RNA Granule Protein 140 (RNG140), a Paralog of RNG105 Localized to Distinct RNA Granules in Neuronal Dendrites in the Adult Vertebrate Brain (original) (raw)
Abstract
RNA granules mediate the transport and local translation of their mRNA cargoes, which regulate cellular processes such as stress response and neuronal synaptic plasticity. RNA granules contain specific RNA-binding proteins, including RNA granule protein 105 (RNG105), which is likely to participate in the transport and translation of mRNAs. In the present report, an RNG105 paralog, RNG140 is described. A homolog of RNG105/RNG140 is found in insects, echinoderms, and urochordates, whereas vertebrates have both of the two genes. RNG140 and RNG105 are similar in that both bind to mRNAs and inhibit translation in vitro, induce the formation of RNA granules, are most highly expressed in the brain, and are localized to dendritic RNA granules, part of which are accumulated at postsynapses. However, they differ in several characteristics; RNG105 is highly expressed in embryonic brains, whereas RNG140 is highly expressed in adult brains. Furthermore, the granules where RNG105 or RNG140 is localized are distinct RNA granules in both cultured cells and neuronal dendrites. Thus, RNG140 is an RNA-binding protein that shows different expression and localization patterns from RNG105. Knockdown experiments in cultured neurons also are performed, which demonstrate that suppression of RNG140 or RNG105 reduces dendrite length and spine density. Knockdown effects of RNG140 were not rescued by RNG105, and vise versa, suggesting distinct roles of RNG105 and RNG140. These results suggest that RNG140 has roles in the maintenance of the dendritic structure in the adult vertebrate brain through localizing to a kind of RNA granules that are distinct from RNG105-containing granules.
Keywords: Cellular Regulation, Neurobiology, Neuron, RNA-binding Protein, RNA Transport, RNA Granule, Dendrite, Stress Granule
Introduction
The transport of specific mRNAs and local control of translation play important roles in local translation in a cell, which contributes to several cellular processes including axis formation, stress responses, and long term synaptic plasticity (1–4). The specific mRNAs are recruited into RNA granules, which are key RNA-protein complexes for controlling the transport, stability, and translation of mRNAs.
There are different types of RNA granules, including neuronal RNA granules, stress granules, and processing bodies (P bodies).3 Neuronal RNA granules store specific mRNAs and repress their translation during the transport of the mRNAs into dendrites. The granules release the mRNAs to be translated in response to synaptic stimulation, which is responsible for long term synaptic plasticity (5, 6). Stress granules appear in the cytoplasm of somatic cells in response to stress such as arsenite treatment. Stress granules recruit mRNAs encoding “housekeeping” proteins and repress their translation, which contributes to stress-induced translational arrest (7). P bodies are cytoplasmic RNA granules containing mRNA decay machinery and are involved in mRNA degradation and translational repression (8).
All of the three types of RNA granules share some protein components such as translation initiation factor 4E (eIF4E) and the fragile X mental retardation protein (FMRP). On the other hand, other components are shared only between neuronal RNA granules and stress granules, e.g. G3BP (RasGAP SH3 domain-binding protein) and ribosomes (6, 7). P bodies are distinguished from the other two granules by the absence of ribosomes and are characterized by the presence of enzymes for the mRNA decay pathway such as DCP1/2 and XRN1. Although stress granules and P bodies are distinct structures, they are linked dynamically and share some enzymes, including XRN1 (7).
RNG105 is an RNA-binding protein highly expressed in the brain and localized to neuronal RNA granules in dendrites (9). RNG105 also is expressed in proliferating cells, so it also is known as caprin-1 (cytoplasmic activation/proliferation-associated protein-1) (10). In proliferating cells, RNG105 is localized to stress granules (9, 11). RNG105 binds to mRNAs nonspecifically and represses translation in vitro or when overexpressed in cells (9, 11). However, endogenous RNG105 binds to specific mRNAs both in neurons and proliferating cells, and loss of RNG105 does not affect the global translation rates in cells (9, 11). RNG105 has two basic domains, N-terminal basic helices (coiled-coil) and C-terminal RGG boxes (RG-rich region), which are responsible for RNA binding and translational repression in vitro and for RNA granule formation (9). In neurons, the localization of RNG105 to neuronal RNA granules coincides with cargo mRNAs and is regulated dynamically by synaptic stimulation, suggesting the role of RNG105 in mRNA transport and local translational control (9).
RNG140, also termed EEG-1L or caprin-2, is reported as a paralog of RNG105 (10, 12). Although similarities in the amino acid sequences between RNG140 and RNG105 have been shown, the functions of RNG140 still remain to be characterized.
In this study, RNG140 was identified as an RNA-binding protein, which was present in RNA granules that were distinct from RNG105-containing RNA granules. RNG140 and RNG105 also were different in their expression patterns in fetal and adult mouse brains. Loss of RNG140 or RNG105 in neurons resulted in the reduction of dendrite length and spine density. The results suggested roles of RNG140 and RNG105 in dendrite organization at different location and times in neurons.
EXPERIMENTAL PROCEDURES
cDNA Sequences of Rng105 and Rng140
cDNA sequences were obtained from the GenBankTM/EMBL/DDBJ databases. Rng105 sequences; Homo sapiens, GenBankTM accession no. NM 005898; Danio rerio, NM 213068; Rngi for Strongylocentrotus purpuratus, XM 00194889; Drosophila melanogaster, NM 144414; Rng140 for H. sapiens, NM 001002259; and D. rerio, NM 001013273. The cDNA sequence for Ciona intestinalis Rngi was assembled from expressed sequence tag sequences: BW 240466, BW 291363, BW 289399, BW 270664, AV 675094, BW 044090, BW 209789, BW 260723, BW 264501, AV 958493, BW 402084, AV 968680, AV 676119, BW 232064, BW 293691, BW 90982, and BW 245018.
The aligned sequences were compared using ClustalW software. Dendrograms were generated using Phylodendron software.
