HIV-1 Protease Mutations and Protease Inhibitor Cross-Resistance (original) (raw)
Abstract
The effects of many protease inhibitor (PI)-selected mutations on the susceptibility to individual PIs are unknown. We analyzed in vitro susceptibility test results on 2,725 HIV-1 protease isolates. More than 2,400 isolates had been tested for susceptibility to fosamprenavir, indinavir, nelfinavir, and saquinavir; 2,130 isolates had been tested for susceptibility to lopinavir; 1,644 isolates had been tested for susceptibility to atazanavir; 1,265 isolates had been tested for susceptibility to tipranavir; and 642 isolates had been tested for susceptibility to darunavir. We applied least-angle regression (LARS) to the 200 most common mutations in the data set and identified a set of 46 mutations associated with decreased PI susceptibility of which 40 were not polymorphic in the eight most common HIV-1 group M subtypes. We then used least-squares regression to ascertain the relative contribution of each of these 46 mutations. The median number of mutations associated with decreased susceptibility to each PI was 28 (range, 19 to 32), and the median number of mutations associated with increased susceptibility to each PI was 2.5 (range, 1 to 8). Of the mutations with the greatest effect on PI susceptibility, I84AV was associated with decreased susceptibility to eight PIs; V32I, G48V, I54ALMSTV, V82F, and L90M were associated with decreased susceptibility to six to seven PIs; I47A, G48M, I50V, L76V, V82ST, and N88S were associated with decreased susceptibility to four to five PIs; and D30N, I50L, and V82AL were associated with decreased susceptibility to fewer than four PIs. This study underscores the greater impact of nonpolymorphic mutations compared with polymorphic mutations on decreased PI susceptibility and provides a comprehensive quantitative assessment of the effects of individual mutations on susceptibility to the eight clinically available PIs.
HIV-1 protease inhibitors (PIs) are the mainstays of salvage therapy. As the number of licensed PIs has increased, it has become important to identify whether and how each PI-selected mutation affects cross-resistance to each of the other PIs. In a previous study (41), we previously applied several data mining approaches to assess associations between HIV-1 protease genotype and phenotype test results for the first-generation PIs: amprenavir (APV), the active component of the prodrug fosamprenavir (FPV), atazanavir (ATV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), and saquinavir (SQV). Specifically, we used a data set containing about 300 susceptibility results for ATV, 500 for LPV, and 800 for FPV, IDV, NFV, and SQV (41). We used a predefined list of PI-selected mutations in this previous study to reduce the number of independent variables influencing PI susceptibility.
Here we analyze a data set that contains between 1,600 and 2,600 isolates tested for susceptibility to the first-generation PIs and about 600 and 1,200 isolates tested for susceptibility to darunavir (DRV) and tipranavir (TPV), respectively. We use two regression methods in tandem: one to identify genotypic predictors of decreased susceptibility and one to quantify the impact of specific mutations on decreased PI susceptibility. We then integrate our findings with previously published data linking protease mutations to decreases in susceptibility to specific PIs.
MATERIALS AND METHODS
HIV-1 isolates.
We analyzed HIV-1 isolates in the HIV Drug Resistance Database (HIVDB) (40) for which cell-based in vitro PI susceptibility testing had been performed by the PhenoSense (Monogram, South San Francisco, CA) (38) or Antivirogram (Virco, Mechelin, Belgium) (19) assays. Seventy percent of the test results were obtained from published studies, and 30% were obtained from virus samples collected at Stanford University or one of several collaborating clinics. Of the test results from published studies, about one-half were obtained in clinical trials performed by Boehringer Ingelheim (3), Tibotec (26), Gilead Sciences (30), and Abbott Laboratories (23). The Stanford University Human Subjects Committee approved this study.
Drug susceptibility results were expressed as the fold change in susceptibility, defined as the ratio of the 50% inhibitory concentration (IC50) of the tested isolate to that of a standard wild-type control isolate. We used the STAR method to assign an HIV-1 subtype to isolates for which nucleotide sequences were available (32). We obtained subtypes for the remaining isolates from the studies from which the phenotype data were obtained or, for the collaborating clinics, from the phenotype report. Mutations were defined as differences from the consensus subtype B amino acid sequence (http://hivdb.stanford.edu/pages/documentPage/consensus_amino_acid_sequences.html). Nonpolymorphic mutations were defined as mutations that occurred at a prevalence of ≤0.5% in the eight most common HIV-1 subtypes (4).
To minimize bias that would result from overrepresenting highly similar viruses, we excluded 200 redundant viruses—defined as viruses obtained from the same person with the same mutations at the following PI resistance positions: positions 30, 32, 46, 47, 54, 76, 82, 84, and 90 (20). Because the presence of mixtures at influential drug resistance positions may confound genotype-phenotype correlations, we excluded 472 viruses with sequences containing electrophoretic mixtures at these protease positions.
Identification of mutations associated with decreased PI susceptibility.
We used two independent regression analyses in tandem: least-angle regression (LARS) was performed to identify protease mutations associated with reduced susceptibility to at least one PI, and least-squares regression was performed to quantify the contribution of protease mutations to reduced PI susceptibility. Protease mutations for which the mean regression coefficient determined by LARS was ≥0.5 or ≤−0.5 and more than 3 standard deviations above or below zero in 10 repeated runs of 5-fold cross-validation were considered significant predictors. All of the significant predictors, including the ones that were significant for only one PI, were retained for our second regression analysis using least-squares regression.
LARS is a model selection algorithm for selecting a parsimonious set for the efficient prediction of a response variable from a large collection of possible explanatory variables (16). We applied LARS to the 200 most common mutations in our data set. Each of these mutations occurred in 20 or more sequences in the data set, although some of these mutations occurred in fewer than 20 sequences for TPV and DRV. Eight regression models—one for each PI—were created. In these models, each of the 200 mutations was an explanatory variable, and the log of the fold change in susceptibility was the response variable. For each 5-fold cross-validation, 80% of data were used for learning regression coefficients and 20% were used for testing. During learning regression coefficients, LARS used four-fifths of the learning data for selecting a model and one-fifth for validating the selected model using the LASSO option. LARS constructs a model by first finding the mutation most correlated with the log fold and then incrementally builds the model by following the equiangular vector until another variable is equally correlated with the residual. The validation set (the one-fifth of learning data) is used to decide when to stop adding variables to the model. The regularization parameter was chosen as the smallest parameter whose mean cross-validation error was less than or equal to the minimum cross-validation error plus 1 standard deviation of the LARS cross-validation error at the minimum.
