High-Density Association Study of 383 Candidate Genes for Volumetric BMD at the Femoral Neck and Lumbar Spine Among Older Men (original) (raw)
Abstract
Genetics is a well-established but poorly understood determinant of BMD. Whereas some genetic variants may influence BMD throughout the body, others may be skeletal site specific. We initially screened for associations between 4608 tagging and potentially functional single nucleotide polymorphisms (SNPs) in 383 candidate genes and femoral neck and lumbar spine volumetric BMD (vBMD) measured from QCT scans among 862 community-dwelling white men ≥65 yr of age in the Osteoporotic Fractures in Men Study (MrOS). The most promising SNP associations (p < 0.01) were validated by genotyping an additional 1156 white men from MrOS. This analysis identified 8 SNPs in 6 genes (APC, DMP1, FGFR2, FLT1, HOXA, and PTN) that were associated with femoral neck vBMD and 13 SNPs in 7 genes (APC, BMPR1B, FOXC2, HOXA, IGFBP2, NFATC1, and SOST) that were associated with lumbar spine vBMD in both genotyping samples (p < 0.05). Although most associations were specific to one skeletal site, SNPs in the APC and HOXA gene regions were associated with both femoral neck and lumbar spine BMD. This analysis identifies several novel and robust genetic associations for volumetric BMD, and these findings in combination with other data suggest the presence of genetic loci for volumetric BMD that are at least to some extent skeletal-site specific.
Key words: osteoporosis, genetics, BMD, men, QCT
INTRODUCTION
Osteoporosis, a condition marked by low BMD and an increased risk of fracture, is a significant health burden in older individuals.(1) Hip and vertebral fractures are major osteoporotic fractures and occur in ∼300,000 and 750,000 individuals, respectively, each year.(2,3) Although osteoporosis is more common in women, it is also a substantial problem among older men. In contrast to our understanding of osteoporosis in women, considerably less is known about the etiology and prevention of osteoporosis in men.
Genetic factors have an established influence on BMD, with heritability estimates indicating that as much as 85% of the population variance in BMD is caused by genetic variants(4) and that some of these variants may be skeletal site and sex specific.(5) Identification of individual genetic variants by candidate gene association studies has had some success, but many studies have been limited because they characterized a single candidate gene at a time, did not comprehensively investigate genetic variation for the candidate gene of interest, and did not validate findings in an independent sample. Further complicating the search for genetic factors contributing to BMD, many studies have had small sample sizes and/or had potential design flaws, such as failing to account for population stratification. Recently, genome-wide association studies have been completed to identify genetic variants that influence DXA measures of areal BMD.(6,7) Although these genome-wide studies have identified several genetic loci of interest, the loci identified explain very little of the variation in BMD.(8) Thus, much of the genetic variation in BMD remains to be explained. Furthermore, the majority of genetic studies have measured areal BMD by DXA, which is confounded by bone size and may be increased erroneously at the lumbar spine by spinal degenerative disease and aortic calcification.(9–13) Volumetric BMD (vBMD) measured by QCT avoids some of the limitations of DXA, but to date, little is known about the genetic determinants of vBMD.
In this study, we assessed and subsequently validated the associations between common genetic variation in 383 biological candidate genes and vBMD of the femoral neck and lumbar spine among 2018 older white men in the Osteoporotic Fractures in Men Study (MrOS).
MATERIALS AND METHODS
Study participants
Participants for this study were selected from the MrOS. MrOS is a prospective, cohort study designed to investigate anthropometric, lifestyle, and medical factors related to bone health in older, community-dwelling men. At study entry, participants were at least 65 yr old, community dwelling, ambulatory, and had not had bilateral hip replacement.(14) In total, 5995 men were recruited from March 2000 through April 2002 primarily using population-based mailings in six geographic regions in the United States: Birmingham, AL; Minneapolis, MN; Palo Alto, CA; Pittsburgh, PA; Portland, OR; and San Diego, CA.(15)
White participants with volumetric BMD (vBMD) were selected for genotyping in this study if they had not reported taking bone-altering medications such as androgens, anti-androgens, or oral corticosteroids and had not reported being on osteoporosis treatment. Medication use was defined as medications taken daily or almost daily for the last 30 days. Prescription medications recorded by the clinics were stored in an electronic medications inventory database, and each medication was matched to its ingredient(s) based on the Iowa Drug Information System (IDIS) Drug vocabulary.
Genotyping was completed in two phases using two independent samples from the MrOS cohort: a discovery sample and a validation sample. Specifically, the discovery sample was comprised of 862 white men with lumbar spine or femoral neck vBMD measures. These men were selected without regard to their BMD level from the Minneapolis and Pittsburgh clinic sites for genotyping. Promising SNPs identified in the discovery sample were tested for replication in 1156 additional men with vBMD measures in a validation sample that was comprised of men from the remainder of the MrOS clinic sites (Birmingham, Palo Alto, Portland, and San Diego).
vBMD
vBMD was measured using QCT of the hip and lumbar spine. Because of cost restraints, only the first 65% of the MrOS cohort and all nonwhite participants were referred for QCT scans. There were few differences between the men who did and did not receive QCT scans except that those with QCT scans were slightly younger and more likely to be from a minority population.(16)
QCT measurement of the lumbar spine was obtained using an anatomical region 5 mm above the L1 superior endplate to 5 mm below the L2 inferior endplate and hip scans were obtained in the anatomical region defined by the femoral head to 3.5 cm below the lesser trochanter. Lumbar spine images were acquired using settings of 120 kVp, 150 mA, 1-mm slice thickness, and 512 × 512 matrices. The region of interest was the second lumbar vertebra excluding the transverse processes. Hip images were acquired at settings of 80 kVp, 280 mA, 3-mm slice thickness, and 512 × 512 matrix in spiral reconstruction mode. The femoral neck region was defined as the region from the minimum cross-sectional area to the point 25% toward the maximum cross-sectional area where cross-sectional area was measured along the neutral axis. Different scanners were used at different clinic sites. Specifically, the images were acquired on a GE Prospeed at the Birmingham clinic, a GE Hispeed Advantage at the Minneapolis clinic, a Philips MX-8000 at the Palo Alto clinic, a Siemans Somatom +4 at the Pittsburgh clinic, either a Phillips CT-Twin or Toshiba Acquilion at the Portland site, and a Picker PQ-5000 in San Diego.
QCT images were processed (by T.F.L.) at the University of California at San Francisco using a standardized protocol. Each participant scan included a calibration standard of three hydroxyapatite concentrations (150, 75, and 0 mg/cm3), and these were used to convert between Hounsfield units and vBMD. Differences between the clinic sites exist and are statistically adjusted for in all analyses.
Other baseline characteristics
Participant characteristics including age, health history, and medication use were obtained by a self-administered questionnaire. Physical characteristics were obtained by clinic staff. Height was measured by Harpenden stadiometer, and weight was measured by balance beam scale except at the Portland clinic, which used a digital scale.
Candidate gene and single nucleotide polymorphism selection
Physiologically defined candidate genes were identified from publicly available resources. Specifically, literature searches were conducted using PubMed, evidence of gene expression in normal human trabecular bone cells was obtained from the Skeletal Gene Database (sgd.nia.nih.gov, no longer available) and the NCBI UniGene database (www.ncbi.nlm.nih.gov/sites/entrez?db=unigene), genes with functions of interest (e.g., “regulation of bone mineralization” or “skeletal development”) were obtained from Entrez Gene (www.ncbi.nlm.nih.gov/sites/entrez?db=gene) and Amigo (amigo.geneontology.org/cgi-bin/amigo/go.cgi), genes with a skeletal phenotype in mice were identified from the Jackson Laboratory Mouse Genome Informatics database (http://www.informatics.jax.org/), and the Online Mendelian Inheritance in Man database (www.ncbi.nlm.nih.gov/omim/) was queried for evidence of genes implicated in skeletal conditions in humans. In total, 383 candidate genes were identified for genotyping (Table 1).
