum secretion ( Fig. 2B ). The character was equivocal in most of the C. salviifolius lineage because, in part, of missing data from two species (C. mumbyi, C. pouzolzii). A medium percentage (5–10%) of secretion per unit leaf dry weight was, however, traced as the most likely ancestral state.
- Upper leaf pubescence ( Fig. 2C ). The character was revealed as very homoplastic within the _C. salviifolius_lineage. Despite the reconstruction was equivocal tracing the state at some nodes, independent acquisition (up to three times) of a dense tomentum is interpreted. Shifts between glabrous and subglabrous leaves appeared dynamic.
- Soil ( Fig. 2D ). The historical reconstruction traced silicolous soils as the ancestral state for the_C. salviifolius_ lineage. It was noteworthy that the only two species inhabiting basic (C. parviflorus) and ultrabasic (C. albanicus) soils within this lineage are sister species.
- Insolation conditions ( Fig. 2E ). Character optimization was equivocal reconstructing the ancestral state in the_C. salviifolius_ lineage. Two sister species groups underwent a dramatic change in insolation conditions (_C. parviflorus_-C. albanicus; _C. populifolius_-C. pouzolzii). Although ancestral character states were poorly optimised for insolation conditions, reversal to high solar exposure (helioxerophyllous) was unequivocally acquired for C. parviflorus.
- Environment ( Fig. 2F ). A high frequency in habitat change was found in the C. salviifolius lineage. Similar environments were shared in a few groups with (C. pouzolzii, C. populifolius) or without (C. psilosepalus, C. parviflorus) sister relationships. In contrast, three habitats were occupied by four closely-related species (C. laurifolius,C. psilosepalus, C. parviflorus, C. albanicus) suggesting a dynamic habitat change in the course of evolution of the _C. salviifolius_lineage.
The BayesTraits analysis of trait evolution was used to test reconstruction uncertainty. Table S2 reports ratedev settings and mean values (±95% confidence intervals) of the log-likelihood and posterior distributions of the rate of coefficients obtained from the reversible jump (RJ) MCMC analysis. The mean of the Bayesian posterior probabilities of each character state at every node (nine nodes) are provided in Table 4 and Fig. 2. The 95% confidence intervals of the posterior probabilities were all lower than±0.004. The Bayesian results mostly supported the MP (MacClade) optimization. Particular points of disagreement between both analyses were: (1) subglabre leaves at the root of the tree in the MP analysis whereas the Bayesian probability (0.52) was higher for glabre leaves; (2) ancestral states at node 2 were reconstructed as linear and subglabre leaves in the MP analysis, while the Bayesian approach estimated a higher probability for linear-lanceolate (0.68) and glabre leaves (0.68) states to be ancestral; (3) the historical reconstruction using the MP optimization traced Quercus suber/ilex and Pinus woodlands as the ancestral state at node 4 (C. ladanifer-C. salviifolius), while _Quercus suber/ilex_woodlands showed the highest posterior probability (0.38); (4) subheliophyllous condition was ancestral at node 5 in the MP optimization but the submesophyllous condition displayed the highest posterior probability (0.70); (5) scrub vegetation was the ancestral state at node 9 (C. parviflorus-C. albanicus) using the MacClade optimization, while _Abies cephalonica_woodlands displayed the highest posterior probability (0.42).
Table 4. Mean of posterior probabilities of Bayesian inference character state evolution of successive iterations (9,000,000) by RJ MCMC (see text) for six characters.
| Leaf shapea | Labdanum secretionb | Leaf pubescencec | Soild | Insolation conditionse | Environmentf | |
|---|---|---|---|---|---|---|
| Root | 0.42/0.42/0.08/0.07 | 0.23/0.49/0.28 | 0.52/0.36/0.12 | 0.77/0.17/0.06 | 0.32/0.31/0.16/0.21 | 0.21/0.10/0.22/0.14/0.11/0.13/0.09 |
| Node 1 | 0.99/0.00/0.01/0.00 | 0.05/0.89/0.06 | 0.01/0.98/0.01 | 0.00/0.99/0.01 | 0.99/0.00/0.00/0.01 | 0.96/0.01/0.01/0.01/0.01/0.00/0.00 |
| Node 2 | 0.18/0.68/0.07/0.07 | 0.30/0.34/0.36 | 0.68/0.19/0.13 | 0.98/0.01/0.01 | 0.05/0.45/0.17/0.33 | 0.06/0.09/0.33/0.18/0.12/0.14/0.08 |
| Node 3 | 0.03/0.86/0.06/0.05 | 0.35/0.22/0.43 | 0.75/0.10/0.15 | 0.98/0.01/0.01 | 0.02/0.51/0.02/0.45 | 0.06/0.09/0.40/0.19/0.12/0.06/0.08 |
| Node 4 | 0.08**/0.60**/0.09/0.23 | 0.32/0.14/0.54 | 0.66/0.09/0.25 | 0.89/0.07/0.04 | 0.06/0.76/0.06/0.12 | 0.09/0.09/0.13/0.38/0.13/0.09/0.09 |
| Node 5 | 0.02**/0.85**/0.11/0.02 | 0.38/0.31/0.31 | 0.70/0.15/0.15 | 0.96/0.02/0.02 | 0.02/0.27/0.01/0.70 | 0.04/0.11/0.57/0.04/0.12/0.04/0.08 |
| Node 6 | 0.04/0.60/0.32/0.04 | 0.09/0.80/0.11 | 0.58/0.06/0.36 | 0.98/0.01/0.01 | 0.05/0.49/0.05/0.41 | 0.03/0.03/0.74/0.04/0.10/0.03/0.03 |
| Node 7 | 0.04/0.73/0.15/0.08 | 0.65/0.09/0.26 | 0.67/0.21/0.12 | 0.80/0.08/0.12 | 0.01/0.02/0.01/0.96 | 0.07/0.33/0.07/0.08/0.18/0.08/0.19 |
| Node 8 | 0.07/0.55/0.31/0.07 | 0.46/0.12/0.42 | 0.53/0.38/0.09 | 0.95/0.02/0.03 | 0.03/0.04/0.04/0.89 | 0.08/0.33/0.08/0.09/0.25/0.08/0.09 |
| Node 9 | 0.07/0.62/0.08/0.23 | 0.90/0.05/0.05 | 0.70/0.07/0.23 | 0.11/0.31/0.58 | 0.03/0.04/0.04/0.89 | 0.08/0.18/0.08/0.08/0.08/0.08/0.42 |
Bayesian analysis of correlated evolution
Table 5 shows the log-Bayes factor calculations and significance following the scale of Bayes factor test presented by Kass & Raftery [29]. The evolution of leaf traits was not closely associated with ancestral changes in environment and insolation conditions. There was evidence against a correlated evolution of insolation conditions to leaf shape (log-Bayes factor = −1.5) and labdanum secretion (log-Bayes factor = −1.8). Additionally, barely evidence against correlated evolution for leaf pubescence and environment has been found (log-Bayes factor = −0.1). In contrast, barely correlated evolution was suggested between three pairs of variables: leaf shape/environment (log-Bayes factor = 0.7), labdanum secretion/environment (log-Bayes factor = 0.8) and leaf pubescence/insolation conditions (log-Bayes factor = 0.6). As already discussed elsewhere for organism radiations [30], estimates of barely correlated evolution have a strong evolutionary significance considering short tree branches.