Quantification of the contributions of protease mutations to decreased PI susceptibility.
In the least-squares regression analyses, the mutations identified by LARS were explanatory variables and the log fold change in susceptibility was the response variable. We used weighted least-squares regression to minimize the potential influence of data obtained by two different phenotypic methods. Weighting was performed by fitting the complete data set (without cross-validation) and then calculating the fitted mean-squared error (MSEfitted) for each of the phenotypic methods (Antivirogram and PhenoSense). We then weighted the contribution of each sequence by 1/MSEfitted for each of the eight regression models. For each weighted regression model, we performed 5-fold cross-validation 10 times on different subdivisions of the complete data set. During each 5-fold cross-validation, 80% of the data were used to derive the coefficient of each genotypic predictor and 20% were used to test the prediction performance of the model.
The mutations for which the mean regression coefficients from the 10 repeated runs of 5-fold cross-validation exceeded 3 standard deviations above or below zero were considered statistically significant. Due to the disparity in the numbers of genotype-phenotype results available for each PI, we also required that the mean regression coefficient equal or exceed 0.5 to be considered significantly associated with decreased susceptibility to a specific PI. Without this additional criterion, there would be more mutations associated with reduced susceptibilities to older PIs simply because the greater numbers of available results for these PIs would make even very weak associations appear significant. We also standardized the mutation regression coefficients by dividing each coefficient by the square root of the sum of squares of the residuals obtained during the learning stages of each cycle of learning and cross-validation. Standardizing the regression coefficients made it possible to obtain unbiased comparisons of the magnitude of the coefficient for a protease mutation across PIs.
Prediction performance was evaluated during cross-validation using continuous and categorical approaches on 20% of the testing data set. The continuous approach involved calculating the mean-squared error between the actual and predicted log-fold (MSE) change in susceptibility. The categorical approach involved determining how often the predicted phenotype correlated with one of three predefined categories of susceptibility that we refer to as susceptible, low/intermediate, and high-level resistance. The predefined susceptibility categories for each PI were derived in advance to approximate the geometric mean of the published estimated clinical cutoffs provided with the PhenoSense and VircoType reports (Table 1).
TABLE 1.
Phenotypic cutoffs used in this study to assess classification accuracy compared with the clinical cutoffs reported by the Antivirogram and PhenoSense assays
| Druga | Antivirogram assayb | PhenoSense assayc | Cutoffs for assessing classification accuracy in our analysis | |||
|---|---|---|---|---|---|---|
| Low cutoff | High cutoff | Low cutoff | High cutoff | Low cutoff | High cutoff | |
| ATV/r | 2.5 | 32.5 | NA | NA | 3.0 | 15 |
| DRV/r | 10.0 | 106.9 | 10.0 | 90 | 10.0 | 90 |
| FPV/r | 1.5 | 19.5 | 4.0 | 11 | 3.0 | 15 |
| IDV/r | 2.3 | 27.2 | NA | NA | 3.0 | 15 |
| LPV/r | 6.1 | 51.2 | 9.0 | 55 | 9.0 | 55 |
| NFV | 2.2 | 9.4 | NA | NA | 3.0 | 6.0 |
| SQV/r | 3.1 | 22.6 | 2.3 | 12 | 3.0 | 15 |
| TPV/r | 1.5 | 7.0 | 2.0 | 8.0 | 2.0 | 8.0 |
Corroboration with other sources of published data.
We compared our findings on PI susceptibility with three sources of published data: (i) previous publications showing that a protease mutation was selected in vitro or in vivo with a specific PI or directly decreased in vitro PI susceptibility in a well-characterized set of virus samples or in a site-directed mutagenesis study; (ii) the published genotypic susceptibility scores for darunavir and tipranavir developed by Tibotec and Boeringer-Ingelheim, respectively (12, 13, 25); and (iii) a linear regression analysis of genotypes and Antivirogram phenotypes in the VircoLab database published in 2007 (46). The VircoLab linear regression analysis generated a table with 39 mutations at 22 protease positions showing the estimated contribution of each mutation to a decrease or increase in the fold change susceptibility (referred to as fold change factor) of either 1 to 1.5, 1.5 to 2, or >2. Associations in our analysis were considered to be corroborated by the VircoLab analysis for protease mutations having the highest possible fold change factor (>2) for a PI.
RESULTS
Summary of PI susceptibility results.
In vitro drug susceptibility results were available for 2,725 isolates, including 2,643 from 2,439 individuals and 82 mutant laboratory isolates. Fifty-six percent (1,525) of isolates had been tested by the PhenoSense assay and 44% (1,200) by the Antivirogram assay. Table 2 shows the number of isolates analyzed by each assay for susceptibility to each of the eight PIs. More than 2,400 genotype-phenotype correlations were available for FPV, IDV, NFV, and SQV. A total of 2,130 genotype-phenotype correlations were available for LPV, 1,644 for ATV, 1,265 for TPV, and 642 for DRV. Fifty-five percent of PhenoSense results were classified as susceptible, 18% were classified as low-intermediate, and 27% were classified as high level resistant. Comparable numbers for the Antivirogram assay were 41% susceptible, 21% low-intermediate, and 38% high level resistant. Ninety-two percent (2,500) of the isolates were categorized as subtype B; 8% (225) were categorized as a non-B subtype.
TABLE 2.