Table 1.
Candidate Genes Screened for Association With BMD in the Discovery Sample
| Chr1 | DVL1, TNFRSF1B, PAX7, ALPL, WNT4, ID3, CSF3R, LEPR, TGFBR3, DNTTIP2, CSF1, HSD3B2, HSD3B1, NOTCH2, GNRHR2, CTSK, IL6R, ZBTB7B, BGLAP, MEF2D, NTRK1, RXRG, ADIPOR1, MYOG, HSD11B1, TGFB2, WNT9A, WNT3A |
|---|---|
| Chr2 | ID2, NCOA1, POMC, LTBP1, CYP1B1, LHCGR, PPP3R1, IL1R2, IL1R1, IL1A, IL1B, IL1RN, EN1, GLI2, LCT, NR4A2, TANK, DLX1, DLX2, ATF2, HOXD13, HOXD12, HOXD11, HOXD10, HOXD9, HOXD8, HOXD4, HOXD3, HOXD1, FRZB, MSTN, STAT1, CASP8, FZD7, BMPR2, FZD5, IGFBP2, IGFBP5, WNT10A, IHH, PAX3, IRS1, TWIST2 |
| Chr3 | IRAK2, GHRL, PPARG, WNT7A, THRB, TGFBR2, MYD88, ACVR2B, CTNNB1, PTHR1, WNT5A, POU1F1, GSK3B, CASR, GATA2, TRH, SHOX2, GHSR, TNFSF10, CHRD, AHSG, ADIPOQ, OSTN, HES1 |
| Chr4 | FGFR3, MSX1, NKX3–2, PPARGC1A, KDR, GNRHR, GC, BMP2K, BMP3, DMP1, IBSP, MEPE, SPP1, BMPR1B, NFKB1, DKK2, LEF1, EGF, FGF2, SMAD1, SFRP2, CASP3 |
| Chr5 | LIFR, PTGER4, GHR, FST, IL6ST, MAP3K1, CRHBP, MEF2C, APC, HSD17B4, CSF2, PDLIM4, TCF7, HDAC3, FGF1, NR3C1, CSF1R, SPARC, FGF18, MSX2, PROP1 |
| Chr6 | RIPK1, SOX4, TNF, CYP21A2, RXRB, PPARD, MAPK14, CDKN1A, VEGFA, RUNX2, OSTM1, WISP3, ENPP1, CTGF, TNFAIP3, ESR1, IGF2R |
| Chr7 | TWIST1, IL6, GPNMB, HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, HOXA10, HOXA11, HOXA13, CRHR2, GHRHR, SFRP4, GLI3, IGFBP1, IGFBP3, EGFR, FZD1, CDK6, CALCR, DLX6, DLX5, CYP3A4, LEP, SMO, NRF1, PTN, TRPV5, CASP2, SHH |
| Chr8 | EGR3, TNFRSF10A, GNRH1, STAR, FGFR1, SFRP1, IKBKB, DKK4, CRH, NCOA2, HEY1, KLF10, FZD6, EXT1, TNFRSF11B, FBXO32, WISP1, CYP11B1 |
| Chr9 | CER1, IFNB1, CNTFR, OSTF1, NTRK2, ROR2, OGN, PTCH1, HSD17B3, TGFBR1, TLR4, TRAF1, WDR5, RXRA, NOTCH1, TRAF2 |
| Chr10 | DKK1, EGR2, BMPR1A, CHUK, CYP17A1, FGFR2 |
| Chr11 | IGF2, CDKN1C, DKK3, PTH, CALCA, SOX6, MYOD1, BDNF, TRAF6, EXT2, CNTF, ESRRA, LTBP3, FOSL1, TCIRG1, LRP5, CCND1, FGF3, FADD, CHRDL2, ARRB1, WNT11, FZD4 |
| Chr12 | WNT5B, ADIPOR2, FGF23, NTF3, TNFRSF1A, LRP6, MGP, SOX5, PTHLH, VDR, WNT10B, WNT1, IGFBP6, SP7, CYP27B1, WIF1, IRAK3, MYF6, MYF5, DCN, IGF1, TBX3, HNF1A, P2RX7 |
| Chr13 | FLT1, KL, POSTN, TNFSF11 |
| Chr14 | NFKBIA, PAX9, BMP4, ESR2, LTBP2, FOS, TGFB3, TSHR, DLK1, AKT1 |
| Chr15 | GREM1, CYP19A1, MAP2K1, SMAD3, CYP11A1, CYP1A1, CYP1A2, NTRK3, IGF1R, MEF2A |
| Chr16 | AXIN1, CLCN7, IGFALS, MAPK3, RBL2, CBFB, TRADD, HSD17B2, FOXC2 |
| Chr17 | ALOX15, ARRB2, DVL2, SHBG, PIK3R5, CSF3, THRA, IGFBP4, HSD17B1, SOST, MAP3K14, CRHR1, PHOSPHO1, DLX3, COL1A1, TOB1, NOG, TBX2, TBX4, GH1, SOX9 |
| Chr18 | MC2R, SMAD2, SMAD4, TCF4, TNFRSF11A, BCL2, NFATC1 |
| Chr19 | KISS1R, MAP2K2, MEF2B, CEBPA, NFKBIB, DLL3, AKT2, TGFB1, FOSB, BAX, LHB, OSCAR |
| Chr20 | GNRH2, BMP2, JAG1, PAX1, ID1, E2F1, GDF5, RBL1, GHRH, SRC, WISP2, MMP9, NCOA3, CEBPB, CYP24A1, BMP7 |
| Chr21 | IFNAR2, IFNAR1, RUNX1, ETS2 |
| Chr22 | COMT, KREMEN1, LIF, CSF2RB, ATF4, MCHR1, PPARA |
| X Chr | STS, RPS6KA3, GATA1, AR, BGN, IRAK1, IKBKG |
Publicly available databases were interrogated for single nucleotide polymorphism (SNP) variation in the region surrounding the candidate gene. For the first phase of genotyping (the discovery sample), two SNP selection strategies were used. In the first strategy, genetic variation in the region spanning 30 kb upstream and 10 kb downstream of each candidate gene was captured by creating a reference SNP panel of variants with a minor allele frequency (MAF) of at least 5% in phase I of the International HapMap Project (www.hapmap.org).(17) Tag SNPs were selected using a pairwise correlation method (_r_2 ≥ 0.80).(18) Candidate genes that were clustered near each other on the chromosome were tagged as a unit spanning all loci of interest. For example, IGFBP2 and IGFBP5 are located only 7.6 kb from each other on chromosome 2. Because the region of interest for these two candidates overlapped, they were tagged as a unit. In the second strategy, potentially functional SNPs that were either nonsynonymous coding variants, predicted to alter a putative transcription factor binding site in the promoter region, or a putative exon splice enhancer with MAF ≥1% were selected for genotyping using the PupaSNP (pupasuite.bioinfo.cipf.es/) and Promolign (polly.wustl.edu/promolign/main.html) databases.(19,20)
Genotyping for the second phase of the project was conducted in a validation sample for promising SNP associations identified from the discovery sample. Specifically, SNPs with p ≤ 0.015 for either femoral neck or lumbar spine vBMD in the discovery sample were genotyped in the validation sample. Additionally, SNPs with p ≤ 0.05 in a gene that also had a SNP with p ≤ 0.015 in the discovery sample were included in the second phase of genotyping.