Table 5. Calculations for log-Bayes factor tests in favour of a dependent model. In the final column, we followed the Bayes factor test [29] in our interpretation of the log-Bayes factor.
| Log-harmonic meana | log-Bayes factor | Significance | ||
|---|---|---|---|---|
| Dependent model | Independent model | |||
| Leaf shape/environment | −12.72 | −13.06 | 0.7 | barely in favour |
| Leaf shape/insolation | −18.12 | −17.36 | −1.5 | against |
| Labdanum secretion/environment | −11.99 | −12.39 | 0.8 | barely in favour |
| Labdanum secretion/insolation | −17.23 | −16.33 | −1.8 | against |
| Leaf pubescence/environment | −11.49 | −11.44 | −0.1 | barely against |
| Leaf pubescence/insolation | −16.25 | −16.58 | 0.6 | barely in favour |
Haplotype analysis of the white-flowered _Cistus_lineage
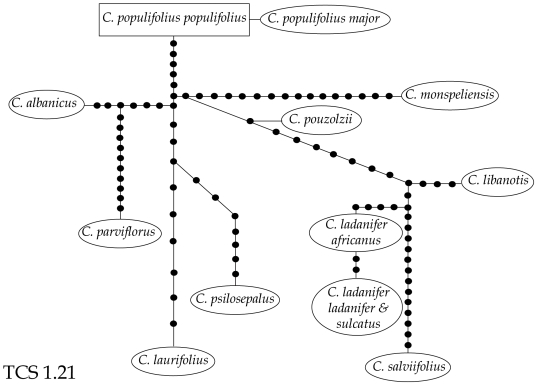
Sequence length of the white-flowered Cistus lineage was 417–461 bp for trnL-trnF, 561–585 for tnrS-trnG, 1309–1357 for_trnK-matK_ and 1378–1379 for_rbcL_ (Table 2). The combined data of plastid sequences for 10 species (13 taxa) of the C. salviifolius lineage distinguished only 12 substitution-based haplotypes (Table S3). Haplotypes were exclusive to a single species or subspecies (Table S3), except for one for both C. ladanifer subspp. ladanifer and_sulcatus_. TCS constructed a single, star-like network (Fig. 3) displaying no loops. This analysis is congruent with a multiple lineage divergence pattern from ancestral haplotypes, as expected in a radiation.
Figure 3. Statistical parsimony network representing relationships of the 12 plastid (trnL-trnF,trnK-matK, rbcL,trnS-trnG) haplotypes of the white-flowered Cistus lineage.
Lines indicate mutation steps (single nucleotide substitutions) and dots (•) represent missing haplotypes (extinct or not found). A star-like shape of the network is congruent with a process of radiation in this group.
Estimates of divergence times
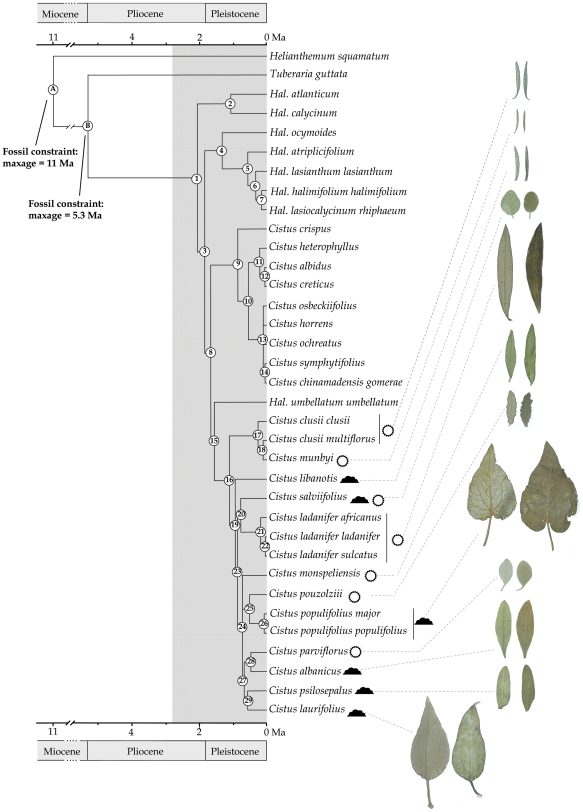
Results of the dating analysis are shown in Table 6 and Fig. 4. In general, the data indicated a Pliocene-Pleistocene (2.11±0.87 Ma) divergence between the basal-most Halimium and_Cistus-Halimium_ groups, followed by a Pleistocene differentiation of the major clades of the latter group. An ancestor shared by Halimium umbellatum and_Cistus_ appeared to have diverged after the Pliocene-Pleistocene boundary (1.47±0.35 Ma). Short branch lengths may reflect a rapid divergence process in the white-flowered lineage. An early divergent lineage of C. clusii and_C. mumbyi_ (C. clusii lineage) at 1.04±0.25 Ma was followed by differentiation of 10 species (C. salviifolius lineage) in the Mid Pleistocene (0.88±0.22 Ma). The average per-lineage species diversification rate for the C. salviifolius lineage was 1.46–2.44 species per million years.