Total number of drug susceptibility results according to PI and susceptibility assays
| Druga | No. of isolates analyzed by the following assay: | Total no. of isolates analyzed | |
|---|---|---|---|
| Antivirogram | PhenoSense | ||
| ATV | 744 | 900 | 1,644 |
| DRV | 277 | 365 | 642 |
| FPV | 1,054 | 1,416 | 2,470 |
| IDV | 1,130 | 1,450 | 2,580 |
| LPV | 973 | 1,157 | 2,130 |
| NFV | 1,153 | 1,488 | 2,641 |
| SQV | 1,159 | 1,457 | 2,616 |
| TPV | 684 | 581 | 1,265 |
| Meanb | 897 | 1,102 | 1,999 |
Mutations identified by least-angle regression as predictive of PI resistance.
Among the 200 mutations that occurred 20 or more times in our data set, LARS identified 46 mutations at 26 positions as statistically significant predictors of decreased susceptibility to one or more PIs. These mutations included L10FI, V11L, K20T, L24FI, D30N, V32I, L33F, E35GN, K43T, M46IL, I47AV, G48MV, I50LV, F53L, I54ALMSTV, Q58E, G73CST, T74PS, L76V, V82AFLST, N83D, I84AV, N88DS, L89V, and L90M. The median number of mutations per sample was 4.0. Figure S1 in the supplemental material shows the number of other mutations occurring with each mutation. Table S1 in the supplemental material lists the number of times each combination of mutations occurred in the data set.
Among the 46 significant mutations, 40 were nonpolymorphic mutations in the eight most common subtypes. In contrast, only 14 of the 154 nonsignificant mutations were not polymorphic in these eight subtypes. The nonpolymorphic significant mutations included L10I (which occurs in 1.8% to 9.3% of all subtypes), L33F (0.7% in subtype A and 1.4% in subtype CRF01_AE), E35G (1.1% in subtype A, 1.2% in subtype G, and 2.4% in subtype CRF02_AG), K43T (0.6% in subtype F), Q58E (0.9% in subtype D), and T74S (0.6% to 8% in all subtypes except subtypes B and D) (40).
The prevalence of each of the 46 mutations in the susceptibility data set was highly correlated with the prevalence of these mutations in sequences from more than 15,000 PI-treated individuals in the Stanford HIV Drug Resistance Database (_R_2 = 0.87; P < 0.001) (40) (see Fig. S2 in the supplemental material).
Least-squares regression prediction performance.
Table 3 summarizes the prediction performance of weighted least-squares regression using the 46 mutations identified by LARS. The MSE of 50 trials (5-fold cross-validation performed 10 times) per PI ranged from 0.10 to 0.19, with standard deviation of 0.01 to 0.02 (Table 3). For the PIs with the lowest MSE (FPV and LPV), the predicted fold value was on average 100.10 (1.3) times higher or lower than the actual fold value. For the PI with the highest MSE (SQV), the predicted fold value was on average 100.19 (1.5) times higher or lower than the actual fold value. The classification accuracy ranged from 0.73 for TPV to 0.90 for NFV. In general, PIs with a low MSE had high classification accuracy. However, this was not always the case. The classification accuracy depended on the proportion of phenotypic results that were close to the classification cutoffs and thus more prone to misclassification. Table 3 also shows the prediction performances using the results of each assay separately. With one exception (PhenoSense LPV results), the performance of the weighted least-squares regression was superior or equal to the results obtained solely using one assay.
TABLE 3.
Mean squared error (MSE) and classification accuracy of least-squares regression (LSR) using PhenoSense and Antivirogram data sets separately and in combination
| Druga | Mean squared errorb | Classification accuracyb,c | ||||
|---|---|---|---|---|---|---|
| PhenoSense and LSR | Antivirogram and LSR | Combined, weighted LSRd | PhenoSense and LSR | Antivirogram and LSR | Combined, weighted LSRd | |
| ATV | 0.14 ± 0.01 | 0.14 ± 0.01 | 0.13 ± 0.01 | 0.83 ± 0.02 | 0.82 ± 0.02 | 0.83 ± 0.02 |
| DRV | 0.15 ± 0.03 | 0.14 ± 0.03 | 0.11 ± 0.02 | 0.85 ± 0.03 | 0.86 ± 0.03 | 0.87 ± 0.03 |
| FPV | 0.11 ± 0.01 | 0.11 ± 0.01 | 0.10 ± 0.01 | 0.82 ± 0.01 | 0.81 ± 0.02 | 0.82 ± 0.02 |
| IDV | 0.12 ± 0.01 | 0.12 ± 0.01 | 0.11 ± 0.01 | 0.81 ± 0.02 | 0.82 ± 0.02 | 0.82 ± 0.02 |
| LPV | 0.12 ± 0.01 | 0.11 ± 0.01 | 0.10 ± 0.01 | 0.78 ± 0.02 | 0.75 ± 0.02 | 0.77 ± 0.02 |
| NFV | 0.14 ± 0.01 | 0.14 ± 0.01 | 0.13 ± 0.01 | 0.89 ± 0.01 | 0.90 ± 0.01 | 0.90 ± 0.01 |
| SQV | 0.23 ± 0.02 | 0.21 ± 0.02 | 0.19 ± 0.02 | 0.82 ± 0.02 | 0.80 ± 0.02 | 0.82 ± 0.02 |
| TPV | 0.19 ± 0.02 | 0.15 ± 0.02 | 0.15 ± 0.02 | 0.69 ± 0.03 | 0.73 ± 0.02 | 0.73 ± 0.02 |