Genotyping
Genomic DNA from frozen whole blood specimens was extracted using the Flexigene protocol (Qiagen, Valencia, CA, USA). Genotyping was completed using the Illumina Golden Gate custom assay. For the discovery sample, 37 participant samples were run as blind duplicates, and 4 internal controls were included per plate to ensure reproducibility. We observed 100% reproducibility among the internal controls and 99.9% reproducibility among replicate participant samples. In the validation sample, 26 participant samples and 4 internal controls per plate were included for quality control. We observed 99.9% reproducibility among internal controls and among duplicate participant samples. To ensure maximum genotyping completeness in the validation sample, loci of interest that could not be genotyped using the Illumina Golden Gate assay were genotyped using one of two platforms: the TaqMan allelic discrimination assay system (Applied Biosystems, Foster City, CA, USA) on a 7900HT Real-time PCR instrument with probes and reagents purchased from Applied Biosystems or Sequenom MassARRAY iPLEX Gold technology (Sequenom, San Diego, CA, USA) with PCR primers purchased from Invitrogen (Carlsbad, CA, USA). A subset of participant samples were run in duplicate for these platforms, and an average reproducibility of 99.8% and 99.9% was observed for TaqMan and Sequenom, respectively.
Loci with a minor allele frequency <1% in the genotyping sample (N = 129) that did not conform to the expectations of Hardy-Weinberg equilibrium (p < 0.0005; N = 123) or that had a low call rate (<85%; N = 248) were excluded from statistical analysis. Individual samples with a low call rate (<85%; N = 14) or that were highly correlated with another sample (indicating relatedness; n = 13) were excluded from the analysis.
Statistical analysis
Although only white individuals were investigated in this study, population stratification is a potential concern in large-scale genomic analyses.(21) Stratification was initially assessed using the program Structure. Structure is a model-based clustering program that parses the participants into subpopulations and assesses whether there are distinct populations or admixed individuals.(22) We found little evidence of population stratification in the discovery, validation, or pooled samples. Nevertheless, we accounted for potential fine scale population substructure by using a principal components method of analysis using uncorrelated SNPs (r < 0.2) to calculate the principal components.(23)
Analyses of association between genotype and vBMD assumed both an additive and recessive model of inheritance. Linear regression was used to test for an additive association between the number of copies of the minor allele and vBMD. For the recessive model, regression methods were implemented to determine whether individuals having two copies of the minor allele differed from those with the other two genotypes. SNPs with 10 or less individuals having the rare genotype were not tested for the recessive model to minimize spurious findings based on small genotype specific sample sizes. All analyses adjusted for participant age and clinic site in addition to the first principal component of the population substructure analysis. SNPs with associations (p < 0.05) in both the discovery and validation sample and that also had the same direction of association (regression coefficient + or − for both genotyping samples) were considered to be replicated. Therefore, although we did not use a strict Bonferoni p value to adjust for multiple testing in the discovery sample, we conservatively required that the same SNP had an equivalent, significant (p < 0.05) effect in the validation sample to be considered a replication.
Replicated SNP associations were examined further in the pooled sample of 2018 individuals from the discovery and validation samples. In addition to the analyses described above, further adjustment for height and weight was conducted in the pooled sample to determine whether body size attenuated the relationship between genotype and vBMD. Linear regression analysis was used to determine the amount of phenotypic variation explained by all of the significant (replicated) SNPs. Because SNPs in the same gene region are often correlated, the colinearity of individual SNPs in the model was assessed. One pair of SNPs in the femoral neck analysis and three in the lumbar spine analysis were highly correlated, and, in those instances, the SNP with the most missing genotypes was dropped from the regression modeling of all significant SNPs.
RESULTS
The average age of men in the pooled analysis was 74 yr (range, 65–100 yr). The men in the discovery and validation samples were similar in age. Men in the validation sample had lower body weight, lower BMI, taller stature, and slightly lower lumbar spine and femoral neck vBMD (p < 0.001 for all; Table 2).
Table 2.
Participant Characteristics (Mean and SD)
| Discovery sample (N = 862) | Validation sample (N = 1156) | Pooled sample (discovery + validation) (N = 2018) | |
|---|---|---|---|
| Age (yr) | 74 (5.8) | 74 (6.0) | 74 (5.9) |
| Weight (kg) | 85.3 (14.1) | 82.9 (12.5)* | 83.9 (13.3) |
| Height (cm) | 173.6 (6.7) | 174.9 (6.7)* | 174.3 (6.7) |
| BMI | 28.3 (4.1) | 27.1 (3.6)* | 27.6 (3.9) |
| Lumbar spine vBMD (g/ml) | 0.243 (0.042) | 0.226 (0.038)* | 0.234 (0.040) |
| Femoral neck vBMD (g/ml) | 0.305 (0.056) | 0.275 (0.052)* | 0.287 (0.056) |
In the discovery sample, 4108 of 4608 attempted SNPs passed quality control criteria and were analyzed for their association with lumbar spine and femoral neck vBMD. The mean SNP density for candidate genes was one SNP per 13.2 kb (range, 1 SNP/3.2 kb to 1 SNP/97 kb). Tag SNPs were selected based on phase I of the International HapMap Project. Nevertheless, the SNPs included in our analysis tagged on average 64% (range per gene, 1–100%) of the SNPs with an MAF >5% in phase II of the International HapMap project. Of the SNPs captured by our tag SNP set, the mean max _r_2 was 0.97.
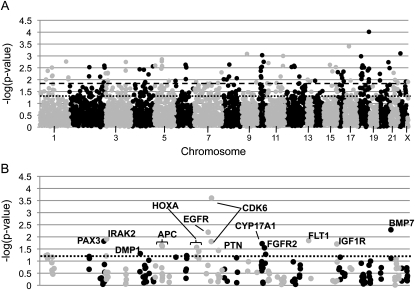
One hundred ninety-three SNPs in 56 genes were associated with femoral neck vBMD and 173 SNPs in 59 genes were associated with lumbar spine vBMD in the discovery sample and were subsequently genotyped in the validation sample (Figs. 1A and 2A). For the femoral neck, there were several SNPs that had significant p values in both the discovery and validation samples, but the direction of association was not the same in the two samples. Specifically, this occurred for one SNP in PAX3 (rs1367408), IRAK2 (rs779905), EGFR (rs2075109), CYP17A1 (rs12219246), IGF1R (rs3784606), and BMP7 (rs6127983) and for two SNPs in CDK6 (rs2374589 and rs3802073) (Fig. 1B). Eight SNPs in six genes (DMP1, APC, HOXA, PTN, FGFR2, and FLT1) were associated with femoral neck vBMD in both the discovery and validation sample, and the association was in the same direction (Table 3). The strongest SNP association with femoral neck vBMD in the pooled analysis was with rs4705573 in APC (p = 0.001). Men who were homozygous for the minor allele (GG genotype) of this SNP had a 3.4% lower vBMD then men homozygous for the major allele (AA genotype). Although rs1381632 in DMP1 was significant in both the discovery and validation sample, different genetic models were significant, and consequently, the pooled analysis was not significant (p = 0.0961). Additional adjustment for weight and height did not attenuate any of the SNP associations for femoral neck vBMD (Table 3). Each SNP only explained a small amount of the variation in femoral neck vBMD in the pooled sample (0.1–0.5%; Table 3). The two SNPs in APC (rs6594646 and rs4705573) were in linkage disequilibrium (LD; _r_2 = 0.979 and D′ = 0.991), and rs6594646 was consequently dropped from the regression modeling. The seven replicated SNPs in APC, DMP1, FGFR2, FLT1, HOXA, and PTN included in the regression modeling explained 1.7% of the variation in femoral neck vBMD after accounting for age, clinic, population substructure, height, and weight.
FIG. 1.