Table 6. Penalized Likelihood (bootstrapping of 100 trees) molecular clock estimates of ages for constrained and unconstrained nodes.
| Node | Mean age (Ma) | SD (Ma) | Maximum age (Ma) | Minimum age (Ma) |
|---|---|---|---|---|
| A (11) | 9.65 | 2.21 | 11.00 | 0.58 |
| B (5.3) | 4.87 | 1.10 | 5.30 | 0.21 |
| 1 | 2.11 | 0.87 | 4.93 | 0.14 |
| 2 | 1.01 | 0.31 | 1.99 | 0.06 |
| 3 | 1.78 | 0.45 | 2.74 | 0.12 |
| 4 | 1.25 | 0.32 | 1.80 | 0.07 |
| 5 | 0.53 | 0.15 | 0.83 | 0.03 |
| 6 | 0.30 | 0.09 | 0.49 | 0.02 |
| 7 | 0.15 | 0.06 | 0.33 | 0.006 |
| 8 | 1.56 | 0.38 | 2.32 | 0.09 |
| 9 | 0.80 | 0.21 | 1.17 | 0.05 |
| 10 | 0.52 | 0.14 | 0.78 | 0.03 |
| 11 | 0.19 | 0.07 | 0.33 | 0.01 |
| 12 | 0.04 | 0.02 | 0.13 | 0.002 |
| 13 | 0.05 | 0.06 | 0.21 | 0.000 |
| 14 | 0.04 | 0.02 | 0.13 | 0.003 |
| 15 | 1.47 | 0.35 | 2.09 | 0.08 |
| 16 | 1.04 | 0.25 | 1.41 | 0.06 |
| 17 | 0.23 | 0.09 | 0.43 | 0.01 |
| 18 | 0.09 | 0.04 | 0.20 | 0.003 |
| 19 | 0.88 | 0.22 | 1.22 | 0.06 |
| 20 | 0.82 | 0.20 | 1.13 | 0.05 |
| 21 | 0.72 | 0.18 | 0.97 | 0.04 |
| 22 | 0.17 | 0.06 | 0.34 | 0.009 |
| 23 | 0.04 | 0.02 | 0.11 | 0.0003 |
| 24 | 0.65 | 0.16 | 0.89 | 0.04 |
| 25 | 0.45 | 0.12 | 0.71 | 0.02 |
| 26 | 0.06 | 0.04 | 0.21 | 0.002 |
| 27 | 0.61 | 0.23 | 0.91 | 0.00 |
| 28 | 0.31 | 0.23 | 0.67 | 0.00 |
| 29 | 0.28 | 0.28 | 0.71 | 0.00 |
Figure 4. Phylogenetic chronogram of the_Cistus-Halimium_ complex based on the Bayesian consensus tree.
Fossil calibration points are indicated on the tree. Shaded area delineates the establishment of the Mediterranean climate 2.8 million years ago [50]. Geological timescales are shown both at the top and the bottom. Photographs illustrate diversity in leaf morphology of the white-flowered Cistus species (only subsp. ladanifer of C. ladanifer, subsp. clusii of_C. clusii_ and subsp.populifolius of C. populifolius are illustrated). Species insolation conditions [77] are plotted on the right side of the tree ( , helioxerophyllous and subhelioxerophyllous;
, helioxerophyllous and subhelioxerophyllous; , subsciophyllous and submesophyllous).
, subsciophyllous and submesophyllous).
Discussion
An adaptive radiation comprises a group of species that inhabit a variety of environments, differ in morphological and other traits important in utilizing these environments, and are descended from a common ancestor that rapidly speciated over a short period of time [1]. Available phylogenetic and ecological evidence suggests that the C. salviifolius lineage of 10 white-flowered species meets the four criteria to strictly test adaptive radiation: common ancestry, rapid speciation, phenotype-environment relationships and trait utility [1], [4].
Monophyly of the white-flowered Cistus lineage is strongly supported irrespective of phylogenetic methods and DNA sequences used (Fig. 1). Accordingly, the 12 white-flowered species form a well-defined natural group and fulfil the common ancestry condition.
The concept of rapid speciation is not very well defined, even though a considerable number of species is needed [1]. Asymmetry between sister clades in their number of descendant species is one of the operational standards to distinguish speciation bursts from stochastic background rates [31]. Compared with the two taxa included in the C. clusii lineage, the remaining taxa (13) form a sister group (C. salviifolius lineage) and can be consider as a significant burst. In fact, asymmetries between both lineages can also be inferred in a temporal pattern. After the split of the most common recent ancestor of the two lineages (1.04±0.25 Ma), a relatively long period of time was necessary to bring about limited (2) extant species in_C. clusii_ lineage, in contrast to the 10 species generated in the C. salviifolius group (Fig. 4). Alternatively, rapid radiation is also interpreted as high rates of differentiation in comparison to those of flowering plants. The estimated rate of diversification in the C. salviifolius lineage was significantly higher (1.46–2.44 species per million years) compared to the median rate of diversification of angiosperm families (0.12 species per million years; with a maximum of 0.39) [32] and to that found in the Andean Valeraniaceae [33], and similar to the explosive radiation described for Andean Lupinus [34]. Rapid diversification in the C. salviifolius lineage was already predicted by a combination of different sources of evidence prior to performing explicit analysis of radiation: (1) lack of resolution and low support values depicted mainly in the parsimony-based tree because of a low number of polymorphisms [20], which is overcome by increasing the number of DNA substitutions (this paper); (2) short branch lengths and low pairwise sequence divergence (Fig. 4, Table 2); (3) low resolution at the core of the haplotype network [35] (Fig. 3).
In addition to evidence for common ancestry and rapid diversification, the fit of the diverse phenotypes observed in a lineage with their environment is necessary in prediction of adaptive radiations [1]. Our character reconstruction suggests that shifts in leaf features allowing the colonization of different habitats have been related with specific speciation events (Fig. 2). Acquisition of diverse leaf features is associated with recent lineage splits, and thus closely related taxa exhibit different leaf morphologies [26]. Our character state optimization reveals that the most common recent ancestors of four sister species diversified in different environmental conditions (Fig. 2D, 2E, 2F) by means of shifts in leaf shape (Fig. 2A) and leaf pubescence (Fig. 2C). In addition, transition in leaf labdanum secretion is observed in the Bayesian inference (Table 4; Fig. 2B). Trends of correlated evolution between leaf traits and at least one ecological trait (environment, insolation conditions) have been found (Table 5). The barely correlated evolution found in three morphological/ecological traits indicates that shifts in environmental conditions must parallel evolutionary changes in Cistus leaf morphology as a whole and not in individual leaf features. Experimental studies testing correlated evolution of all leaf traits should be further performed to analyse compensatory effects (trade-off). Alternatively to sister species approaches, another strong indication of the adaptive value of a trait is when phylogenetically separate, but ecologically similar, species converge or show parallel patterns of variation along similar ecological gradients [36]. Multiple leaf morphological character-states studied across the white-flowered lineage (shape, labdanum secretion, pubescence) have been independently acquired at least twice (Fig. 2).