| Avg | 0.15 | 0.14 | 0.13 | 0.81 | 0.81 | 0.82 |
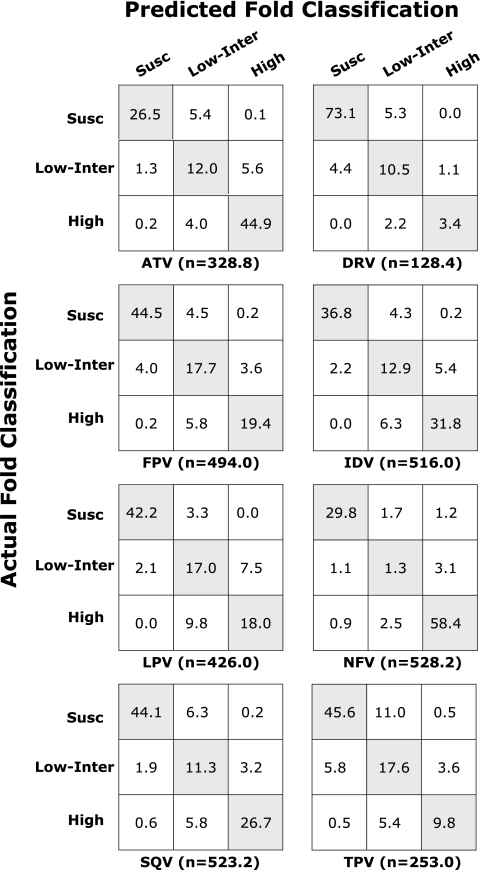
Figure 1 illustrates the classification accuracy for each PI. It contains eight tables (also known as “confusion matrices”) in which each cell contains the mean proportion of results for which the actual and predicted phenotypes were concordant in 10× 5-fold cross-validation. The numbers shown beneath each table represent the mean number of results in each test set (e.g., one-fifth the total number of genotype-phenotype correlations for each PI). For example, the test sets for ATV averaged 328.8 genotype-phenotype correlations of which 32% (sum of values in the first row) were susceptible, 18.9% (sum of values in the second row) were low-intermediate, and 49.1% (sum of values in the third row) were high level resistant. Of the 328.8 genotype-phenotype correlations, the weighted least-squares regression (LSR) model's predictions were concordant with the actual results for 83.4% (the values in the shaded diagonal cells); the predictions and actual results were completely discordant for 0.3% (i.e., susceptible versus high level resistant) and were partially discordant for 16.3% (i.e., intermediate versus susceptible or intermediate versus high-level). The classification accuracy shown in Table 3 was calculated as the sum of the values in the shaded diagonal.
FIG. 1.
Confusion matrices of six PIs containing the number of folds according to the classification of actual fold and the classification of predicted fold. The y axis indicates the susceptible (Susc), intermediate-low (Low-Inter), and high resistance (High) levels of the actual fold. The x axis indicates the susceptible (Susc), intermediate-low (Low-Inter), and high resistance (High) levels of the predicted fold. Each cell contains the average number of folds belonging to corresponding classifications of the actual and predicted fold from 50 test sets (5-fold cross-validation 10 times). The average number of genotype-phenotype correlations in 50 test sets for each drug is indicated (n). The classification accuracy for each test was calculated by the number of correctly predicted fold classification (which is the sum of the diagonal cells) divided by the total number of folds (which is the sum of all the cells). The average classification accuracy over 50 test sets is shown in Table 3. Drug abbreviations: ATV, atazanavir; DRV, darunavir; FPV, fosamprenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; SQV, saquinavir; TPV, tipranavir.
Contributions of protease mutations to decreased PI susceptibility.
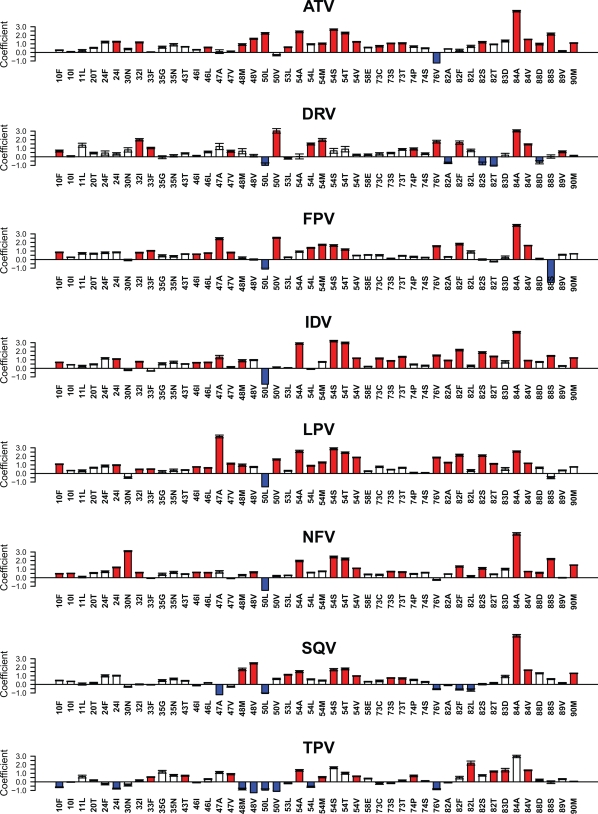
In the least-squares regression analysis, the 46 mutations at 26 positions had a regression coefficient of ≥0.5 for one or more PIs. The median number of mutations associated with decreased susceptibility to each PI was 28 (range, 19 to 32): 19 for DRV, 19 for TPV, 21 for SQV, 27 for FPV, 29 for NFV, 31 for LPV, 31 for IDV, and 32 for ATV. The median number of mutations associated with increased susceptibility to each PI was 2.5 (range, 1 to 8): one each for NFV, ATV, and IDV; two for FPV; three for LPV; four for DRV; five for SQV; and eight for TPV. Figure 2 shows the regression coefficients for each of the 46 mutations with each of the PIs. Mutations with corroborative data supporting an association with decreased susceptibility are indicated with red bars. Table S2 in the supplemental material contains the mean regression coefficients and the standard deviations for each of the 46 mutations for each of the eight drugs.
FIG. 2.
Graphical representation of the regression coefficients of the weighted least-squares regression (LSR) model for predicting changes in protease inhibitor susceptibility using genotypic predictors. For each mutation, the y axis indicates the magnitude of the mean coefficient of 50 LSR runs (10 repetitions of 5-fold cross-validation), and the error bar indicates the standard deviation from the mean. Positive coefficients indicate mutations that decrease drug susceptibility. Negative coefficients indicate mutations that increase drug susceptibility. The y axis has no units because coefficients were normalized. The red bars indicate mutations for which we identified corroborative data supporting the association between the mutation and decreased PI susceptibility. Drug abbreviations: ATV, atazanavir; DRV, darunavir; FPV, fosamprenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; SQV, saquinavir; TPV, tipranavir.