SNP association results for femoral neck vBMD. Association results for the femoral neck are presented for the first phase of genotyping (discovery sample) in A and in the validation sample in B. Specifically, the –log of the p value observed is presented on the _y_-axis. The most significant result of the two models tested (either additive or recessive) is presented for each SNP. The SNPs are ordered across the _x_-axis by chromosome and the base pair position on the chromosome. Odd numbered chromosomes and the X chromosome are presented in light gray. Even numbered chromosomes are presented in dark gray. In A, the dark dashed line represents p = 0.015 and the dotted line represents p = 0.05. The dotted line in B represents p = 0.05, and SNPs with p ≤ 0.05 are labeled with the gene symbol that they lie in.
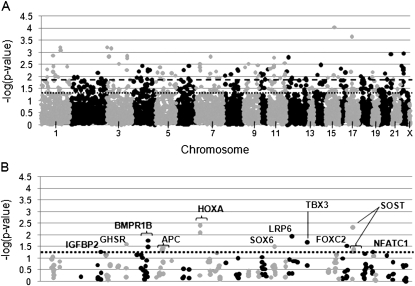
FIG. 2.
SNP association results for lumbar spine vBMD. Association results for the lumbar spine are presented for the discovery (A) and validation sample (B). Specifically, the –log of the p value observed is presented on the _y_-axis, and SNPs are ordered across the _x_-axis by chromosome and base pair position. The most significant result of the two models tested (either additive or recessive) is presented for each SNP. Odd numbered chromosomes and the X chromosome are presented in light gray. Even numbered chromosomes are presented in dark gray. In A, the dark dashed line represents p = 0.015 and the dotted line represents p = 0.05. The dotted line in B represents p = 0.05, and SNPs with p ≤ 0.05 are labeled with the gene symbol that they lie in.
Table 3.
Replicated Associations for Femoral Neck vBMD
| Gene | SNP | Allele | Location | Frequency* | Discovery sample (adjustment 1)† | Validation sample (adjustment 1)† | Pooled sample | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adjustment 1† | Adjustment 2‡ | ||||||||||||||||
| β | p | β | p | β | p | β | p | _r_2§ | |||||||||
| APC | rs459552 | T→A | Exon (Val->Asp) | 0.228 | 0.007 | 0.044 | add | 0.014 | 0.024 | rec | 0.005 | 0.011 | add | 0.005 | 0.014 | add | 0.003 |
| APC | rs4705573 | A→G | Upstream | 0.480 | −0.008 | 0.004 | add | −0.008 | 0.022 | rec | −0.009 | 0.001 | rec | −0.009 | 0.001 | rec | 0.005 |
| APC | rs6594646 | A→G | Intron | 0.497 | −0.006 | 0.021 | add | −0.008 | 0.023 | rec | −0.008 | 0.003 | rec | −0.008 | 0.005 | rec | 0.004 |
| DMP1 | rs1381632 | A→T | Upstream | 0.228 | 0.007 | 0.037 | add | 0.014 | 0.049 | rec | 0.003 | 0.096 | add | 0.008 | 0.122 | rec | 0.001 |
| FLT1 | rs1408245 | C→G | Intron | 0.166 | −0.032 | 0.010 | rec | −0.007 | 0.014 | add | −0.021 | 0.003 | rec | −0.020 | 0.004 | rec | 0.004 |
| FGFR2 | rs7916940 | G→A | Downstream | 0.299 | −0.013 | 0.050 | rec | −0.005 | 0.052 | add | −0.004 | 0.019 | add | −0.004 | 0.018 | add | 0.003 |
| HOXA¶ | rs6951180 | A→G | Upstream of HOXA13 | 0.133 | 0.010 | 0.018 | add | 0.024 | 0.039 | rec | 0.027 | 0.008 | rec | 0.024 | 0.015 | rec | 0.003 |
| PTN | rs322297 | A→C | Intron | 0.116 | 0.010 | 0.014 | add | 0.007 | 0.037 | add | 0.008 | 0.002 | add | 0.008 | 0.003 | add | 0.004 |
Four SNPs in four genes (GHSR, rs558572; SOX6, rs1354329; LRP6, rs4477532; TBX3, rs6489968) had significant associations with lumbar spine vBMD in both the discovery and validation sample, but the direction of the association was in different directions (Fig. 2B). For the lumbar spine, there were 13 SNPs in seven genes (APC, BMPR1B, FOXC2, HOXA, IGFBP2, NFATC1, and SOST), which had a consistent direction of association in both genotyping samples (Table 4). Each individual SNP explained 0.03–0.89% of the variation in lumbar spine vBMD in the pooled sample. The SNP explaining the most variation in lumbar spine vBMD was rs1877632 in the SOST gene region, which explained 0.89% of the phenotypic variation in vBMD. Men with the less common AA genotype for rs1877632 had 6% higher lumbar spine vBMD than men with the more common GG and GA genotypes. As in the femoral neck analysis, rs6594646 and rs4705573 were in LD and rs6594646 was consequently dropped from the regression modeling of significant SNPs for lumbar spine vBMD. In addition, two SNPs in HOXA (rs6951180 and rs6964896; _r_2 = 0.939 and D′ = 1.000) and two SNPs in BMPR1B (rs3796443 and rs1434536; _r_2 = 1.000 and D′=0.992) were in high LD, and thus rs6964896 and rs1434536 were dropped from the regression modeling. Collectively, the 10 replicated SNP associations in APC, BMPR1B, FOXC2, HOXA, IGFBP2, NFATC1, and SOST explained 3.5% of the variation in lumbar spine vBMD after accounting for age, clinic, population substructure, height, and weight.
Table 4.