Evidence that some morphological and/or physiological traits of species are particularly useful is the fourth necessary condition to support the most strict concept of adaptive radiation: trait utility [1]. The adaptive implications of leaf size and shape differences are well documented[37], [38]. In absence of explicit experimental studies (plant translocation, common garden conditions) for all species involved in this adaptive radiation [39], the body of knowledge for particular traits is analysed. Although our six DNA sequence data set rendered certain phylogenetic uncertainty for some sister species relationships because of moderate support (Fig. 1), the most plausible hypothesis allows assessing low character-state reconstruction uncertainty of leaf morphological utility using MacClade optimization and BayesTraits analysis of trait evolution. Leaf size and shape are implicated in important aspects as thermoregulation[40], [41], efficiency of water use [23], [42], photosynthetic potential [43], branching and rooting strategies [44], among others. Moreover, comparative studies have revealed the existence of well-marked ecological and leaf morphological trends [45]. Small leaf size (specifically narrow leaves) are generally favoured under high exposure and/or low water availability as they help to maintain favourable leaf temperature and improves water use efficiency [42], [44]. Indeed, small-leaved species are concentrated at the high exposure end on south-facing slopes in Mediterranean garrigue and Californian chaparral [46]. Although our character reconstruction hypothesis indicates dynamic shifts of leaf shapes, the ancestral state (narrow leaves) appears to have evolved early into linear-lanceolate to elliptic, and then into ovate (plus ovate-lanceolate) leaves independently four times. In fact, helioxerophyllous species (C. clusii, C. munbyi, C. libanotis) show ancestral linear revolute leaves while submesophyllous species with broad, flat leaves inhabit shadier environments (C. laurifolius, C. populifolius, C. psilosepalus) (Table 3, Fig. 2). Leaf shape is not, however, the only phenotypic trait associated with adaptation to dry conditions. Leaf pubescence is reported to be an adaptation to sunnier and hotter environments by reducing transpiration, increasing the probability of water uptake by leaves, maintaining favourable leaf temperature, and protecting against UV-B radiation responsible for photosynthetic inhibition[47], [48]. Accordingly, a combination of leaf trait strategies meets in unrelated species of Cistus. Sister species within the white-flowered_Cistus_ lineage have different leaf traits related to leaf transpiration. For instance, the ovate leaves of C. parviflorus unsuitable for xeric environments are protected by a dense tomentum of stellate hairs. The same is true at a lower extent in_C. salviifolius_. In addition, leaves can be highly reflective in the visible spectrum by covering the upper surface with labdanum, and then decreasing transpiration [49]. The high leaf secretion of resins (labdanum) in the linear-lanceolate leaves of C. monspeliensis and C. ladanifer may confer a trade-off compared to the narrower leaves of C. clusii,C. mumbyi and C. libanotis, which display linear leaves and lower labdanum concentration (Fig. 2). Further studies are needed to pinpoint whether combination of multiple leaf strategies are equally fit in dry, Mediterranean habitats suffering from dry hot summers and high solar radiation.
In summary, the evolutionary history of the 10 species (13 taxa) of the_C. salviifolius_ lineage fits into utilization of the niche space in a novel manner far after the Mediterranean climate establishment [50]. A Mediterranean_Cistus_ ancestor with linear, medium labdanum content and glabrous or subglabrous leaves may have spawned new lines of evolution exploiting six pre-existing Mediterranean habitats. Multiple leaf strategies were successfully essayed in the course of speciation to occupy particular environments and become part of the dominant element in the Mediterranean scrub. As far as we know, this is the first documented plant group involved in an adaptive radiation process in the Mediterranean region.
Materials and Methods
Sample strategy and DNA sequencing
A total of 36 individuals representing the 21 species of_Cistus_, one of Fumana, eight of_Halimium_, one of _Helianthemum_and one of Tuberaria was sequenced for four plastid (trnL-trnF spacer,trnS_-trnG spacer,trnK_-matK spacer,rbcL exon) and two nuclear (ITS, ncp_GS) DNA regions (Table 1; Table S1) to perform phylogenetic analyses and estimate divergence times of Cistus and related lineages. In addition, a data set comprising only the white-flowered_Cistus species (plus C. parviflorus) was used to infer character evolution, correlated evolution and haplotype analyses.
Standard primers were used for amplification of the ITS region [51 for 17SE], [52 for ITS4], the trnL(UAA)-trnF(GAA) [53], the trnK-matK [trnK-3914F and matK-1470R, 54] and the_trnS_ (GCU)-trnG (UCC) [55] spacers. The_rbcL_ exon was amplified in two overlapping segments using the following primer combination: 1F-724R and 636F-1460R [56]. A portion of the glutamine synthetase (ncp_GS_) was amplified for the first time in 11 Cistus species with the universal primers Gscp687f and Gscp856r [57]. To ensure a homogeneous amplification reaction we design two 24-nucleotide-long primers specific for amplifying and sequencing_Cistus_ species (CIS-687f: 5′GTAGCTGGAATCAACATCAGTGG3′, CIS-856r: 5′GCTTGTTCAGTGATTCTCTGTCAG3′).
After 1–3 min pretreatment at 94°C, PCR conditions for amplification were: 24–39 cycles of 1 min at 94°C, 30 s-1 min at 48–50–55°C and 1–4 min at 72°C (for details see 19). PCR primers were used for cycle sequencing of the spacers, the rbcL exon and the ncp_GS_ gene while the ITS 5 and ITS 4 [52] primers were used for cycle sequencing the ITS region. Additionally, due to mononucleotide repeat stretches (poly-T, poly-A) the internal primer trnSGpolyTf (5′TTAGATTCTATTTACATTCT3′) was used to sequence the trnS-trnG spacer in the purple-flowered species. Sequenced data were assembled and edited using the program Seqed (Applied Biosystems, California). The limits of the regions were determined by position of flanking primers. IUPAC symbols were used to represent nucleotide ambiguities.