Eleven of the 32 mutations associated with decreased ATV susceptibility were previously reported to be selected by ATV in vivo or in vitro or to decrease ATV susceptibility: V32I, M46L, G48V, I50L, I54V, G73ST, I84AV, N88S, and L90M (5, 6, 18, 28, 31, 45, 51). Additionally, L24I, G48M, F53L, I54AMST, G73C, V82S, and N88D had a fold change factor of >2 in the VircoLab analysis (46).
Ten of the 19 mutations associated with decreased DRV susceptibility were previously reported to be selected in vivo by DRV or to decrease the virological response to DRV salvage therapy: V32I, L33F, I47V, I50V, I54LM, T74P, L76V, I84V, and L89V (11, 13). Two additional mutations (L10F and V82F) were shown to decrease DRV susceptibility (44). Additionally, I84A had a fold change factor of >2 in the VircoLab analysis (46).
Fourteen of the 27 mutations associated with decreased FPV susceptibility were previously reported to have been selected by APV in vivo or in vitro or to decrease APV susceptibility: L10F, V32I, L33F, M46IL, I47AV, I50V, I54LM, L76V, V82F, and I84AV (1, 10, 21, 27, 29, 31, 34, 36). Additionally, I54ST had a fold change factor of >2 in the VircoLab analysis (46).
Fifteen of the 31 mutations associated with decreased IDV susceptibility were previously reported to have been selected by IDV in vivo or in vitro or to decrease IDV susceptibility: L10F, L24I, V32I, M46IL, I54V, G73S, L76V, V82ATF, I84AV, N88S, and L90M (7, 31, 40, 43, 50). Additionally, I47A, G48M, I54ATS, G73CT, and V82S had a fold change factor of >2 in the VircoLab analysis (46).
Nineteen of the 31 mutations associated with decreased LPV susceptibility were previously reported to be selected by LPV in vitro or in vivo or to decrease LPV susceptibility: L10F, L24I, V32I, L33F, M46IL, I47AV I50V, I54LMV, L76V, V82AFST, and I84AV (10, 14, 17, 21, 23, 31, 34, 35). Additionally, G48M, and I54AST had a fold change factor of >2 in the VircoLab analysis (46).
Fourteen of the 30 mutations associated with decreased NFV susceptibility were previously reported to be selected by NFV in vitro or in vivo or to decrease NFV susceptibility: L10FI, D30N, M46IL, G48V, I54V, G73S, V82F, I84AV, N88DS, and L90M (2, 31, 37, 49, 50). Additionally, L24I, V32I, I54AST, G73T, and V82S had a fold change factor of >2 in the VircoLab analysis (46).
Eight of the 21 mutations associated with decreased SQV susceptibility were previously reported to be selected by SQV in vitro or in vivo or to decrease SQV susceptibility: G48V, F53L, I54TV, G73S, I84AV, and L90M (9, 31, 40, 47, 49). Additionally, G48M, I54AS, and G73T had a fold change factor of >2 in the VircoLab analysis (46).
Eleven of the 19 mutations associated with decreased TPV were previously reported to be selected by TPV in vitro or in vivo or to decrease the virological response to TPV salvage therapy (L33F, K43T, I47V, I54AMV, T74P, V82LT, N83D, and I84V) (3, 15, 25).
PI cross-resistance.
Of the mutations with a regression coefficient above 1.5, I84AV were associated with decreased susceptibility to eight PIs; V32I, G48V, I54ALMSTV, V82F, and L90M were associated with decreased susceptibility to six or seven PIs; I47A, G48M, I50V, L76V, V82ST, and N88S were associated with decreased susceptibility to four or five PIs; and D30N, I50L, and V82AL were associated with decreased susceptibility to fewer than four PIs.
At seven protease positions, positions 10, 47, 50, 54, 82, 84, and 88, different mutations at the same position had markedly different effects on PI susceptibility—defined here as a difference in regression coefficients of >1.0 for at least one PI. In some cases, one mutation was associated with resistance to a PI, while a different amino acid change at the same position was associated with increased susceptibility to that PI. Conversely, mutations at six protease positions—positions 24, 35, 46, 48, 73, and 74—conferred similar effects on PI susceptibility regardless of the specific amino acid change. At positions 11, 20, 30, 32, 33, 43, 53, 58, 76, 89, and 90, only a single mutation was associated with decreased PI susceptibility.
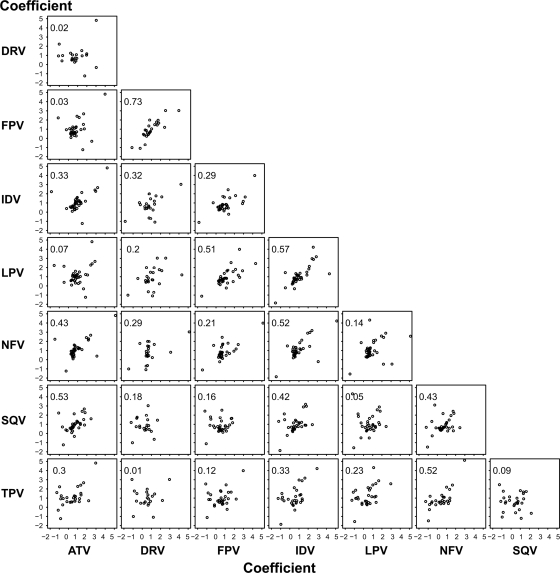
Figure 3 shows the correlation between the regression coefficients for each pair of PIs. The regression coefficients of the mutations associated with decreased susceptibility to DRV and FPV (_R_2 = 0.73; P < 0.001), IDV and LPV (_R_2 = 0.57; P < 0.001), and ATV and SQV (_R_2 = 0.53; P < 0.001) were most strongly correlated. The regression coefficients of the mutations associated with decreased susceptibility to DRV and TPV had the lowest correlation (_R_2 = 0.01; not statistically significant).