Replicated Findings for Lumbar Spine vBMD
| Gene | SNP | Allele | Location | Frequency* | Discovery sample (adjustment 1)† | Validation sample (adjustment 1)† | Pooled sample | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adjustment 1† | Adjustment 2‡ | ||||||||||||||||
| β | p | β | p | β | p | β | p | _r_2§ | |||||||||
| APC | rs4705573 | A→G | Upstream | 0.477 | −0.005 | 0.007 | add | −0.005 | 0.049 | rec | −0.006 | 0.005 | rec | −0.005 | 0.007 | rec | 0.004 |
| APC | rs6594646 | A→G | Intron | 0.494 | −0.004 | 0.035 | add | −0.005 | 0.044 | rec | −0.005 | 0.015 | rec | −0.004 | 0.023 | rec | 0.003 |
| BMPR1B | rs1434536 | A→G | 3′ UTR | 0.443 | 0.005 | 0.006 | add | 0.006 | 0.032 | rec | 0.007 | 0.001 | rec | 0.007 | 0.001 | rec | 0.005 |
| BMPR1B | rs3796443 | A→G | Intron | 0.439 | 0.006 | 0.005 | add | 0.007 | 0.017 | rec | 0.008 | 3.2 × 10−4 | rec | 0.008 | 3.0 × 10−4 | rec | 0.006 |
| FOXC2 | rs3751797 | T→A | Upstream | 0.260 | −0.016 | 0.007 | rec | −0.004 | 0.029 | add | −0.004 | 0.005 | add | −0.004 | 0.003 | add | 0.004 |
| HOXA¶ | rs6951180 | A→G | Upstream of HOXA13 | 0.134 | 0.009 | 0.003 | add | 0.025 | 0.004 | rec | 0.008 | 3.7 × 10−5 | add | 0.007 | 2.5 × 10−4 | add | 0.007 |
| HOXA¶ | rs6964896 | C→A | Upstream of HOXA13 | 0.127 | 0.008 | 0.006 | add | 0.006 | 0.008 | add | 0.007 | 8.1 × 10−5 | add | 0.006 | 0.001 | add | 0.006 |
| IGFBP2 | rs10932669 | C→A | Upstream | 0.134 | 0.007 | 0.026 | add | 0.004 | 0.053 | add | 0.005 | 0.002 | add | 0.006 | 0.002 | add | 0.005 |
| NFATC1 | rs177820 | A→G | Intron | 0.365 | −0.005 | 0.027 | add | −0.006 | 0.052 | rec | −0.008 | 0.003 | rec | −0.007 | 0.008 | rec | 0.004 |
| SOST | rs1534401 | A→G | Downstream | 0.380 | −0.004 | 0.030 | add | −0.006 | 0.050 | rec | −0.006 | 0.012 | rec | −0.003 | 0.012 | add | 0.003 |
| SOST | rs1877632 | G→A | Downstream | 0.313 | 0.015 | 0.001 | rec | 0.011 | 0.005 | rec | 0.013 | 1.5 × 10−5 | rec | 0.006 | 1.4 × 10−5 | add | 0.009 |
| SOST | rs851054 | A→G | Upstream | 0.381 | −0.004 | 0.054 | add | −0.006 | 0.046 | rec | −0.006 | 0.008 | rec | −0.006 | 0.008 | rec | 0.004 |
| SOST | rs851056 | C→G | Upstream | 0.380 | −0.004 | 0.054 | add | −0.006 | 0.043 | rec | −0.006 | 0.008 | rec | −0.006 | 0.008 | rec | 0.004 |
DISCUSSION
Although highly heritable, little is known about the genetic variants contributing to BMD in men and vBMD in general.(24,25) This study used a two-staged genotyping strategy to investigate the association between SNPs in 383 biologically defined candidate genes and vBMD at the lumbar spine and femoral neck in a large sample of older men. We identified associations between SNPs in 11 genes and vBMD that were validated in a separate sample of older men. To our knowledge, the associations with SNPs in APC, BMPR1B, DMP1, FLT1, HOXA, IGFBP2, NFATC1, and PTN have not yet been described. We also confirmed previously identified associations in the sclerostin (SOST) and forkhead box C2 (FOXC2) genes.(26–30) Importantly, although two of the gene associations were shared between the femoral neck and lumbar spine, most were distinct for one skeletal site. These observations underscore the importance of measuring BMD at multiple skeletal sites in studies aimed at identifying osteoporosis susceptibility genes.
Although most of the candidate gene associations identified were specific to either the femoral neck or lumbar spine, SNPs in the gene encoding APC were associated with vBMD at both sites. The minor alleles of rs4705573 in the 5′ flanking region and rs6594646 in intron 1 of APC were associated with lower vBMD at both the femoral neck and lumbar spine. These SNPs are not known or predicted to influence APC function or expression and may be in LD with the causal variation. In contrast, a third SNP rs459552, which is associated only with femoral neck vBMD, is a nonsynonymous coding variant. This variant is located in the β-catenin downregulation domain of APC and may be potentially functional.(31) More commonly known for its role in cancer biology, APC targets β-catenin for degradation in the WNT signaling pathway, an important pathway in bone metabolism.(32) APC is expressed in osteoblasts and osteoclasts from adult human bone(33) and mice with osteoblast-specific APC deletions have increased bone deposition.(34)
Two SNPS upstream of the HOXA gene cluster in the 5′ flanking region of HOXA13 were also associated with femoral neck and lumbar spine vBMD. The HOXA genes are involved in the normal development of the axial skeleton and limbs.(35) The role of HOXA genes in determining BMD is less well characterized, but HOXA10 contributes to osteoblastogenesis by regulating target genes for osteoblast differentiation and bone formation including RUNX2, alkaline phosphatase, bone sialoprotein, and osteocalcin.(36) Activation of HOXA genes has also been described during fracture repair, and a re-examination of HOXA gene function in adult bone and BMD regulation may be warranted.(37) The HOXA genes are sufficiently close to each other and oriented in such a way to enable enhancer sharing, and it is possible that SNPs in the 5′ region of the gene cluster may directly or indirectly influence HOXA gene expression.
Four genes were uniquely associated with femoral neck vBMD. DMP1 encodes dentin matrix protein 1, an extracellular matrix protein that regulates osteoblast gene expression and mineralization of bone matrix.(38) _DMP1_-null mice have impaired bone mineralization, mutations in DMP1 are known to cause autosomal recessive hypophosphatemia, and there is evidence for a BMD quantitative trait locus (QTL) in mice (Bmd2) that encompasses the DMP1 gene.(39–43) FGFR2 encodes a transmembrane receptor for fibroblast growth factor that is involved in bone growth and development. Activating and dominant negative mutations in FGFR2 are associated with altered bone mineralization and familial craniosynostosis syndromes.(44,45) FLT1 encodes the cell surface receptor for vascular endothelial growth factor that is involved in osteoclastogenesis and osteoblast differentiation.(46–50) _FLT1_-null mice have decreased bone mineral apposition and lower trabecular bone volume.(51) Pleiotrophin (PTN) is an extracellular matrix protein released by osteoblasts that recruits, promotes adhesion of, and increases proliferation of osteoprogenitor cells.(52,53) PTN transgenic mice have greater bone calcium content compared with controls.(54)
Five gene associations were specific to vBMD at the lumbar spine. Bone morphogenetic protein receptor, type IB (BMPR1B) encodes a receptor for BMPs, which are involved in osteoblast commitment and differentiation.(55) Transgenic mice with a truncated form of BMPR1B display reduced bone formation rates and BMD.(55,56) FOXC2 encodes forkhead box C2, a member of the forkhead/winged helix transcription factor family that serves as a key regulator of embryogenesis. Mutant mice null for FOXC2 show defects in axial skeletogenesis.(57) Association between a promoter SNP in FOXC2 and vBMD of the radius was reported in a study of Japanese men and women.(26) IGFBP2, which encodes an insulin-like growth factor binding protein, is thought to target IGFs to bone and is associated with long bone growth; overexpression of this gene product results in shorter bones.(58,59) Furthermore, male Igfbp2 knockout mice have reduced cortical and trabecular bone because of thinner trabeculae than controls.(60) Additionally, levels of circulating IGFBP2 are negatively correlated with BMD in postmenopausal women.(61) Nuclear factor of activated T-cells 1 (NFATC1) is a transcription factor induced by TNF superfamily, member 11 (RANKL) that is involved in both osteoclast and osteoblast regulation.(62–64) Mutations in SOST have been associated with sclerosteosis and Van Buchem disease, and polymorphisms in SOST have been associated with normal variation in BMD.(27–29) Both rs851054 and rs851056 are predicted to lie in the promoter region of SOST, and rs851054 is predicted to abolish a sex determining region Y (SRY) binding site, whereas rs851056 is predicted to change a transcription factor binding site from the TAL1/TCF3 complex to c-MYC or RUNX1.(20,65)
Several significant associations were observed in both the discovery and validation samples, but the direction of the association was different. The inconsistency in direction of association may indicate spurious findings. However, others have noted a similar “flip-flop” phenomenon and have described biologically plausible scenarios that may account for this discordant pattern of association.(66,67) Some of these inconsistent associations occur in previously identified candidate genes. For example, a promoter variant (rs743572) in CYP17A1 has been associated with BMD in some studies.(68–72) Although rs743572 was not directly genotyped in our study, it is in LD with the SNP identified in our study (D′ = 0.915 and _r_2 = 0.670). A nonsynonymous coding polymorphism in LRP6 (rs2302685) has been associated with areal BMD and fracture.(73,74) This polymorphism was not associated with vBMD in our study, but an intronic variant (rs4477532) was associated with lumbar spine vBMD. Although there was inconsistent evidence for association in our study, these candidate genes may warrant further examination.