Antibodies
The following antibodies were used in the present study: anti-ribosomal protein S6 (RPS6) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-FMRP (Millipore, Billerica, MA), anti-microtubule-associated protein 2 (MAP2) (Sigma), anti-PSD-95 (BD Biosciences, San Jose, CA), anti-GFP (Nacalai Tesque, Kyoto, Japan), anti-DCP2 (generous gift from Dr. M. Kiledjian), anti-XRN1 (generous gift from Dr. W.-D. Heyer), anti-staufen (generous gift from Dr. J. Ortin), anti-RNG105 (9), anti-digoxigenin Fab fragments (Roche Applied Science), cyanine 3 (Cy3)-labeled anti-mouse IgG, Cy3-labeled anti-rabbit IgG, Cy3-labeled anti-goat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), biotinylated anti-rabbit IgG, and alkaline phosphatase-conjugated streptavidin (GE Healthcare).
Generation of Polyclonal Antibodies
Rat cDNAs encoding amino acids 884–1,031 of RNG140 and the full length of G3BP were obtained by reverse transcription-PCR from rat brain RNA. These fragments were cloned into a pGEX-5X-3 vector (GE Healthcare) to produce fusion proteins with glutathione _S_-transferase (GST). The GST fusion proteins were expressed in Escherichia coli (BL21) and purified using glutathione-Sepharose 4B columns (GE Healthcare). The GST tag was removed by factor Xa cleavage, and then RNG140 and G3BP proteins were purified in accordance with the manufacturer's protocol. The purified proteins were used as antigens to generate polyclonal antibodies in rabbits. The antibodies were affinity-purified on Affi-Gel 10 gel (Bio-Rad), which had been conjugated with the respective antigens.
Immunoblotting
Extracts from mouse tissues or cultured A6 cells were prepared by homogenization in 150 mm NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris (pH 8.0), 1 mm dithiothreitol, and protease inhibitors (10 μg/ml leupeptin, 10 μg/ml pepstatin, 100 μg/ml aprotinin, and 1 mm phenylmethylsulfonyl fluoride) followed by centrifugation for 10 min at 10,000 × g at 4 °C. Extracts were loaded on polyacrylamide gels (30 μg protein per lane), transferred to polyvinylidene fluoride membranes and probed with the primary antibodies. Biotinylated secondary antibodies and alkaline phosphatase-conjugated streptavidin were used for detection with bromochloroindolyl phosphate/nitro blue tetrazolium solution.
In Vitro RNA Binding and in Vitro Translation Assays
To construct the expression vectors for GST-RNG140 and GST-RNG140 deletion mutants, Rng140 cDNA was obtained by reverse transcription-PCR from rat brain RNA and cloned into the SmaI/NotI sites of the pGEX-5X-3 vector. The primers were 5′-TCCCCCGGGTCATGAAGTCAGCCAAGTCCCAAG-3′ (N-SmaI) and 5′-ATTTGCGGCCGCTTAATCTTGATAAAGAAGATAGCCT-3′ (C-NotI) for GST-RNG140, 5′-TCCCCCGGGTCGAAGCAGTAGAAAAGTATGAAG-3′ (ΔN-SmaI) and C-NotI for GST-RNG140ΔN, N-SmaI and 5′-ATTTGCGGCCGCTCAACTACTGATAAATGGCTGAGCAG-3′ (ΔC-NotI) for GST-RNG140ΔC, and ΔN-SmaI and ΔC-NotI for GST-RNG140ΔNC. The recombinant GST fusion proteins were expressed in E. coli (BL21) and subjected to RNA-binding assay as described previously (9). The recombinant proteins also were subjected to translation assays in rabbit reticulocyte lysates (Promega, Madison, WI) using luciferase mRNA as a template as described previously (9).
Expression of GFP Fusion Proteins in Cultured Cells
To construct the expression vectors for RNG140-green fluorescent protein (RNG140-GFP) and RNG140-GFP deletion mutants, rat Rng140 cDNA was amplified by PCR and cloned into the XhoI and KpnI sites of pEGFP-N1 vector (Clontech). The primers were 5′-CCGCTCGAGATGAAGTCAGCCAAGTCCCAAG-3′ (N-XhoI) and 5′-GGGGTACCCCACCTCCACCTCCATCTTGATAAAGAAGATAGCCTGAAA-3′ (C-KpnI) for RNG140-GFP, 5′-CCGCTCGAGGAAGCAGTAGAAAAGTATGAAG-3′ and C-KpnI for RNG140ΔN-GFP, and N-XhoI and 5′-GGGGTACCCCCCCACCCCCCCCACTACTGATAAATGGCTG-3′ for RNG140ΔC-GFP. Cultured Chinese hamster ovary and A6 cells were transfected with the vectors according to the Lipofectin protocol (Invitrogen). Stable transfectants were selected by treatment with Geneticin (Invitrogen) as described previously (13).
Cell Staining
Immunofluorescence staining was performed as described previously (9). For in situ hybridization, A6 cells or rat hippocampal slices were subjected to fluorescence in situ hybridization with a 3′-digoxigenin-labeled poly(dT) probe (55-mer) as described previously (9).
The specimens were imaged using an FV500 confocal laser-scanning microscope (Olympus Optical, Tokyo, Japan) with a PlanApo 60× oil objective lens or a Delta Vision optical sectioning microscope (Applied Precision, Inc., Issaquah, WA) equipped with an IX70 microscope (Olympus Optical) with a PlanApo 60× oil objective lens or a UPlanApo 20× objective lens.
Time-lapse Imaging
A6 cells co-transfected with RNG140-GFP and RNG105-monomeric red fluorescent protein (mRFP) (9) vectors were cultured on glass bottom dishes (MatTek, Ashland, MA). Time-lapse images of the cells were taken with an inverted fluorescence microscope (IX71; Olympus Optical) using a LUC Plan FL 40× objective lens and a cooled CCD camera (ORCA-ER; Hamamatsu, Shizuoka, Japan). Cells with low expression levels of RNG140-GFP and RNG105-mRFP were selected for the imaging. The cells had very few RNG140-GFP-localizing and RNG105-mRFP-localizing granules before arsenite treatment.