Molecular analyses
Phylogenetic analyses
Maximum Parsimony (MP) and Bayesian Inference (BI) analyses were performed on a combined molecular data set of_trnL-trnF_,_trnS_-trnG,trnK_-matK,rbcL, ITS and ncp_GS sequences. Sequences were aligned using Clustal X 1.62b [58], with further adjustments by visual inspection. All parsimony analyses were conducted using Fitch parsimony [59]with equal weighting of all characters and of transitions/transversions. Heuristic searches were replicated 1000 times with random taxon–addition sequences, tree–bisection–reconnection (TBR) branch swapping, the options MulTrees and Steepest Descent in effect and holding 10 trees per replicate. Internal support was assessed using 5,000,000 bootstrap (BS) replicates [fast stepwise-addition, 60].
To determine the simplest model of sequence evolution that best fits the sequence data, the Hierarchical Likelihood Ratio Test (hLRT) and Akaike Information Criterion (AIC) were implemented using MrModeltest 1.1b [61], [62]in each data set. A Bayesian Inference analysis (BI) was conducted in MrBayes 3.0b4 [63]using two identical searches with two million generations each (four MCMC, chain temperature = 0.2; sample frequency = 100). In both runs probabilities converged at the same stable value after generation 100,000 approximately. A 50% majority-rule consensus tree was calculated using the sumt command to yield the final Bayesian estimate of phylogeny. We used posterior probability (PP) as an estimate of robustness.
Molecular dating and diversification rates
Divergence dates were estimated for nodes of the Bayesian consensus tree. To check the constancy of substitution rates we used the Langley and Fitch (LF) test [64]. We rejected the null hypothesis of constant rate (χ2 = 5204.26; d.f. = 34) and, then, divergence times were estimated using the r8S 1.71 program [65] with a Penalized Likelihood (PL) approach. Penalized Likelihood was implemented with the Truncated Newton (TN) algorithm. Initial results were obtained under the following parameters: cvstart = 0.5; cvinc = 0.5; cvnum = 10 with cross-validation enforced to estimate the rate smoothing parameter (measure of the rate variation and autocorrelation of rates from clade to clade). The rate smoothing with the lowest crossvalidation score was selected and the dating procedure was repeated with the following parameters: collapse; num_time_guesses = 5 and num_restarts = 5. Crossvalidation suggested 10 as the best smooth parameter. Branching order and branch lengths from 100 Bayesian trees sampled every 10,000 generations after stationary were analyzed to obtain means and standard deviations of clade ages [34]. To convert relative divergence times into absolute time units we used two maximum-age fossil constraints. Palynological studies identified_Helianthemum_ pollen in Upper Miocene formations (11 Ma) from France [66] and_Tuberaria_ pollen in Pliocene formations (5.3 Ma) from Germany [67].
Species diversification rates, assuming an equal rate of random speciation Yule model, were calculated using the formula SR = [(loge(N)–loge(N0)]/T[34], [68],[69], where N is the total number of extant species in the clade of interest, N0is the initial species diversity, usually taken as 1, and T is the inferred age of the clade (million years). Upper and lower standard deviations of age estimates were used in calculations of speciation rates.
Character evolution
Patterns of evolution of six key traits (leaf shape, leaf labdanum secretion, leaf pubescence, soil requirements, insolation conditions, habitat) were explored in the white-flowered_Cistus_ lineage using the Bayesian consensus tree (calculated using the same parameters as above). Optimizations were performed in MacClade 4.06 [70] assuming Fitch Parsimony, equal weighting of all characters, transitions among all states equally probable and treating characters as unordered. Character states were determined from literature and personal observations. Samples of Cistus crispus and_C. heterophyllus_ were used as outgrop sequences.
In addition, to account for values of phylogenetic mapping uncertainty, probabilities of ancestral states for the six traits were estimated individually using the BayesMultiState program, contained in the BayesTraits 1.0 package [71], under Montecarlo Markov Chain (MCMC) method and allowing transitions between character states in both directions. To reduce the autocorrelation of successive samples, 1000 trees were drawn from the distribution of 1.9×106 trees, which equates to sampling every 1900th generation of the chains used in the phylogenetic analysis. As suggested in BayesMultiState manual, to reduce some of the uncertainty and arbitrariness of choosing prior in MCMC studies, we used the hyperprior approach, in concrete the reversible-jump (RJ) hyperprior with a gamma prior (mean and variance seeded from uniform distributions on the interval 0 to 10). Preliminary analyses were run to adjust the_ratedev_ parameter until the acceptance rates of proposed changes was around 20–40%. Using_ratedev_ settings (Table S2), we ran the RJ MCMC analyses for each trait three times independently for 1.0×107iterations, sampling every hundredth iteration (to produce 90,000 sampled points) and discarding the first 1,000,000 iterations. All runs gave mostly the same results and we report one of them here. We use the “Addnode” command to find the proportion of the likelihood associated with each of the possible states at each node.
Testing correlated evolution
We modelled correlated evolution of discrete binary traits (leaf shape/insolation conditions, leaf shape/habit, labdanum secretion/insolation conditions, labdanum secretion/habit, leaf pubescence/insolation conditions, leaf pubescence/habit) on 1000 Bayesian trees using the BayesDiscrete program, contained in the BayesTraits 1.0 package [71]and the same parameters described above. The method compares the statistical likelihood of a model in which two binary traits are allowed to evolve independently on the tree, with a model in which the two traits are allowed to evolve in a correlated fashion. Evidence for correlated evolution arises if the dependent or correlated model shows significantly better fit to the data than the independent model. As the independent and dependent models are estimated by MCMC, their goodness of fit is compared using the log-Bayes Factor test: 2*log[harmonic mean(dependent model)] – log[harmonic mean (independent model)].
We used one sample per species of the white-flowered lineage given the monophyly of all species [72]. As binary traits are required, we coded traits as followed: leaf shape, 0 linear to elliptic, 1 ovate-lanceolate to ovate; labdanum secretion, 0 zero to eight percent, 1 nine to fifteen; upper leaf pubescence, 0 glabre to subglabre, 1 dense tomentum; insolation conditions, 0 helioxerophyllous to subheliophyllous, 1 subsciophyllous to submesophyllous; environment, 0 bushy and scrub vegetation, 1 woodlands.