We were unable to document an association with SNPs in several widely studied candidate genes (COL1A1, ESR1, LRP5, and VDR) and vBMD. There are several reasons why we may not have been able to replicate these associations. First, our study includes only men, whereas many of the past candidate gene studies have focused on women. In addition, most of these studies investigated areal and not volumetric BMD. Finally, our study had 70% statistical power to detect a SNP association that explained 1% of the variation in vBMD in the screening stage at α = 0.01. Thus, we cannot exclude the possibility of a weaker association between SNPs in the ESR1, COL1A1, VDR, or LRP5 genes and vBMD at the femoral neck or lumbar spine in older men.
Three SNPs associated with vBMD in our study were also genotyped in a genome-wide association study of areal BMD but were not significantly associated with BMD in that study (p > 0.05).(7) Although not directly genotyped in our study, other SNPs in the gene regions associated with vBMD in our study (APC, BMPR1B, DMP1, FGFR2, FOXC2, HOXA, NFATC1, and PTN) also showed associations (p < 0.05) in this genome-wide association study but did not achieve genome-wide significance. Most notably, rs11984297 located just downstream of the HOXA13 gene was associated with both hip (p = 0.0002) and spine (p = 0.0014) BMD, but this SNP is not in high LD with rs6951180 (_r_2 = 0.015) or rs6964896 (_r_2 = 0.008).(7)
The amount of phenotypic variation in vBMD explained by SNPs in this study was small: 1.7% for the femoral neck and 3.5% for the lumbar spine. Although small, these findings are comparable to recent genome-wide studies of BMD and other quantitative traits like height.(6,7,75) For example, in a recent genome-wide association study of areal BMD, 0.6% and 0.2% of the phenotypic variation in lumbar spine and femoral neck BMD was explained by two SNPs in two genes.(6) Given the high heritability of BMD, much of the variation in BMD explained by genetic factors remains to be identified. Future studies not only examining SNP associations, but also investigating insertion deletion mutations, copy number variants, rare variants (<5% MAF), and interactions between genetic factors and between genetic and environmental factors may explain more of the variation in BMD.
Our study has potential limitations. First, our reference SNP panel for tag SNP selection was based on phase I of the International Haplotype Map Project (HapMap), and consequently does not provide as comprehensive a set of tag SNPs as current projects based on phase II. However, the SNPs included in this analysis tagged an average of 64% of the SNPs with >5% MAF in phase II of HapMap with a mean maximum _r_2 of 0.97. To put this in perspective, our tag SNP set captured 82% and 95% of the SNPs with MAF >5% in the HOXA and APC gene regions, whereas a recent genome wide study of BMD examining >300,000 SNPs captured 68% and 74% for these gene regions.(7) Our analysis also focused on SNPs with a MAF of ≥5% (or ≥1% for potentially functional SNPs) and thus cannot exclude the possible contributions of less common variants. Our study is also limited to white men ≥65 yr of age, and our results may not be generalizable to other ethnic groups or to women. Our analysis of vBMD was cross-sectional, and future studies of bone loss may give further insight into the genetics of osteoporosis in men. Furthermore, a genome-wide association study, which has the advantage of being hypothesis free, may yield additional insight on the genetics of volumetric BMD. Additional genotyping in the gene regions of interest will be needed in this and in other ethnically diverse populations to refine the association signals and to inform future in vitro functional studies.
Although limited to studying only older white men, this study provided the first large-scale assessment of the genetic contributions to a unique skeletal trait (QCT volumetric BMD), assessed and adjusted for potential population stratification, identified a number of novel and robust genetic associations by including a two-stage internal replication design and collectively, and explained more of the phenotypic variance in BMD than other genetic association studies to date. We identified several novel genetic associations with vBMD, and our findings suggest that distinct genetic factors may contribute to lumbar spine and femoral neck vBMD in older men. Additional studies will be needed to confirm and extend these findings.
ACKNOWLEDGMENTS
Genotyping was supported by Grant R01-AR051124 from The National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Institute on Aging (NIA), the National Center for Research Resources (NCRR), and NIH Roadmap for Medical Research under the following grants: U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, U01-AG027810, and UL1 RR024140. L.M.Y. was supported by NIA Grant T32-AG00181. Additional support was provided by Grant UL1 RR024153 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Riggs BL, Melton LJ., III The worldwide problem of osteoporosis: Insights afforded by epidemiology. Bone. 1995;17(Suppl):505S–511S. doi: 10.1016/8756-3282(95)00258-4. [DOI] [PubMed] [Google Scholar]
- 2.Silverman SL. Quality-of-life issues in osteoporosis. Curr Rheumatol Rep. 2005;7:39–45. doi: 10.1007/s11926-005-0007-x. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285:785–795. doi: 10.1001/jama.285.6.785. [DOI] [PubMed] [Google Scholar]
- 4.Ralston SH, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 2006;20:2492–2506. doi: 10.1101/gad.1449506. [DOI] [PubMed] [Google Scholar]
- 5.Jones G, Nguyen TV. Associations between maternal peak bone mass and bone mass in prepubertal male and female children. J Bone Miner Res. 2000;15:1998–2004. doi: 10.1359/jbmr.2000.15.10.1998. [DOI] [PubMed] [Google Scholar]
- 6.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, Valdes AM, Arp P, Whittaker P, Verlaan DJ, Jhamai M, Kumanduri V, Moorhouse M, van Meurs JB, Hofman A, Pols HA, Hart D, Zhai G, Kato BS, Mullin BH, Zhang F, Deloukas P, Uitterlinden AG, Spector TD. Bone mineral density, osteoporosis, and osteoporotic fractures: A genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 8.Zmuda JM, Kammerer CM. Snipping away at osteoporosis susceptibility. Lancet. 2008;371:1479–1480. doi: 10.1016/S0140-6736(08)60600-5. [DOI] [PubMed] [Google Scholar]
- 9.Carter DR, Bouxsein ML, Marcus R. New approaches for interpreting projected bone densitometry data. J Bone Miner Res. 1992;7:137–145. doi: 10.1002/jbmr.5650070204. [DOI] [PubMed] [Google Scholar]
- 10.Reid IR, Evans MC, Ames R, Wattie DJ. The influence of osteophytes and aortic calcification on spinal mineral density in postmenopausal women. J Clin Endocrinol Metab. 1991;72:1372–1374. doi: 10.1210/jcem-72-6-1372. [DOI] [PubMed] [Google Scholar]
- 11.Yu W, Gluer CC, Fuerst T, Grampp S, Li J, Lu Y, Genant HK. Influence of degenerative joint disease on spinal bone mineral measurements in postmenopausal women. Calcif Tissue Int. 1995;57:169–174. doi: 10.1007/BF00310253. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Peacock M, Eilam O, Dorulla G, Braunstein E, Johnston CC. Effect of osteoarthritis in the lumbar spine and hip on bone mineral density and diagnosis of osteoporosis in elderly men and women. Osteoporos Int. 1997;7:564–569. doi: 10.1007/BF02652563. [DOI] [PubMed] [Google Scholar]