RNA Interference
Mixtures of siRNAs for RNG105, RNG140, and control were purchased from Dharmacon (Lafayette, CO) (siGENOME SMART pool M-056618-00-0005, M-057521-01-0005, and D-001206-13-05, respectively). Cerebral cortical neurons from embryonic day 15.5 mice were cultured in minimum essential medium (Sigma) containing N2 supplement (Invitrogen) and 2% fetal bovine serum. The neurons were co-transfected with the siRNAs (0.5 μg each for 7.1 cm2) and pEGFP-N1 vector (0.5 μg for 7.1 cm2) at 7 days in vitro using the conventional calcium-phosphate precipitation method. In rescue experiments, the expression vector for RNG105-GFP or RNG140-GFP (2 μg each for 7.1 cm2) was introduced to the neurons. Three days later, the neurons were subjected to immunofluorescence studies.
RESULTS
RNG140 Is a Paralog of RNG105 That Is Conserved in Vertebrates
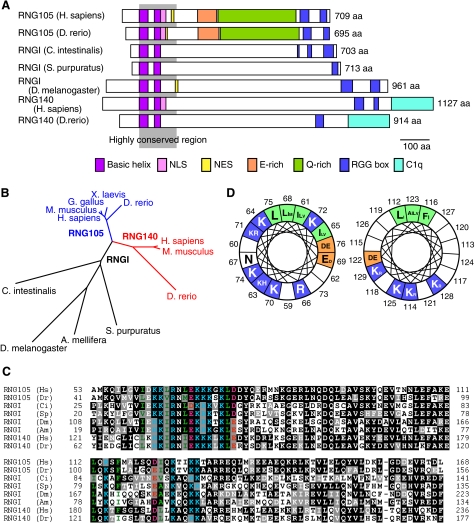
RNG105 has been shown previously to contain specific domains such as basic helices (coiled-coil), a glutamic acid-rich region, a glutamine-rich region, and RGG boxes (9). The N-terminal basic helices and the C-terminal RGG boxes are responsible for the strong and weak RNA-binding abilities, respectively. Structural analysis of RNG140 revealed that RNG140 also contains basic helices and RGG boxes, suggesting that RNG140 is an RNA-binding protein (Fig. 1A). However, RNG140 does not contain the glutamic acid-rich region or glutamine-rich region. RNG140 is characterized by the C-terminal domain, which is homologous to the globular domain of complement component C1q of the immune system (Fig. 1A) (14).
FIGURE 1.
RNG140 is a paralog of RNG105 that is conserved in vertebrates. A, schematic diagram showing the domain structures of RNG105, RNGI, and RNG140 from the indicated organisms. The gray shadow shows highly conserved region among them. NLS, nuclear localization signal; NES, nuclear export signal; E-rich, glutamic acid-rich region; Q-rich, glutamine-rich region; C1q, C1q-related domain. B, a dendrogram of RNG105, RNGI, and RNG140 from the indicated organisms. C, alignment of the highly conserved region from RNG105, RNGI, and RNG140. Black boxes indicate identities; gray boxes, similarity. Colored amino acids are predicted to form the basic helices. Two basic helices are contained in the highly conserved region. Blue, basic; red, acidic; green, hydrophobic amino acids. Numbers indicate residue numbers. Hs, H. sapiens; Dr, D. rerio; Ci, C. intestinalis; Sp, S. purpuratus; Dm, D. melanogaster; and Am, A. mellifera. D, Schiffer-Edmundson wheel diagrams for the two basic helices. Numbers indicate the amino acid residues of human RNG105. The size of the amino acid letters corresponds to their frequency among the eight sequences in C. Blue, basic; red, acidic; green, hydrophobic amino acids (aa).
Database homology searches found that an RNG105/RNG140 homolog is conserved in urochordates, echinoderms, and insects (Fig. 1, A and B). Only one gene for RNG105/RNG140 homolog was found in the invertebrate animals, suggesting that duplication and divergence of the gene had occurred in the early evolution of vertebrates. We designated the invertebrate homolog RNGI (RNG invertebrate).
Sequence alignment of RNG105, RNG140, and RNGI revealed that the basic helix-containing region was the most highly conserved region across animal phyla (Fig. 1, A and C). This region had two peaks of basic charge, corresponding to two basic helices (Fig. 1, A and C). In each helix, the amino acids are arranged to form a hydrophobic stripe along one side of the α-helix and basic clusters on the other side (Fig. 1D). The hydrophobic stripe may contribute to protein-protein interactions, such as those found in coiled-coil domains, and the basic clusters may serve as a docking surface for nucleic acids (9).
RNG140 Binds Directly to RNA
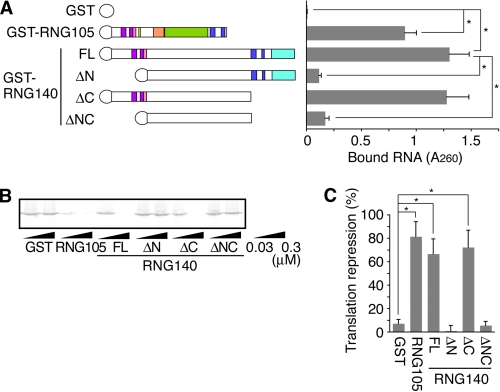
Recombinant GST-RNG140 was expressed in E. coli and purified, and then its RNA-binding ability was examined in vitro. RNG140, as well as RNG105, bound directly to RNA (Fig. 2A). Deletion of the N terminus containing the basic helices (ΔN and ΔNC) markedly reduced the binding ability. In contrast, deletion of the C terminus containing the RGG boxes (ΔC) did not appear to affect the RNA-binding ability (Fig. 2A). These results indicated that the N-terminal region of RNG140 is a major RNA-binding site, consistent with findings in RNG105 (9). In addition, RNA binding of RNG140 was sequence-independent, which was similar to that of RNG105 (data not shown).
FIGURE 2.
RNG140, as well as RNG105, binds directly to RNA and inhibits translation in vitro. A, recombinant GST, GST-RNG105, GST-RNG140, and GST-RNG140 deletion mutants were incubated with RNA isolated from rat brains. RNA bound to the recombinant proteins was extracted and quantified. FL, full length; ΔN, deletion of N terminus (from N-terminal end to NLS); ΔC, deletion of C terminus (from RGG box to C-terminal end); ΔNC, deletion of both N and C termini. n = 3, *, p < 0.05, Student's t test. Error bars are S.E. B, luciferase mRNA was translated in rabbit reticulocyte lysates in the presence of 0.03 or 0.3 μm recombinant GST, GST-RNG105, GST-RNG140, and GST-RNG140 deletion mutants. Translated luciferase protein was detected by SDS-PAGE. C, intensities of the bands in B were quantified, and translational repression activity of each recombinant protein (0.3 μm) was calculated. n = 3, *, p < 0.05, Student's t test. Error bars are S.E.