FIG. 3.
Correlation between the regression coefficients for 46 protease mutations for each pair of PIs. Each plot shows the regression coefficient for the 46 protease mutations. The x axis indicates the magnitude of regression coefficient of the PI denoted along the bottom of the figure. The y axis indicates the magnitude of regression coefficient of the PI denoted on the left side of the figure. The correlation coefficient (_R_2) for the values of the 46 regression coefficients between each pair of PIs is also shown on each plot. Drug abbreviations: ATV, atazanavir; DRV, darunavir; FPV, fosamprenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; SQV, saquinavir; TPV, tipranavir.
DISCUSSION
We used two independent regression analyses to assess the contributions of individual HIV-1 protease mutations to PI susceptibility. The first regression analysis, LARS, employs a parsimonious feature selection algorithm. It identified a set of 46 mutations significantly associated with decreased PI susceptibility of which 40 were not polymorphic in the eight most common group M subtypes. The second analysis relied on least-squares regression to quantify the effects of these 46 mutations on susceptibility to each of the PIs. We used LARS for the first regression analysis because it is optimal for feature selection. We used least-squares regression for the second analysis because the theoretical basis for weighting data produced by different assays is established for least-squares regression but not for LARS. In addition, the use of one regression method for feature selection and another for model optimization reduces the likelihood of overfitting the regression coefficients.
This study differs in several ways from a study of genotype-phenotype correlations we published in 2006 (41). First, the current study includes nearly three times as much data for the first-generation PIs. Second, the current study contains more than 600 genotype-phenotype correlations for TPV and DRV—PIs that were not included in our previous analysis. Third, the current study uses an unbiased feature selection approach to identify genotypic predictors of resistance. In contrast, our 2006 study used a predefined list of nonpolymorphic treatment-selected mutations (39). Fifteen mutations in this study were not significantly independently associated with decreased susceptibility to one or more PIs in the 2006 study; these included L10I, V11L, L24F, E35GN, I47A, G48M, I54AST, G73C, V82SL, N83D, and I84A. L10I was not evaluated in the 2006 study because it did not include mutations that were polymorphic in subtype B viruses. The remaining additional mutations were not included in the 2006 study because of their infrequent occurrence in that data set.
Despite the increase in the number of genotype-phenotype correlations in this study over the 2006 study, predictions of fold decreased susceptibility (mean-squared error) and classification accuracies for the PIs ATV, FPV, IDV, LPV, NFV, and SQV were only slightly improved. The classification accuracies increased on average from 0.81 to 0.83. There are several likely explanations for what appears to be a plateau in prediction accuracy despite an increasing number of genotype-phenotype correlations. First, our analysis did not include three types of mutations that influence PI susceptibility: (i) rare mutations present too infrequently to be included in our regression analysis, including several highly unusual mutations at known PI resistance positions, such as G48AST, V82CM, and I84C (42); (ii) common polymorphic mutations, which alone have little if any effect on PI susceptibility but which influence susceptibility in combination with other PI resistance mutations; and (iii) gag and gag cleavage site mutations, which often have a significant effect on in vitro PI susceptibility (8, 24, 33). Second, our analysis did not use combinations of mutations (i.e., interaction terms) as predictors of reduced susceptibility because the use of interaction terms and of more complex models relating mutations to decreased susceptibility (e.g., neural networks or support vector machines) would have made it more difficult to understand the contributions of individual mutations.
The mutations identified in our analysis were consistent with data from other publications. An average of 13 mutations per PI had a regression coefficient of ≥1.0; 88% of these associations were reported in other publications. An average of 15 mutations per PI had a regression coefficient between 0.5 and 1.0; 43% of these associations were reported in other publications. The less frequent published support for mutations with lower regression coefficients may reflect the lesser effect of these mutations on decreased susceptibility and/or the lower frequency of these mutations. Collinearity may also inflate or distort the regression coefficients associated with some mutations. The most striking example in this data set was L24F which occurred with L90M in 26 of 27 instances. Similarly, I54S or T occurred with V82A in 69 of 81 instances and with G48V in 65 of 81 instances. Because of their relative infrequency and their linkage with other mutations, independent support for I54S or T was present for six PIs only in the VircoLab regression analysis (46).
Our analysis found that several mutations originally identified because of their association with one PI were equally or more strongly associated with a different PI. L76V, which was originally recognized because of its association with decreased virological response to DRV (11, 13), was also strongly associated with decreased susceptibility to FPV, IDV, and LPV. F53L, originally identified because of its association with decreased virological response to LPV (22, 23), was more strongly associated with decreased susceptibility to ATV and SQV. N83D, which was identified because of its association with decreased virological response to TPV (25), was also associated with decreased susceptibility to ATV, NFV, and SQV. L89V, which was identified because of its association with decreased virological response to DRV (12, 13), was found to also be associated with decreased susceptibility to FPV.
In addition to identifying each of the previously reported associations between the protease mutations L24I, I47A, I50L, I50V, I54L, L76V, and N88S and increased PI susceptibility (21, 25, 48, 50), our analysis identified another eight mutations associated with increased PI susceptibility, including L10F and G48MV for TPV, V82AST for DRV, and V82FL for SQV. Two of these new associations, G48V increasing TPV susceptibility and V82F increasing SQV susceptibility, were also detected in the VircoLab analysis (46). PI “hypersusceptibility” mutations provide insight into the mechanisms of PI activity. However, the use of hypersusceptibility mutations to guide therapy has been approached warily out of concern that the potential benefit of the PIs made more active by these mutations would be negated by the emergence of residual wild-type variants within a patient.