- 13.Orwoll ES, Oviatt SK, Mann T. The impact of osteophytic and vascular calcifications on vertebral mineral density measurements in men. J Clin Endocrinol Metab. 1990;70:1202–1207. doi: 10.1210/jcem-70-4-1202. [DOI] [PubMed] [Google Scholar]
- 14.Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K, Lewis C, Cawthon PM, Marcus R, Marshall LM, McGowan J, Phipps K, Sherman S, Stefanick ML, Stone K. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study–a large observational study of the determinants of fracture in older men. Contemp Clin Trials. 2005;26:569–585. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Blank JB, Cawthon PM, Carrion-Petersen ML, Harper L, Johnson JP, Mitson E, Delay RR. Overview of recruitment for the osteoporotic fractures in men study (MrOS) Contemp Clin Trials. 2005;26:557–568. doi: 10.1016/j.cct.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Marshall LM, Lang TF, Lambert LC, Zmuda JM, Ensrud KE, Orwoll ES. Dimensions and volumetric BMD of the proximal femur and their relation to age among older U.S. men. J Bone Miner Res. 2006;21:1197–1206. doi: 10.1359/jbmr.060506. [DOI] [PubMed] [Google Scholar]
- 17.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 18.Roeder K, Bacanu SA, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genet Epidemiol. 2005;28:207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- 19.Zhao T, Chang LW, McLeod HL, Stormo GD. PromoLign: A database for upstream region analysis and SNPs. Hum Mutat. 2004;23:534–539. doi: 10.1002/humu.20049. [DOI] [PubMed] [Google Scholar]
- 20.Conde L, Vaquerizas JM, Santoyo J, Al-Shahrour F, Ruiz-Llorente S, Robledo M, Dopazo J. PupaSNP finder: A web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–W248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 22.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Havill LM, Mahaney MC, Binkley TL, Specker BL. Effects of genes, sex, age, and activity on BMC, bone size, and areal and volumetric BMD. J Bone Miner Res. 2007;22:737–746. doi: 10.1359/jbmr.070213. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Kammerer CM, Wheeler VW, Patrick AL, Bunker CH, Zmuda JM. Genetic and environmental determinants of volumetric and areal BMD in multi-generational families of African ancestry: The Tobago Family Health Study. J Bone Miner Res. 2007;22:527–536. doi: 10.1359/jbmr.070106. [DOI] [PubMed] [Google Scholar]
- 26.Yamada Y, Ando F, Shimokata H. Association of polymorphisms in forkhead box C2 and perilipin genes with bone mineral density in community-dwelling Japanese individuals. Int J Mol Med. 2006;18:119–127. [PubMed] [Google Scholar]
- 27.Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, Paes-Alves AF, Hill S, Bueno M, Ramos FJ, Tacconi P, Dikkers FG, Stratakis C, Lindpaintner K, Vickery B, Foernzler D, Van Hul W. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum Mol Genet. 2001;10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- 28.Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, Alisch RS, Gillett L, Colbert T, Tacconi P, Galas D, Hamersma H, Beighton P, Mulligan J. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uitterlinden AG, Arp PP, Paeper BW, Charmley P, Proll S, Rivadeneira F, Fang Y, van Meurs JB, Britschgi TB, Latham JA, Schatzman RC, Pols HA, Brunkow ME. Polymorphisms in the sclerosteosis/van Buchem disease gene (SOST) region are associated with bone-mineral density in elderly whites. Am J Hum Genet. 2004;75:1032–1045. doi: 10.1086/426458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tacconi P, Ferrigno P, Cocco L, Cannas A, Tamburini G, Bergonzi P, Giagheddu M. Sclerosteosis: Report of a case in a black African man. Clin Genet. 1998;53:497–501. doi: 10.1111/j.1399-0004.1998.tb02603.x. [DOI] [PubMed] [Google Scholar]
- 31.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 33.Monaghan H, Bubb VJ, Sirimujalin R, Millward-Sadler SJ, Salter DM. Adenomatous polyposis coli (APC), beta-catenin, and cadherin are expressed in human bone and cartilage. Histopathology. 2001;39:611–619. doi: 10.1046/j.1365-2559.2001.01287.x. [DOI] [PubMed] [Google Scholar]
- 34.Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL, Williams BO. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 35.Newman SA. Sticky fingers: Hox genes and cell adhesion in vertebrate limb development. Bioessays. 1996;18:171–174. doi: 10.1002/bies.950180302. [DOI] [PubMed] [Google Scholar]
- 36.Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gersch RP, Lombardo F, McGovern SC, Hadjiargyrou M. Reactivation of Hox gene expression during bone regeneration. J Orthop Res. 2005;23:882–890. doi: 10.1016/j.orthres.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Ling Y, Rios HF, Myers ER, Lu Y, Feng JQ, Boskey AL. DMP1 depletion decreases bone mineralization in vivo: An FTIR imaging analysis. J Bone Miner Res. 2005;20:2169–2177. doi: 10.1359/JBMR.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, Yu X, Rauch F, Davis SI, Zhang S, Rios H, Drezner MK, Quarles LD, Bonewald LF, White KE. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38:1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin C, D'Souza R, Feng JQ. Dentin matrix protein 1 (DMP1): New and important roles for biomineralization and phosphate homeostasis. J Dent Res. 2007;86:1134–1141. doi: 10.1177/154405910708601202. [DOI] [PubMed] [Google Scholar]
- 41.Rios HF, Ye L, Dusevich V, Eick D, Bonewald LF, Feng JQ. DMP1 is essential for osteocyte formation and function. J Musculoskelet Neuronal Interact. 2005;5:325–327. [PubMed] [Google Scholar]
- 42.Beamer WG, Shultz KL, Churchill GA, Frankel WN, Baylink DJ, Rosen CJ, Donahue LR. Quantitative trait loci for bone density in C57BL/6J and CAST/EiJ inbred mice. Mamm Genome. 1999;10:1043–1049. doi: 10.1007/s003359901159. [DOI] [PubMed] [Google Scholar]
- 43.Xiong Q, Han C, Beamer WG, Gu W. A close examination of genes within quantitative trait loci of bone mineral density in whole mouse genome. Crit Rev Eukaryot Gene Expr. 2008;18:323–343. doi: 10.1615/critreveukargeneexpr.v18.i4.20. [DOI] [PubMed] [Google Scholar]
- 44.Wilkie AO. Bad bones, absent smell, selfish testes: The pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Ratisoontorn C, Fan GF, McEntee K, Nah HD. Activating (P253R, C278F) and dominant negative mutations of FGFR2: Differential effects on calvarial bone cell proliferation, differentiation, and mineralization. Connect Tissue Res. 2003;44(Suppl 1):292–297. [PubMed] [Google Scholar]
- 46.Matsumoto Y, Tanaka K, Hirata G, Hanada M, Matsuda S, Shuto T, Iwamoto Y. Possible involvement of the vascular endothelial growth factor-Flt-1-focal adhesion kinase pathway in chemotaxis and the cell proliferation of osteoclast precursor cells in arthritic joints. J Immunol. 2002;168:5824–5831. doi: 10.4049/jimmunol.168.11.5824. [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa M, Kaneda T, Arakawa T, Morita S, Sato T, Yomada T, Hanada K, Kumegawa M, Hakeda Y. Vascular endothelial growth factor (VEGF) directly enhances osteoclastic bone resorption and survival of mature osteoclasts. FEBS Lett. 2000;473:161–164. doi: 10.1016/s0014-5793(00)01520-9. [DOI] [PubMed] [Google Scholar]