It was shown previously that RNG105 represses the translation of bound mRNAs in vitro, and the N terminus is required for the repression activity (9). In Fig. 2, B and C, the effect of RNG140 on translation was examined in vitro. Full-length RNG140 repressed translation in a dose-dependent manner. Deletion of the C terminus (ΔC) did not alter the translation repression activity, whereas deletion of the N terminus (ΔN and ΔNC) resulted in decreases in the repression activity. These results indicated that RNG140 has translation repression activity in vitro and that the basic helix region was responsible for the activity. These results were similar to the case of RNG105 (9).
RNG140 Induces the Formation of Cytoplasmic RNA Granules
RNG105 induces the formation of cytoplasmic RNA granules when overexpressed in cultured cells, and both the N-terminal basic helices and the C-terminal RGG boxes are required for granule formation (9). It was examined whether RNG140 also has ability to induce cytoplasmic RNA granules in cultured cells. Overexpression of RNG140-GFP in Chinese hamster ovary and A6 cultured cells induced the formation of cytoplasmic granules (Fig. 3 and supplemental Fig. 1). The granules contained mRNAs as judged by in situ hybridization with poly(dT) (Fig. 3, top panels), indicating that they were RNA granules. Deletion of the N terminus (ΔN) markedly reduced granule formation (supplemental Fig. 1). The effect of the C-terminal deletion (ΔC) was more severe, i.e. the granules were not formed at all (supplemental Fig. 1). These results indicated that RNG140 induces the formation of cytoplasmic RNA granules and that both the N- and the C- terminal regions are required for the granule formation.
FIGURE 3.
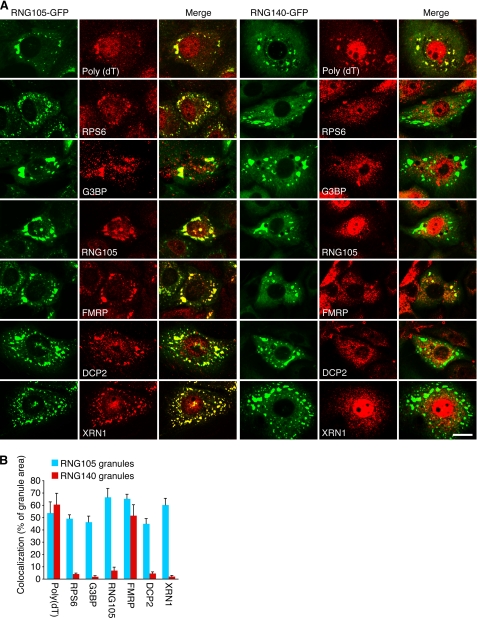
The RNG140-induced granules are distinct RNA granules from RNG105-induced ones. A, A6 cells expressing RNG105-GFP or RNG140-GFP were stained by in situ hybridization with poly(dT) for mRNA detection or immunostained with anti-ribosomal protein S6 (RPS6), anti-G3BP, anti-RNG105, anti-FMRP, anti-DCP2, and anti-XRN1 antibodies. Scale bar, 10 μm. B, quantification of co-localization of RNG105-GFP or RNG140-GFP granules with the stains in A. Shown are percentages of RNG105-GFP or RNG140-GFP granule areas that overlap with the stains. n = 10. Error bars are S.E.
It is reported that cytoplasmic RNA granules such as stress granules are dynamic structures containing translationally inactive mRNAs which are in equilibrium with polysomes. Granules formed by trapping translationally inactive mRNAs are disassembled by cycloheximide that stabilizes polysomes (15). RNG140-induced RNA granules as well as RNG105-induced granules were disassembled by cycloheximide (supplemental Fig. 2), which suggested that these granules are related to repression of translation.
RNG140-induced RNA Granules Are Distinct from RNG105-induced Granules
RNG105-induced RNA granules contain ribosomes, G3BP, and T cell intracellular antigen-1, and they also are induced by stress, indicating that they are stress granules (9, 11). RNG105-GFP-expressing cultured cells were immunostained, and it was confirmed that RNG105-induced RNA granules contained the components of stress granules such as ribosomes, G3BP, and FMRP (Fig. 3). The granules also contained P body components DCP2 and XRN1, which were consistent with the report that stress granules and P bodies are dynamically linked and share some components (7, 8).
RNG140-GFP-expressing cells were immunostained with the same antibodies (Fig. 3). In contrast to the RNG105-induced RNA granules, the RNG140-induced granules were not co-localized with ribosomes, G3BP, or RNG105, suggesting that they were not stress granules. FMRP, which reportedly is localized in all the three types of RNA granules, was contained in the RNG140-induced granules.
The results that RNG140-induced RNA granules do not contain ribosomes led to an experiment to determine whether the granules are identical to P bodies. Immunostaining of RNG140-GFP-expressing cells revealed that the RNG140-induced RNA granules did not contain DCP2 or XRN1, indicating that the granules were not P bodies (Fig. 3).
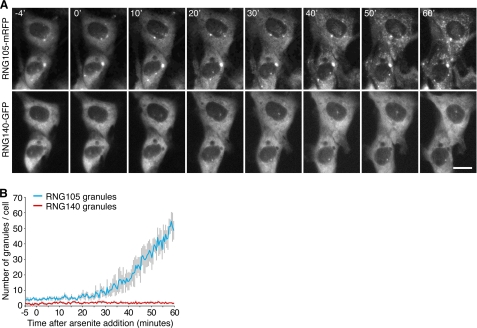
Time-lapse observation of stress-induced granule formation after arsenite treatment was performed in cultured cells co-expressing RNG105-mRFP and RNG140-GFP (Fig. 4). Cells that expressed low amounts of RNG105-mRFP and RNG140-GFP were used so that no or few granules were formed in the cytoplasm before arsenite treatment. After arsenite treatment, stress granules where RNG105-mRFP was concentrated were induced in the cytoplasm. In contrast, RNG140-GFP remained distributed diffusely in the cytoplasm and was not concentrated into RNG105-containing stress granules (Fig. 4). Taken together, these results indicated that RNG140 was not recruited into the RNG105-containing RNA granules and that RNG140-induced RNA granules were not stress granules or P bodies.