Haplotype data analysis
Sequences of plastid DNA (trnL-trnF,trnK_-matK trnS-trnG and_rbcL) were combined to analyze relationships among the white-flowered Cistus(plus C. parviflorus) plastid haplotypes. We used the software TCS 1.21 to infer plastid haplotype ancestry[73]. The program implements a statistical parsimony approach using the algorithm described in Templeton et al. [74] to construct haplotype networks. The maximum number of differences among haplotypes, as a result of single substitutions, was calculated with 95% confidence limits and treating gaps as missing data.
Supporting Information
Table S1
GenBank accession numbers.
(0.10 MB DOC)
Table S2
Bayesian inference of trait evolution of successive iterations of the chain (9,000,000) in the white-flowered_Cistus_ lineage by reversible jump Markov chain Monte Carlo. Means±confidence intervals (95%) of the log-likelihoods (Lh) and rate coefficients are shown.
(0.07 MB DOC)
Table S3
List of haplotypes found in 16 species and subspecies of the white-flowered Cistus lineage. Variable sites of the sequences of four plastid DNA regions (_trnL_-trnF,rbcL,_trnK_-matK,_trnS_-trnG) are shown. Nucleotide position for each data set is numbered from the 5′ to the 3′ DNA ends.
(0.20 MB DOC)
Acknowledgments
We thank E. Cano and N. Rønsted for lab assistance; P. Escobar, A. Fernández, O. Filippi, J. Leralta, J. Martínez and R. G. Page for plant materials; I. Sanmartín for advice on BayesTraits analyses; J. J. Aldasoro for computer assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research has been supported by the Spanish Direccion General de Investigacion y Ciencia (DGICYT) through the project CGL2005-06017-C02 and by the Comunidad de Madrid through the project GR/AMB/0759/2004 and a PhD scholarship to B. G.. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schluter D. New York: Oxford University Press Inc; 2000. The ecology of adaptive radiation. [Google Scholar]
- 2.Gavrilets S, Losos JB. Adaptive radiation: contrasting theory with data. Science. 2009;323:732–737. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin BG, Robichaux RH. Historical biogeography and ecology of the Hawaiian silversword alliance (Asteraceae): new molecular phylogenetic perspectives. In: Wagner WL, Funk VA, editors. Hawaiian biogeography: evolution on a hotspot archipelago. Washington, D.C.: Smithsonian Institution Press; 1995. pp. 257–287. [Google Scholar]
- 4.Meimberg H, Abele T, Brauchler C, McKay JK, Perez de Paz PL, et al. Molecular evidence for adaptive radiation of Micromeria Benth. (Lamiaceae) on the Canary Islands as inferred from chloroplast and nuclear DNA sequences and ISSR fingerprint data. Mol Phylogenet Evol. 2006;41:566–578. doi: 10.1016/j.ympev.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 5.Hodges SA. Rapid radiation due to a key innovation in columbines (Ranunculaceae: Aquilegia). In: Givnish TJ, Sytsma KJ, editors. Molecular evolution and adaptive radiation. New York, USA: Cambridge University Press; 1997. pp. 391–405. [Google Scholar]
- 6.MacArthur RH, Wilson EO. Princeton, New Jersey: Princeton University Press; 1967. The theory of island biogeography. [Google Scholar]
- 7.Baldwin BG, Crawford DJ, Francisco-Ortega J, Kim S-C, Sang T, et al. Molecular phylogenetics insights on the origin and evolution of oceanic islands plants. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II DNA sequencing. Boston/Dordretch/London: Kluwer Academic Publishers; 1998. pp. 410–441. [Google Scholar]
- 8.Kim S-C, Crawford DJ, Francisco-Ortega J, Santos-Guerra A. A common origin for woody Sonchus and five related genera in the Macaronesian islands: evidence for extensive radiation. Proc Natl Acad Sci USA. 1996;93:7743–7748. doi: 10.1073/pnas.93.15.7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Givnish TJ, Millam KC, Mast AR, Patterson TB, Theim TJ, et al. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc R Soc Lond [Biol] 2008 doi: 10.1098/rspb.2008.1204. doi: 10.1098/rspb.2008.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinthal PN, Meyer A. Molecular phylogenetic tests of speciation models in lake Malawi cichlid fishes. In: Givnish TJ, Sytsma KJ, editors. Molecular evolution and adaptive radiation. Melbourne, Australia: Cambridge University Press; 1997. pp. 375–390. [Google Scholar]
- 11.Verboom GA, Linder HP, Stock WD. Testing the adaptive nature of radiation: Growth form and life history divergence in the African grass genus Ehrharta (Poaceae: Ehrhartoideade). Am J Bot. 2004;91:1364–1370. doi: 10.3732/ajb.91.9.1364. [DOI] [PubMed] [Google Scholar]
- 12.Daget P. Le bioclimat méditerranéen caractères generaux, mode de caracterisation. Vegetatio. 1977;31:1–20. [Google Scholar]
- 13.Nahal I. The Mediterranean climate from a biological viewpoint. In: de Castri F, Goodall DW, Specht RL, editors. Ecosystems of the World 11, Mediterranean-type shrublands. Amsterdam: Elsevier Scientific Publising Company; 1981. pp. 63–86. [Google Scholar]
- 14.Mooney HA, Dunn EL. Photosynthetic systems of Mediterranean-climate shrubs and trees of California and Chile. Am Nat. 1970;104:447–453. [Google Scholar]
- 15.Read J, Sanson GD. Characterizing sclerophylly: the mechanical properties of a diverse range of leaf types. New Phytol. 2003;160:81–99. doi: 10.1046/j.1469-8137.2003.00855.x. [DOI] [PubMed] [Google Scholar]
- 16.Ackerly DD. Adaptation, niche conservatism, and convergence: comparative studies of leaf evolution in the California chaparral. Am Nat. 2004;163:654–671. doi: 10.1086/383062. [DOI] [PubMed] [Google Scholar]
- 17.Herrera CM. Historical effects and sorting processes as explanations for contemporary ecological patterns: character syndromes in Mediterranean woody plants. Am Nat. 1992;140:421–446. [Google Scholar]