- 48.Niida S, Kaku M, Amano H, Yoshida H, Kataoka H, Nishikawa S, Tanne K, Maeda N, Kodama H. Vascular endothelial growth factor can substitute for macrophage colony-stimulating factor in the support of osteoclastic bone resorption. J Exp Med. 1999;190:293–298. doi: 10.1084/jem.190.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren WP, Markel DC, Zhang R, Peng X, Wu B, Monica H, Wooley PH. Association between UHMWPE particle-induced inflammatory osteoclastogenesis and expression of RANKL, VEGF, and Flt-1 in vivo. Biomaterials. 2006;27:5161–5169. doi: 10.1016/j.biomaterials.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Deckers MM, Karperien M, van der Bent C, Yamashita T, Papapoulos SE, Lowik CW. Expression of vascular endothelial growth factors and their receptors during osteoblast differentiation. Endocrinology. 2000;141:1667–1674. doi: 10.1210/endo.141.5.7458. [DOI] [PubMed] [Google Scholar]
- 51.Otomo H, Sakai A, Uchida S, Tanaka S, Watanuki M, Moriwaki S, Niida S, Nakamura T. Flt-1 tyrosine kinase-deficient homozygous mice result in decreased trabecular bone volume with reduced osteogenic potential. Bone. 2007;40:1494–1501. doi: 10.1016/j.bone.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Tare RS, Partridge KA, Roach HI, Clarke NM, Howdle SM, Shakesheff KM, Oreffo RO. Induction of human osteoprogenitor chemotaxis, proliferation, differentiation, and bone formation by osteoblast stimulating factor-1/pleiotrophin: Osteoconductive biomimetic scaffolds for tissue engineering. J Bone Miner Res. 2003;18:47–57. doi: 10.1359/jbmr.2003.18.1.47. [DOI] [PubMed] [Google Scholar]
- 53.Tare RS, Oreffo RO, Clarke NM, Roach HI. Pleiotrophin/Osteoblast-stimulating factor 1: Dissecting its diverse functions in bone formation. J Bone Miner Res. 2002;17:2009–2020. doi: 10.1359/jbmr.2002.17.11.2009. [DOI] [PubMed] [Google Scholar]
- 54.Tare RS, Oreffo RO, Sato K, Rauvala H, Clarke NM, Roach HI. Effects of targeted overexpression of pleiotrophin on postnatal bone development. Biochem Biophys Res Commun. 2002;298:324–332. doi: 10.1016/s0006-291x(02)02456-7. [DOI] [PubMed] [Google Scholar]
- 55.Zhao M, Harris SE, Horn D, Geng Z, Nishimura R, Mundy GR, Chen D. Bone morphogenetic protein receptor signaling is necessary for normal murine postnatal bone formation. J Cell Biol. 2002;157:1049–1060. doi: 10.1083/jcb.200109012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 57.Nifuji A, Miura N, Kato N, Kellermann O, Noda M. Bone morphogenetic protein regulation of forkhead/winged helix transcription factor Foxc2 (Mfh1) in a murine mesodermal cell line C1 and in skeletal precursor cells. J Bone Miner Res. 2001;16:1765–1771. doi: 10.1359/jbmr.2001.16.10.1765. [DOI] [PubMed] [Google Scholar]
- 58.Fisher MC, Meyer C, Garber G, Dealy CN. Role of IGFBP2, IGF-I and IGF-II in regulating long bone growth. Bone. 2005;37:741–750. doi: 10.1016/j.bone.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 59.Conover CA, Johnstone EW, Turner RT, Evans GL, John Ballard FJ, Doran PM, Khosla S. Subcutaneous administration of insulin-like growth factor (IGF)-II/IGF binding protein-2 complex stimulates bone formation and prevents loss of bone mineral density in a rat model of disuse osteoporosis. Growth Horm IGF Res. 2002;12:178–183. doi: 10.1016/s1096-6374(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 60.DeMambro VE, Clemmons DR, Horton LG, Bouxsein ML, Wood TL, Beamer WG, Canalis E, Rosen CJ. Gender-specific changes in bone turnover and skeletal architecture in igfbp-2-null mice. Endocrinology. 2008;149:2051–2061. doi: 10.1210/en.2007-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugimoto T, Nishiyama K, Kuribayashi F, Chihara K. Serum levels of insulin-like growth factor (IGF) I, IGF-binding protein (IGFBP)-2, and IGFBP-3 in osteoporotic patients with and without spinal fractures. J Bone Miner Res. 1997;12:1272–1279. doi: 10.1359/jbmr.1997.12.8.1272. [DOI] [PubMed] [Google Scholar]
- 62.Shinohara M, Takayanagi H. Novel osteoclast signaling mechanisms. Curr Osteoporos Rep. 2007;5:67–72. doi: 10.1007/s11914-007-0005-1. [DOI] [PubMed] [Google Scholar]
- 63.Stern PH. The calcineurin-NFAT pathway and bone: Intriguing new findings. Mol Interv. 2006;6:193–196. doi: 10.1124/mi.6.4.4. [DOI] [PubMed] [Google Scholar]
- 64.Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell. 2006;10:771–782. doi: 10.1016/j.devcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. FASTSNP: An always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: The flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaykin DV, Shibata K. Genetic flip-flop without an accompanying change in linkage disequilibrium. Am J Hum Genet. 2008;82:794–796. doi: 10.1016/j.ajhg.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zmuda JM, Cauley JA, Kuller LH, Ferrell RE. A common promotor variant in the cytochrome P450c17alpha (CYP17) gene is associated with bioavailability testosterone levels and bone size in men. J Bone Miner Res. 2001;16:911–917. doi: 10.1359/jbmr.2001.16.5.911. [DOI] [PubMed] [Google Scholar]
- 69.Tofteng CL, Abrahamsen B, Jensen JE, Petersen S, Teilmann J, Kindmark A, Vestergaard P, Gram J, Langdahl BL, Mosekilde L. Two single nucleotide polymorphisms in the CYP17 and COMT Genes–relation to bone mass and longitudinal bone changes in postmenopausal women with or without hormone replacement therapy. The Danish Osteoporosis Prevention Study. Calcif Tissue Int. 2004;75:123–132. doi: 10.1007/s00223-004-0176-z. [DOI] [PubMed] [Google Scholar]
- 70.Yamada Y, Ando F, Shimokata H. Association of polymorphisms in CYP17A1, MTP, and VLDLR with bone mineral density in community-dwelling Japanese women and men. Genomics. 2005;86:76–85. doi: 10.1016/j.ygeno.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 71.Chen HY, Chen WC, Hsu CM, Tsai FJ, Tsai CH. Tumor necrosis factor alpha, CYP 17, urokinase, and interleukin 10 gene polymorphisms in postmenopausal women: Correlation to bone mineral density and susceptibility to osteoporosis. Eur J Obstet Gynecol Reprod Biol. 2005;122:73–78. doi: 10.1016/j.ejogrb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 72.Somner J, McLellan S, Cheung J, Mak YT, Frost ML, Knapp KM, Wierzbicki AS, Wheeler M, Fogelman I, Ralston SH, Hampson GN. Polymorphisms in the P450 c17 (17-hydroxylase/17,20-Lyase) and P450 c19 (aromatase) genes: Association with serum sex steroid concentrations and bone mineral density in postmenopausal women. J Clin Endocrinol Metab. 2004;89:344–351. doi: 10.1210/jc.2003-030164. [DOI] [PubMed] [Google Scholar]
- 73.van Meurs JB, Rivadeneira F, Jhamai M, Hugens W, Hofman A, van Leeuwen JP, Pols HA, Uitterlinden AG. Common genetic variation of the low-density lipoprotein receptor-related protein 5 and 6 genes determines fracture risk in elderly white men. J Bone Miner Res. 2006;21:141–150. doi: 10.1359/JBMR.050904. [DOI] [PubMed] [Google Scholar]
- 74.van Meurs JB, Trikalinos TA, Ralston SH, Balcells S, Brandi ML, Brixen K, Kiel DP, Langdahl BL, Lips P, Ljunggren O, Lorenc R, Obermayer-Pietsch B, Ohlsson C, Pettersson U, Reid DM, Rousseau F, Scollen S, Van Hul W, Agueda L, Akesson K, Benevolenskaya LI, Ferrari SL, Hallmans G, Hofman A, Husted LB, Kruk M, Kaptoge S, Karasik D, Karlsson MK, Lorentzon M, Masi L, McGuigan FE, Mellstrom D, Mosekilde L, Nogues X, Pols HA, Reeve J, Renner W, Rivadeneira F, van Schoor NM, Weber K, Ioannidis JP, Uitterlinden AG. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299:1277–1290. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]