FIGURE 4.
RNG105, but not RNG140, is recruited into stress-induced cytoplasmic granules. A, A6 cells co-expressing low levels of RNG105-mRFP and RNG140-GFP were stressed with 0.5 mm arsenite, and time-lapse recordings were made. The time (min) after arsenite addition is indicated. Scale bar, 10 μm. B, profile of changes in the number of RNG105-mRFP granules and RNG140-GFP granules per cell during the experiment in A (n = 4). Error bars are S.E.
RNG140 Is Highly Expressed in Adult Brain
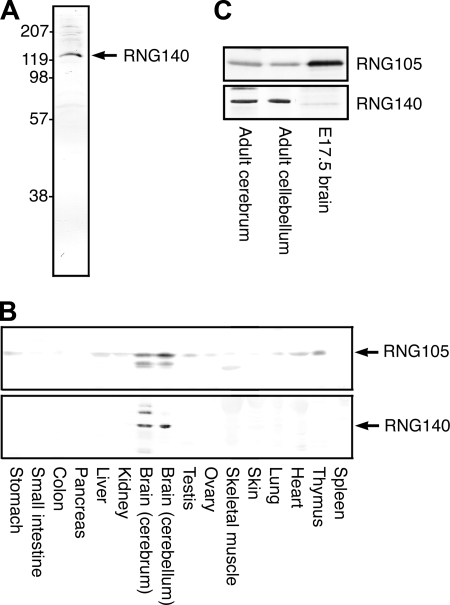
To examine the expression and localization of RNG140 in tissues, a polyclonal antibody against RNG140 was generated. The antibody recognized recombinant RNG140 specifically, and absorption of the antibody with recombinant RNG140 diminished the reaction as judged by immunoblotting (data not shown). Immunoblotting of extracts from adult mouse brain showed that the antibody recognized a 140-kDa protein (Fig. 5A).
FIGURE 5.
RNG140 is highly expressed in the adult brain. A, immunoblotting of mouse brain (cerebrum) extracts with an antibody generated against RNG140. B, immunoblotting of mouse tissue extracts with anti-RNG105 and anti-RNG140 antibodies. Both RNG105 and RNG140 are most highly expressed in the brain (cerebrum and cerebellum). In cerebrum, the anti-RNG140 antibody detected two upper bands and a lower band in addition to the RNG140 band after long exposure to detection reagents, although the bands were much fainter with shorter exposure (cf. A). These bands may be RNG140 with some modifications or nonspecific cross-reactants highly expressed in the brain except the possibility that the bottom band is a splice form of RNG140 encoding a predicted 812-amino acid protein (protein ID ENSMUSP00000072165 on the Ensembl Web site). The lower band detected by RNG105 antibody in brain extracts is an RNG105 degradation product, and the band in skeletal muscle may be a nonspecific cross-reactant specifically expressed in skeletal muscle. C, extracts from adult cerebrum and cerebellum and from brain of embryonic day 17.5 embryo were immunoblotted with anti-RNG105 and anti-RNG140 antibodies. 30 μg proteins are loaded on each lane.
Extracts from adult mouse tissues were immunoblotted with the antibodies against RNG105 and RNG140 (Fig. 5B). RNG105 was expressed highly in the brain, and it also was expressed at low levels in other tissues such as thymus, heart, liver, and testis. On the other hand, RNG140 was expressed specifically in the brain and was not detectable in other tissues (Fig. 5B).
Expression of RNG105 in the brain was abundant in embryos compared with adult mice (Fig. 5C). In contrast, RNG140 was abundant in the brain in adult mice but markedly less abundant in embryos (Fig. 5C). Thus, RNG140 appeared to be expressed mainly in adult brain.
RNG140 Is Localized to Dendritic RNA Granules Distinct from RNG105-containing RNA Granules in Neurons
The subcellular localizations of RNG105 and RNG140 were examined in the brain by immunostaining of slices of adult mouse brains (Fig. 6 and supplemental Fig. 3). In hippocampal neurons, RNG140, as well as RNG105, was localized to granular structures in dendrites (supplemental Fig. 3_A_). In the cerebellar molecular layers, RNG140 was localized to granular structures in the dendrites of Purkinje cells, but RNG105 was not detectable in the dendrites and was limited to the nucleus (supplemental Fig. 3_B_).
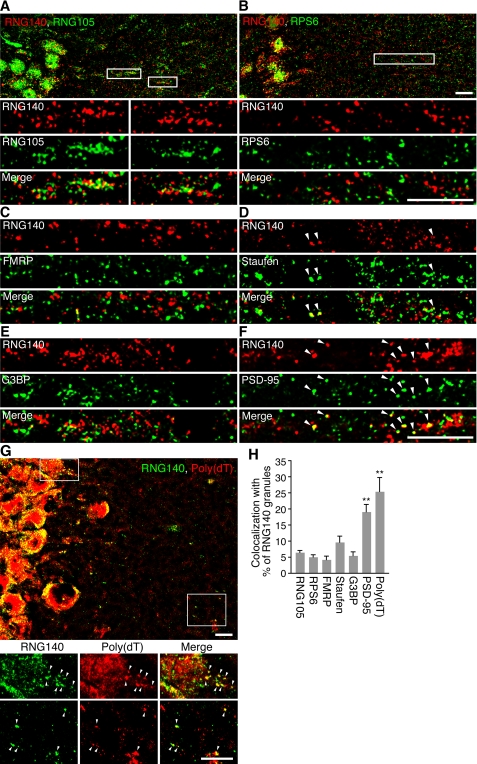
FIGURE 6.
The RNG140-localizing RNA granules in neuronal dendrites are distinct from the RNG105-localizing RNA granules. A–F, rat hippocampal slices were immunostained with anti-RNG140 antibody and co-stained with anti-RNG105 (A), anti-RPS6 (B), anti-FMRP (C), anti-staufen (D), anti-G3BP (E), or anti-PSD-95 (F) antibody. The lower panels in A and B show higher magnification of the boxed areas. G, rat hippocampal slices were immunostained with anti-RNG140 antibody and co-stained by in situ hybridization with poly(dT) for mRNA detection. The lower panels show magnified images of the boxed areas. Arrowheads denote co-localization. Scale bars, 10 μm. H, quantification of co-localization of RNG140 granules with the antibody or poly(dT) staining in A–G. Shown are percentages of RNG140 granules that overlap with the indicated antibody or poly(dT) staining. n = 5, **, p < 0.01, significantly different from the left five bars in Tukey-Kramer test. Error bars are S.E.