- 18.Pausas JG, Verdu M. Plant persistence traits in fire-prone ecosystems of the Mediterranean basin: A phylogenetic approach. Oikos. 2005;109:196–202. [Google Scholar]
- 19.Arrington JM, Kubitzki K. Cistaceae. In: Kubitzki K, editor. The Families and Genera of Vascular Plants IV Flowering Plants Dicotyledons Malvales, Capparales and Non-betalain Caryophyllales. Berlin: Springer; 2003. pp. 62–70. [Google Scholar]
- 20.Guzmán B, Vargas P. Systematics, character evolution, and biogeography of Cistus L. (Cistaceae) based on ITS, trnL-trnF, and matK sequences. Mol Phylogenet Evol. 2005;37:644–660. doi: 10.1016/j.ympev.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Guzmán B, Vargas P. Long-distance colonization of the Western Mediterranean by Cistus ladanifer (Cistaceae) despite the absence of special dispersal mechanisms. J Biogeogr. 2009;36:954–968. [Google Scholar]
- 22.Guzmán B, Vargas P. Historical biogeography and character evolution of Cistaceae (Malvales) based on analysis of plastid rbcL and trnL-trnF sequences. Org Divers Evol. 2009;9:83–99. [Google Scholar]
- 23.Cunningham SA, Summerhayes BA, Westoby M. Evolutionary divergences in leaf structure and chemistry, comparing rainfall and soil nutrient gradients. Ecol Monogr. 1999;69:569–588. [Google Scholar]
- 24.Givnish TJ. Leaf and canopy adaptations in tropical forests. In: Medina E, Mooney HA, Vázquez-Yanes C, editors. Physiological ecology of plants of the wet tropics. The Hague: Junk; 1984. pp. 51–84. [Google Scholar]
- 25.Skelton PW. Adaptive radiation: definition and diagnostic tests. In: Lees DR, Edwards D, editors. Evolutionary patterns and processes. London: Academic Press; 1993. pp. 45–58. [Google Scholar]
- 26.Givnish TJ, Sytsma KJ, Smith JF, Hahn WJ. Molecular evolution, adaptive radiation, and geographic speciation in Cyanea (Campanulaceae, Lebelioideae). In: Wagner WL, Funk VA, editors. Hawaiian biogeography. Washington: Smithsonian Institution Press; 1995. pp. 288–337. [Google Scholar]
- 27.Lowrey TK. Phylogeny, adaptive radiation, and biogeography of Hawaiian Tetramolopium (Asteraceae, Astereae). In: Wagner WL, Funk VA, editors. Hawaiian biogeography. Washington: Smithsonian Institution Press; 1995. pp. 195–220. [Google Scholar]
- 28.Robichaux RH, Carr GD, Liebman M, Pearcy RW. Adaptive radiation of the Hawaiian silversword alliance (Compositae-Madiinae): ecological, morphological, and physiological diversity. Ann Missouri Bot Gard. 1990;77:64–72. [Google Scholar]
- 29.Kass R, Raftery A. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 30.Whitfield JB, Lockhart PJ. Deciphering ancient rapid radiations. Trends Ecol Evol. 2007;22:258–265. doi: 10.1016/j.tree.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Rüber L, Van Tassell JL, Zardoya R. Rapid speciation and ecological divergence in the American seven-spined Gobies (Gobiidae, Gobiosomatini) inferred from a molecular phylogeny. Evolution. 2003;57:1584–1598. doi: 10.1111/j.0014-3820.2003.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson O, Bremer B. Pollination systems, dispersal modes, life forms, and diversification rates in angiosmperm families. Evolution. 1992;46:258–266. doi: 10.1111/j.1558-5646.1992.tb02000.x. [DOI] [PubMed] [Google Scholar]
- 33.Bell CD, Donoghue MJ. Phylogeny and biogeography of Valerianaceae (Dipsacales) with special reference to the South American valerians. Org Divers Evol. 2005;5:147–159. [Google Scholar]
- 34.Hughes C, Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc Nat Acad Sci. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avise JC. Phylogeography: retrospect and prospect. J Biogeogr. 2009;36:3–15. [Google Scholar]
- 36.Endler JA. Princeton, New Jersey: Princeton University Press; 1986. Natural selection in the wild. [Google Scholar]
- 37.Givnish TJ. On the adaptive significance of leaf form. In: Solbrig OT, Jain S, Johnson GB, Raven PH, editors. Topics in plant population biology. New York: Columbia University Press; 1979. pp. 375–407. [Google Scholar]
- 38.Givnish TJ, Montgomery RA, Goldstein G. Adaptive radiation of photosynthetic physiology in the Hawaiian Lobeliads: light regimes, static light responses, and whole-plant compensation points. Am J Bot. 2004;91:228–246. doi: 10.3732/ajb.91.2.228. [DOI] [PubMed] [Google Scholar]
- 39.Nuñez-Olivera E, Martinez-Abaigar J, Escudero JC. Adaptability of leaves of Cistus ladanifer to widely varying environmental conditions. Funct Ecol. 1996;10:636–646. [Google Scholar]
- 40.Gates DM, Alderfer R, Taylor SE. Leaf temperature of desert plants. Science. 1968;159:994–995. doi: 10.1126/science.159.3818.994. [DOI] [PubMed] [Google Scholar]
- 41.Szwarcbaum I. Influence of leaf morphology and optical properties on leaf temperature and survival in three Mediterranean shrubs. Plant Sci Lett. 1982;26:47–56. [Google Scholar]
- 42.Parkhurst D, Loucks O. Optimal leaf size in relation to environment. J Ecol. 1972;60:505–537. [Google Scholar]
- 43.Cunningham GL, Strain BR. An ecological significance of seasonal leaf variability in a desert shrub. Ecology. 1969;50:400–408. [Google Scholar]
- 44.Givnish TJ, Vermeji GJ. Sizes and shapes of liane leaves. Am Nat. 1976;110:743–778. [Google Scholar]
- 45.Givnish TJ. Comparative studies of leaf form: assessing the relative roles of selective pressures and phylogenetic constraints. New Phytol. 1987;106(Suppl.):131–160. [Google Scholar]
- 46.Ackerly DD, Knight CA, Weiss SB, Barton K, Starmer KP. Leaf size, specific leaf area and microhabitat distribution of chaparral woody plants: contrasting patterns in species level and community level analyses. Oecologia. 2002;130:449–457. doi: 10.1007/s004420100805. [DOI] [PubMed] [Google Scholar]