Two types of data other than genotype-phenotype associations contribute to our understanding of the genetic basis for HIV-1 PI resistance: (i) associations between protease mutations and selective PI pressure such as occurs during in vitro passage experiments and in viruses from PI-treated individuals; and (ii) associations between protease mutations and the subsequent virological response to a PI-containing regimen. However, with the increasing number of antiretroviral (ARV) treatment options, fewer PI-containing salvage regimens are unsuccessful and fewer virus sequences from PI-treated individuals are available. Therefore, genotype-phenotype associations have become increasingly important for unraveling the genetic basis of HIV-1 PI resistance.
In conclusion, this study is the first comprehensive quantitative analysis of protease mutations associated with decreased susceptibility to each of the eight licensed PIs. It is also the first analysis in which previously published corroborative support for each genotype-phenotype association is provided. Our analysis shows that the protease mutations with the greatest impact on PI susceptibility are those that are not polymorphic in the absence of PI therapy. Our analysis also shows that protease mutations found in the course of investigating resistance to one PI should be evaluated for their effects on susceptibility to other PIs. Without such an evaluation, the extent of cross-resistance among PIs will be underestimated.
Supplementary Material
[Supplemental material]
Acknowledgments
S.-Y.R., J.T., and R.W.S. were supported in part by two NIH grants (AI06858 and 5P01GM066524-08).
Footnotes
▿
Published ahead of print on 26 July 2010.
‡
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ait-Khaled, M., A. Rakik, P. Griffin, C. Stone, N. Richards, D. Thomas, J. Falloon, and M. Tisdale. 2003. HIV-1 reverse transcriptase and protease resistance mutations selected during 16-72 weeks of therapy in isolates from antiretroviral therapy-experienced patients receiving abacavir/efavirenz/ amprenavir in the CNA2007 study. Antivir. Ther. 8**:**111-120. [PubMed] [Google Scholar]
- 2.Atkinson, B., J. Isaacson, M. Knowles, E. Mazabel, and A. K. Patick. 2000. Correlation between human immunodeficiency virus genotypic resistance and virologic response in patients receiving nelfinavir monotherapy or nelfinavir with lamivudine and zidovudine. J. Infect. Dis. 182**:**420-427. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80**:**10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, D. E., R. J. Camacho, D. Otelea, D. R. Kuritzkes, H. Fleury, M. Kiuchi, W. Heneine, R. Kantor, M. R. Jordan, J. M. Schapiro, A. M. Vandamme, P. Sandstrom, C. A. Boucher, D. van de Vijver, S. Y. Rhee, T. F. Liu, D. Pillay, and R. W. Shafer. 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4**:**e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bristol-Myers Squibb Company. 2009. Reyetaz prescribing information. Bristol-Myers Squibb Company, New York, NY. http://www.bms.com.
- 6.Colonno, R., R. Rose, C. McLaren, A. Thiry, N. Parkin, and J. Friborg. 2004. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J. Infect. Dis. 189**:**1802-1810. [DOI] [PubMed] [Google Scholar]
- 7.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70**:**8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75**:**589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig, C., E. Race, J. Sheldon, L. Whittaker, S. Gilbert, A. Moffatt, J. Rose, S. Dissanayeke, G. W. Chirn, I. B. Duncan, and N. Cammack. 1998. HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS 12**:**1611-1618. [DOI] [PubMed] [Google Scholar]
- 10.De Meyer, S., H. Azijn, D. Surleraux, D. Jochmans, A. Tahri, R. Pauwels, P. Wigerinck, and M. P. de Bethune. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49**:**2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Meyer, S., I. Dierynck, E. Lathouwers, B. Baalen, T. Vangeneugden, S. Spinosa-Guzman, G. Picchio, and M. P. de Bethune. 2008. Identification of mutations predictive of a diminished response to darunavir/ritonavir: analysis of data from treatment-experienced patients in POWER 1, 2, 3, and DUET-1 and DUET-2, abstr. 54. Sixth European HIV Drug Resistance Workshop, Budapest, Hungary.
- 12.De Meyer, S., I. Dierynck, E. Lathouwers, B. Van Baelen, T. Vangeneugden, S. Spinosa-Guzman, M. Peeters, G. Picchio, and M. de Bethune. 2008. Phenotypic and genotypic determinants of resistance to darunavir: analysis of data from treatment experienced patients in POWER 1, 2, 3 and DUET-1 and 2 [abstract 33]. Antivir. Ther. 13(Suppl. 3)**:**A33. [Google Scholar]
- 13.De Meyer, S., A. Hill, G. Picchio, R. DeMasi, E. De Paepe, and M. P. de Bethune. 2008. Influence of baseline protease inhibitor resistance on the efficacy of darunavir/ritonavir or lopinavir/ritonavir in the TITAN trial. J. Acquir. Immune Defic. Syndr. 49**:**563-564. [DOI] [PubMed] [Google Scholar]
- 14.De Meyer, S., E. Lathouwers, I. Dierynck, E. De Paepe, B. Van Baelen, T. Vangeneugden, S. Spinosa-Guzman, E. Lefebvre, G. Picchio, and M. P. de Bethune. 2009. Characterization of virologic failure patients on darunavir/ritonavir in treatment-experienced patients. AIDS 23**:**1829-1840. [DOI] [PubMed] [Google Scholar]
- 15.Doyon, L., S. Tremblay, L. Bourgon, E. Wardrop, and M. G. Cordingley. 2005. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antiviral Res. 68**:**27-35. [DOI] [PubMed] [Google Scholar]
- 16.Efron, B., T. Hastie, I. Johnstone, and R. Tibshirani. 2004. Least angle regression. Ann. Stat. 32**:**407-499. [Google Scholar]
- 17.Ghosn, J., P. Flandre, I. Cohen-Codar, P. M. Girard, M. L. Chaix, F. Raffi, P. Dellamonica, P. Ngovan, M. Norton, and J. F. Delfraissy. 2010. Long-term (96-week) follow-up of antiretroviral-naive HIV-infected patients treated with first-line lopinavir/ritonavir monotherapy in the MONARK trial. HIV Med. 11**:**137-142. [DOI] [PubMed] [Google Scholar]