To compare the subcellular localizations of RNG105 and RNG140 in hippocampal dendrites, brain slices were double stained with RNG105 and RNG140 antibodies (Fig. 6A). Although both RNG140 and RNG105 were localized to granular structures, the granules did not show any significant co-localization (Fig. 6A). It was shown previously that RNG105 co-localized well with ribosomes in hippocampal dendrites (9). In contrast to RNG105, RNG140 did not show significant co-localization with ribosomes in dendrites (Fig. 6B). In addition, RNG140 did not show significant co-localization with markers of neuronal and stress granules such as FMRP, staufen, or G3BP, except that a little fraction of RNG140 granules were co-localized with staufen (Fig. 6, C–E and H). In contrast, RNG140 granules were significantly co-localized with mRNAs as judged by in situ hybridization with poly(dT) (Figs. 6, G and H, and supplemental Fig. 4). In RNG140 granules with low fluorescence intensity of RNG140 (<40 arbitrary units), poly(dT) fluorescence intensity was significantly low (supplemental Fig. 4B). This may be partly due to low sensitivity of poly(dT) staining in small RNG140 granules and resulted in underestimation of co-localization of mRNA and RNG140 (Fig. 6H, ∼25%). Co-localization of RNG105 with mRNA, judged by the same method in Fig. 6H, was similar (∼29%) to that of RNG140. Some of RNG140 granules were accumulated near the postsynaptic sites stained with anti-PSD-95 antibody (Fig. 6, F and H), as reported for neuronal RNA granules (9, 16, 17). Thus, RNG140 is localized to dendritic RNA granules in neurons, but the granules were distinct from RNG105-containing RNA granules.
Suppression of RNG105 and RNG140 Reduces Dendrite Length and Spine Density in Neurons
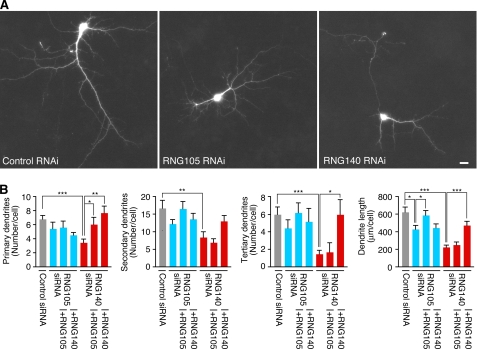
Next, to examine the physiological significance of RNG105 and RNG140 in neurons, RNAi of RNG105 and RNG140 was performed. Cultured neurons from mouse cerebral cortex were co-transfected with siRNA for RNG105 or RNG140 and a GFP reporter to trace the affected cells. Expression of RNG105 or RNG140 was suppressed in neurons expressing the GFP reporter (supplemental Fig. 5). RNG140 siRNA-transfected GFP-positive neurons apparently reduced their dendrite area (Fig. 7A). Quantification revealed that dendrites of RNG140 RNAi neurons were reduced in number, length, and branching (Fig. 7B). RNG105 RNAi also reduced dendrite length, but RNAi of RNG140 had more severe effects on dendrites than RNG105 RNAi (Fig. 7). Rescue experiments showed that RNG105 knockdown was rescued by not RNG140 but RNG105 expression, whereas RNG140 knockdown was rescued by not RNG105 (except the number of primary dendrites) but RNG140 (Fig. 7B). These results suggested that RNG105 and RNG140 play distinct roles in the maintenance and/or development of dendrites.
FIGURE 7.
Knockdown of RNG105 and RNG140 reduces dendrite length in cultured neurons. A, primary cultured neurons from mouse cerebral cortex were co-transfected with a GFP reporter and siRNA for control, RNG105, or RNG140. GFP images of transfected neurons are shown. Scale bar, 10 μm. B, quantification of dendrites in A. Shown are the number of primary, secondary, and tertiary dendrites and the total dendrite length per cell. Cultured neurons were also co-transfected with RNG105 siRNA or RNG140 siRNA with RNG105 or RNG140 for rescue experiments. n = 9, *, p < 0.05, **, p < 0.01, ***, p < 0.001, Student's t test. Error bars are S.E.
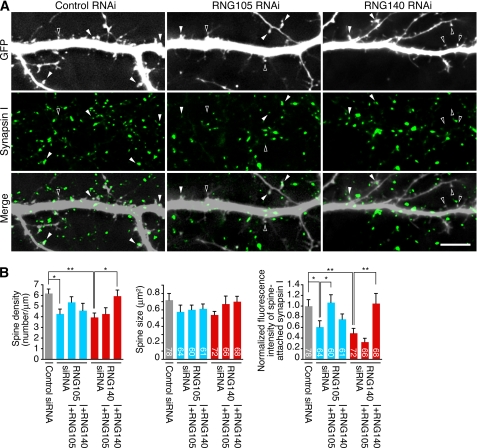
RNAi of RNG105 and RNG140 also affected dendritic spines. Although the size of spines was not affected, spine density was reduced by RNG105 and RNG140 RNAi (Fig. 8). Furthermore, RNAi of RNG105 and RNG140 in postsynaptic neurons resulted in the decrease of the attachment of synapsin I-containing axon terminals from GFP-negative normal neurons to spines (Fig. 8). Although the phenotype was similar between RNG105 and RNG140 RNAi, RNG140 knockdown was not rescued by RNG105 and vice versa (Fig. 8B). These results suggested that RNG105 and RNG140 play distinct roles, but both of them participate in the maintenance and/or development of dendrites and dendritic spines.
FIGURE 8.