- 47.Savé R, Biel C, de Herralde F. Leaf pubescence, water relations and chlrorophyll fluorescence in two species of Lotus creticus L. Biol Plant. 2000;43:239–244. [Google Scholar]
- 48.Ehleringer JR, Clark C. Evolution and adaptation in Encelia (Asteraceae). In: Gottlieb LD, Subodh KJ, editors. Plant evolutionary biology. New York: Chapman and Hall Ltd; 1988. pp. 221–248. [Google Scholar]
- 49.Gülz PG, Herrmann T, Hangst K. Leaf trichomes in the genus Cistus. Flora. 1996;191:85–104. [Google Scholar]
- 50.Suc JP, Bertini A, Combourieu-Nebout N, Diniz F, Leroy S, et al. Structure of West Mediterranean vegetation and climate since 5.3 Ma. Acta Zool Cracov. 1995;38:3–16. [Google Scholar]
- 51.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Geldfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 52.Sun Y, Skinner DZ, Liang GH, Hulbert SH. Phylogenetic analysis of Sorghum and related taxa using Internal Transcribed Spacer of nuclear ribosomal DNA. Theor Appl Genet. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- 53.Taberlet P, Gielly L, G P, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 54.Johnson LA, Soltis DE. matK DNA and phylogenetic reconstruction in Saxifragaceae s. str. Syst Bot. 1994;19:143–156. [Google Scholar]
- 55.Hamilton M. Four primers pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol. 1999;8:521–523. [PubMed] [Google Scholar]
- 56.Savolainen V, Chase MW, Hoot SB, Morton CM, Soltis DE, et al. Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst Biol. 2000;49:306–362. doi: 10.1093/sysbio/49.2.306. [DOI] [PubMed] [Google Scholar]
- 57.Emshwiller E, Doyle JJ. Chloroplast-expressed Glutamine synthetase (ncpGS): Potencial utility for phylogenetic studies with an example from Oxalis (Oxalidaceae). Mol Phylogenet Evol. 1999;12:310–319. doi: 10.1006/mpev.1999.0613. [DOI] [PubMed] [Google Scholar]
- 58.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swofford D. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer; 2002. PAUP*. [Google Scholar]
- 60.Mort ME, Soltis PS, Soltis DE, Mabry M. Comparison of three methods for estimating internal support on phylogenetic trees. Syst Biol. 2000;49:160–171. doi: 10.1080/10635150050207456. [DOI] [PubMed] [Google Scholar]
- 61.Nylander JAA. Uppsala: Department of Systematic Zoology. Uppsala University; 2002. MrModeltest v1.0b. [Google Scholar]
- 62.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 63.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 64.Magallón SA, Sanderson MJ. Angiosperm divergence times: The effect of genes, codon positions, and time constraints. Evolution. 2005;59:1653–1670. doi: 10.1554/04-565.1. [DOI] [PubMed] [Google Scholar]
- 65.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 66.Naud G, Suc JP. Contribution à l'étude paléofloristique des Coirons (Ardèche). Première analyses polliniques dans les alluvions sous-basaltique et interbasaltiques de Mirabel (Miocène supérieur). Bull Soc Géol France. 1975;17:820–827. [Google Scholar]
- 67.Menke B. Pliozäne und ältestquartäre Sporen- und pollenflora von Schleswig-Holstein. Geologisches Jahrbuch Reihe A. 1976;32:3–197. [Google Scholar]
- 68.Kendall DG. Stochastic processes and population growth. J Roy Stat Soc B. 1949;11:230–264. [Google Scholar]
- 69.Moran PA. Estimation methods for evolutive processes. J Roy Stat Soc B. 2000;13:141–146. [Google Scholar]
- 70.Maddison WP, Maddison DR. Sunderland, MA: Sinauer Associates; 1992. MacClade: Analysis of Phylogeny and Character Evolution, version 3.01. [Google Scholar]
- 71.Pagel M, Meade A. BayesTraits.Version 1.0 2007 [Google Scholar]
- 72.Guzmán B. Madrid: Universidad Complutense. PhD Thesis; 2008. Variabilidad morfológica y nucleotídica en el género Cistus: análisis macro- y microevolutivos. [Google Scholar]
- 73.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 74.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grosser W. Cistaceae. In: Engler A, editor. Das Pflanzenreich. Berlin: Breitkopf & Härtel; 1903. p. 161. [Google Scholar]
- 76.Martín Bolaños M, Guinea E. 1949. Jarales y jaras (cistografia hispánica). Madrid: Ministerio de Agricultura. Direccion General de Montes, Caza y Pesca Fluvial. Instituto Forestal de Investigaciones y Experiencias.
- 77.Dansereau P. Notes sur les Cistes. III: Les conditions de la distribution géographique. C R Seances Soc Biogeogr. 1958;304:22–25. [Google Scholar]
- 78.Warburg EF. Cistus L. In: Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, editors. Flora Europaea. Cambridge: Cambridge University Press; 1968. pp. 282–285. [Google Scholar]
- 79.Demoly JP, Montserrat P. Cistus. In: Castroviejo S, Aedo C, Cirujano S, Laínz M, Montserrat P, et al., editors. Flora iberica. Madrid: Consejo de Investigaciones Científicas; 1993. pp. 319–337. [Google Scholar]
- 80.Greuter W. Proposal to conserve the name Cistus albanicus (Cistaceae). Taxon. 1996;45:715–716. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
GenBank accession numbers.
(0.10 MB DOC)
Table S2
Bayesian inference of trait evolution of successive iterations of the chain (9,000,000) in the white-flowered_Cistus_ lineage by reversible jump Markov chain Monte Carlo. Means±confidence intervals (95%) of the log-likelihoods (Lh) and rate coefficients are shown.
(0.07 MB DOC)
Table S3
List of haplotypes found in 16 species and subspecies of the white-flowered Cistus lineage. Variable sites of the sequences of four plastid DNA regions (_trnL_-trnF,rbcL,_trnK_-matK,_trnS_-trnG) are shown. Nucleotide position for each data set is numbered from the 5′ to the 3′ DNA ends.
(0.20 MB DOC)