Knockdown of RNG105 and RNG140 reduces spine density and affects the termination of axons from normal neurons to spines. A, cultured neurons were co-transfected with a GFP reporter and siRNA for control, RNG105, or RNG140. The neurons were then immunostained for presynaptic marker synapsin I. White arrowheads indicate postsynaptic spines of GFP-positive dendrites innervated by synapsin I-containing axon terminals from nontransfected GFP-negative neurons. Black arrowheads indicate spines without synapsin I staining. Scale bar, 10 μm. B, quantification of spine density, spine size, and fluorescence intensity of synapsin I on the GFP-positive spines in A. Rescue experiments also were performed. The fluorescence intensity of synapsin I on spines is normalized to that in GFP-negative areas in the same field. Spine density and synapsin I fluorescence intensity at spines were significantly reduced in RNG105 and RNG140 knockdown neurons. n = 6 for spine density. Numbers inside the bars are the number of spines analyzed. *, p < 0.05, **, p < 0.01, Student's t test. Error bars are S.E.
DISCUSSION
This study characterized RNG140, a paralog of RNG105 in vertebrates. RNG140 was revealed to be an RNA-binding protein, whose in vitro properties were similar to those of RNG105. RNG140 and RNG105 share two RNA-binding domains: N-terminal basic helices and C-terminal RGG boxes. The basic helices were the most highly conserved region between the two proteins and were responsible for the RNA-binding ability and the formation of RNA granules.
RNG140, as well as RNG105, was most highly expressed in the brain. However, the time of their expression was different; expression of RNG105 was high in embryos but that of RNG140 was high in adults.
RNA granules that contain RNG105 or RNG140 were also distinct structures: RNG140-localizing RNA granules did not contain ribosomes, G3BP, or RNG105 in proliferating cells and in neurons. Furthermore, RNG140-induced granules were not stress-inducible, unlike RNG105-induced granules. These results indicated that RNG140-localizing RNA granules were distinct from stress granules or neuronal RNA granules where RNG105 was localized. In addition, RNG140-localizing RNA granules did not contain DCP2 or XRN1, suggesting that they were not P bodies either.
Thus, it was found that RNG140 and RNG105 are localized to distinct RNA granules and are different in spatiotemporal expression patterns in the brain. Present results that the suppression of RNG105 and RNG140 in cultured neurons reduced dendrite length and spine density suggested that these paralogs are involved in the maintenance and/or development of dendrites and spines at distinct location and time in vertebrate neurons.
Domain Structures of RNG105 and RNG140
In the highly conserved basic helices among RNG105, RNG140, and RNGI, hydrophobic amino acids are arranged in a stripe on one side, and basic amino acids are arranged in clusters on the other side. The basic clusters may serve as a docking surface for nucleic acids, as in the case of aminoacyl-tRNA synthetases, signal recognition particle, and basic helix-loop-helix proteins (18–20). In fact, deletion of the basic helices of RNG105 and RNG140 resulted in a marked decrease in their RNA-binding abilities (Fig. 2A) (9).
RGG box is an RNA-binding motif conserved among a variety of RNA-binding proteins (21). The RGG boxes of RNG105 and RNG140 appear to play minor roles in RNA-binding in vitro (Fig. 2A) (9) but major roles in the formation of RNA granules in cultured cells (supplemental Fig. 1) (9).
Glutamic acid-rich and glutamine-rich regions are contained in RNG105 but not conserved in RNG140 or RNGI, although acidic and glutamine-clusters are present in RNGI. Glutamine-rich regions have been reported in some of RNA- and DNA-binding proteins to participate in protein-protein interaction (22, 23). The glutamic acid-rich region is highly acidic and may be involved in electrostatic interaction with some proteins.
The C1q globular domain is contained only in RNG140. The C1q globular domain is found in many extracellular proteins, playing roles in homo- or heterotrimerization and in targeting to specific extracellular receptors (24, 25). In addition to the extracellular proteins, three intracellular proteins containing the C1q globular domain have been reported, including RNG140 (EEG-1L) (12, 24). The C1q globular domain inside the cell is suggested to play a role as a scaffolding element and be involved in the formation of complexes to regulate signaling cascades (24).
Thus, glutamic acid-rich and glutamine-rich regions are specific to RNG105, whereas the C1q globular domain is specific to RNG140. As all of these domains are likely to participate in protein-protein interactions, it is possible that the RNG105- and the RNG140-specific domains may mediate the formation of distinct complexes for RNG105 and RNG140.
Translation Inhibition in Vitro and in Vivo
RNG140 as well as RNG105 bound to mRNAs in a sequence-independent manner and inhibited translation in vitro. However, it has been reported that knock-out of RNG105 (caprin-1) in chicken DT40 cells does not increase global rates of translation (11). The knock-out phenotype that was explained by the effects of RNG105 on translation may be regulated by posttranslational modification or may be dependent on mRNA and associated proteins in vivo (11). Although translation inhibition activity was a common in vitro feature of RNG105 and RNG140, detailed studies on the effects of RNG105 and RNG140 on mRNAs in vivo are required to answer the question of whether RNG105 and RNG140 act as translational repressors.
RNG105- and RNG140-containing RNA Granules
The expression of RNG105 and RNG140 in cultured cells induced the formation of RNA granules. The granule induction was likely to be dependent on the amount of the exogenously expressed proteins. Although ∼85% of RNG105-expressing cells contained RNA granules, only ∼8% of RNG140-expressing cells contained RNA granules. In the other cells, RNG105 and RNG140 showed diffuse cytoplasmic distributions. A possible reason for the low frequency of the cells containing RNG140-induced RNA granules was that the overexpression of RNG140 increased the level of apoptosis (12). Because the expression level of RNG140 was high in cells containing RNG140-induced RNA granules, such cells may be susceptible to cell death. However, in the adult brain, the RNG140 expression level was high, and RNG140-localizing RNA granules were formed without apparent cell death. A possible notion was that the formation of RNG140-localizing RNA granules and the susceptibility to cell death may depend on the state of the cells, e.g. the cells were differentiated or not.
The present study showed that RNG140-localizing granules were mRNA-containing RNA granules that were related to repression of translation but were distinct from RNG105-localizing stress granules or neuronal RNA granules. The RNG140-localizing granules may not be P bodies because they did not contain the enzymes for mRNA decay. RNG140-localizing RNA granules may be like “transport particles” in yeast that do not contain ribosomes (6) or a novel kind of RNA granule. Detailed studies such as the identification of RNG140-associated proteins and mRNAs would be required.
Functional Aspects of RNG105 and RNG140
Expression of RNG105 (caprin-1) reportedly was up-regulated when T and B lymphoblasts are activated (10). Expression of RNG140 (EEG-1L) was reported to be up-regulated when erythroblasts shift from a proliferative state toward their terminal phase of differentiation (12). The expression timings of RNG105 and RNG140 suggested a possibility that they may be involved in the regulation of translation at the onset of differentiation by binding to specific mRNAs. On the other hand, there is a report that RNG140 (caprin-2) participates in the Wnt signaling pathway by regulating the phosphorylation of low density lipoprotein receptor-related proteins 5 and 6 (26). This report may suggest another possible role of RNG140 besides acting as an RNA-binding protein.
RNG105/RNG140/RNGI is conserved in deuterostomes and insects but was not found in yeast or Caenorhabditis elegans. In vertebrates, e.g. frogs and rodents, both RNG105 and RNG140 were most highly expressed in the brain (this study and 9). These facts suggested a possibility that RNG105, RNG140, and possibly RNGI are involved in functions of the central nervous system. In fact, in the present study, RNG105 and RNG140 were demonstrated to be localized to dendrites and postsynapses in neurons and play roles in the maintenance and/or development of dendrites and postsynaptic spines, suggesting their involvement in the proper networking of neurons.
RNG105 knock-out mice were generated previously, and they were shown to die soon after birth because of respiratory failure, suggesting defects in the brainstem.4 Further lines of evidence were obtained that RNG105 was responsible for the formation of synapses and neuronal networks.4 These results are consistent with the idea that RNG105 is involved in the function of embryonic brain. In contrast to RNG105, RNG140 was highly expressed in the brain of adults, suggesting a possibility that RNG140 has roles that are specific in the brain of adult vertebrates. Moreover, the effects of RNG140 knockdown on dendrites and spines were more severe than those of RNG105 knockdown and were not rescued by RNG105 expression, suggesting a possibility that RNG140 plays roles in the maintenance of dendritic structure through different mechanisms from RNG105. Further studies such as the identification of RNG140-associated mRNAs and RNG140 knock-out in mice would further elucidate these possibilities.
Supplementary Material
Supplemental Data
Acknowledgments
We thank K. Shinkura and K. Takada for assistance, Dr. M. Kiledjian for anti-DCP2 antibody, Dr. W. D. Heyer for anti-XRN1 antibody, and Dr. J. Ortin for anti-staufen antibody.
*
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N. S. and M. T.).
4
N. Shiina and M. Tokunaga, unpublished observations.
3
The abbreviations used are:
P bodies
processing bodies
RNG
RNA granule protein
RNGI
RNG invertebrate
GST
glutathione _S_-transferase
FMRP
fragile X mental retardation protein
GFP
green fluorescent protein
mRFP
monomeric red fluorescent protein
siRNA
small interfering RNA
RNAi
RNA interference.
REFERENCES
- 1.Kloc M., Zearfoss N. R., Etkin L. D. (2002) Cell 108, 533–544 [DOI] [PubMed] [Google Scholar]
- 2.Kindler S., Wang H., Richter D., Tiedge H. (2005) Annu. Rev. Cell Dev. Biol. 21, 223–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamasaki S., Anderson P. (2008) Curr. Opin. Cell Biol. 20, 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiebler M. A., Bassell G. J. (2006) Neuron 51, 685–690 [DOI] [PubMed] [Google Scholar]
- 6.Sossin W. S., DesGroseillers L. (2006) Traffic 7, 1581–1589 [DOI] [PubMed] [Google Scholar]
- 7.Anderson P., Kedersha N. (2006) J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker R., Sheth U. (2007) Mol. Cell 25, 635–646 [DOI] [PubMed] [Google Scholar]
- 9.Shiina N., Shinkura K., Tokunaga M. (2005) J. Neurosci. 25, 4420–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grill B., Wilson G. M., Zhang K. X., Wang B., Doyonnas R., Quadroni M., Schrader J. W. (2004) J. Immunol. 172, 2389–2400 [DOI] [PubMed] [Google Scholar]
- 11.Solomon S., Xu Y., Wang B., David M. D., Schubert P., Kennedy D., Schrader J. W. (2007) Mol. Cell. Biol. 27, 2324–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aerbajinai W., Lee Y. T., Wojda U., Barr V. A., Miller J. L. (2004) J. Biol. Chem. 279, 1916–1921 [DOI] [PubMed] [Google Scholar]
- 13.Shiina N., Tsukita S. (1999) Mol. Biol. Cell 10, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid K. B., Colomb M., Petry F., Loos M. (2002) Trends Immunol. 23, 115–117 [DOI] [PubMed] [Google Scholar]
- 15.Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. (2000) J. Cell Biol. 151, 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles R. B., Sabry J. H., Martone M. E., Deerinck T. J., Ellisman M. H., Bassell G. J., Kosik K. S. (1996) J. Neurosci. 16, 7812–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steward O., Levy W. B. (1982) J. Neurosci. 2, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cahuzac B., Berthonneau E., Birlirakis N., Guittet E., Mirande M. (2000) EMBO J. 19, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupert P. B., Ferré-D'amaré A. R. (2000) Structure 8, R99–R104 [DOI] [PubMed] [Google Scholar]
- 20.Nelson H. C. M. (1995) Curr. Opin. Genet. Dev. 5, 180–189 [DOI] [PubMed] [Google Scholar]
- 21.Burd C. G., Dreyfuss G. (1994) Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 22.Ström A. C., Forsberg M., Lillhager P., Westin G. (1996) Nucleic Acids Res. 24, 1981–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Förch P., Puig O., Martínez C., Séraphin B., Valcárcel J. (2002) EMBO J. 21, 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Innamorati G., Bianchi E., Whang M. I. (2006) Cell Signal. 18, 761–770 [DOI] [PubMed] [Google Scholar]
- 25.Tacnet P., Cheong E. C., Goeltz P., Ghebrehiwet B., Arlaud G. J., Liu X. Y., Lesieur C. (2008) Biochim. Biophys. Acta. 1784, 518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Y., Xi Y., Chen T., Wang J. Y., Tao D. L., Wu Z. L., Li Y. P., Li C., Zeng R., Li L. (2008) J. Cell Biol. 182, 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data