- 18.Gong, Y. F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P. F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44**:**2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42**:**269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2009. Update of the drug resistance mutations in HIV-1: December 2009. Top. HIV Med. 17**:**138-145. [PubMed] [Google Scholar]
- 21.Kagan, R. M., M. D. Shenderovich, P. N. Heseltine, and K. Ramnarayan. 2005. Structural analysis of an HIV-1 protease I47A mutant resistant to the protease inhibitor lopinavir. Protein Sci. 14**:**1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, J. Sylte, B. Richards, B. Bernstein, R. Rode, and E. Sun. 2002. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir. Ther. 7**:**165-174. [PubMed] [Google Scholar]
- 23.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75**:**7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolli, M., S. Lastere, and C. A. Schiffer. 2006. Co-evolution of nelfinavir-resistant HIV-1 protease and the p1-p6 substrate. Virology 347**:**405-409. [DOI] [PubMed] [Google Scholar]
- 25.Llibre, J. M., J. M. Schapiro, and B. Clotet. 2010. Clinical implications of genotypic resistance to the newer antiretroviral drugs in HIV-1-infected patients with virological failure. Clin. Infect. Dis. 50**:**872-881. [DOI] [PubMed] [Google Scholar]
- 26.Madruga, J. V., P. Cahn, B. Grinsztejn, R. Haubrich, J. Lalezari, A. Mills, G. Pialoux, T. Wilkin, M. Peeters, J. Vingerhoets, G. de Smedt, L. Leopold, R. Trefiglio, and B. Woodfall. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370**:**29-38. [DOI] [PubMed] [Google Scholar]
- 27.Maguire, M., D. Shortino, A. Klein, W. Harris, V. Manohitharajah, M. Tisdale, R. Elston, J. Yeo, S. Randall, F. Xu, H. Parker, J. May, and W. Snowden. 2002. Emergence of resistance to protease inhibitor amprenavir in human immunodeficiency virus type 1-infected patients: selection of four alternative viral protease genotypes and influence of viral susceptibility to coadministered reverse transcriptase nucleoside inhibitors. Antimicrob. Agents Chemother. 46**:**731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malan, D. R., E. Krantz, N. David, V. Wirtz, J. Hammond, and D. McGrath. 2008. Efficacy and safety of atazanavir, with or without ritonavir, as part of once-daily highly active antiretroviral therapy regimens in antiretroviral-naive patients. J. Acquir. Immune Defic. Syndr. 47**:**161-167. [DOI] [PubMed] [Google Scholar]
- 29.Marcelin, A. G., D. Affolabi, C. Lamotte, H. A. Mohand, C. Delaugerre, M. Wirden, D. Voujon, P. Bossi, N. Ktorza, F. Bricaire, D. Costagliola, C. Katlama, G. Peytavin, and V. Calvez. 2004. Resistance profiles observed in virological failures after 24 weeks of amprenavir/ritonavir containing regimen in protease inhibitor experienced patients. J. Med. Virol. 74**:**16-20. [DOI] [PubMed] [Google Scholar]
- 30.Margot, N. A., E. Isaacson, I. McGowan, A. K. Cheng, R. T. Schooley, and M. D. Miller. 2002. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 16**:**1227-1235. [DOI] [PubMed] [Google Scholar]
- 31.Mo, H., N. Parkin, K. D. Stewart, L. Lu, T. Dekhtyar, D. J. Kempf, and A. Molla. 2007. Identification and structural characterization of I84C and I84A mutations that are associated with high-level resistance to human immunodeficiency virus protease inhibitors and impair viral replication. Antimicrob. Agents Chemother. 51**:**732-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers, R. E., C. V. Gale, A. Harrison, Y. Takeuchi, and P. Kellam. 2005. A statistical model for HIV-1 sequence classification using the subtype analyser (STAR). Bioinformatics 21**:**3535-3540. [DOI] [PubMed] [Google Scholar]
- 33.Nijhuis, M., N. van Maarseveen, D. de Jong, P. Schipper, I. Goedegebuure, and C. Boucher. 2006. Substitutions within Gag but outside the cleavage sites can cause protease inhibitor resistance. Antivir. Ther. 11**:**S149. [Google Scholar]
- 34.Nijhuis, M., A. M. Wensing, W. F. Bierman, D. de Jong, R. Kagan, A. Fun, C. A. Jaspers, K. A. Schurink, M. A. van Agtmael, and C. A. Boucher. 2009. Failure of treatment with first-line lopinavir boosted with ritonavir can be explained by novel resistance pathways with protease mutation 76V. J. Infect. Dis. 200**:**698-709. [DOI] [PubMed] [Google Scholar]
- 35.Parkin, N. T., C. Chappey, and C. J. Petropoulos. 2003. Improving lopinavir genotype algorithm through phenotype correlations: novel mutation patterns and amprenavir cross-resistance. AIDS 17**:**955-961. [DOI] [PubMed] [Google Scholar]
- 36.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, et al. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69**:**5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patick, A. K., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40**:**292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44**:**920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee, S. Y., W. J. Fessel, A. R. Zolopa, L. Hurley, T. Liu, J. Taylor, D. P. Nguyen, S. Slome, D. Klein, M. Horberg, J. Flamm, S. Follansbee, J. M. Schapiro, and R. W. Shafer. 2005. HIV-1 protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J. Infect. Dis. 192**:**456-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee, S. Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31**:**298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee, S. Y., J. Taylor, G. Wadhera, A. Ben-Hur, D. L. Brutlag, and R. W. Shafer. 2006. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 103**:**17355-17360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahriar, R., S. Y. Rhee, T. F. Liu, W. J. Fessel, A. Scarsella, W. Towner, S. P. Holmes, A. R. Zolopa, and R. W. Shafer. 2009. Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrob. Agents Chemother. 53**:**4869-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tisdale, M., R. E. Myers, B. Maschera, N. R. Parry, N. M. Oliver, and E. D. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 39**:**1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Marck, H., I. Dierynck, G. Kraus, S. Hallenberger, T. Pattery, G. Muyldermans, L. Geeraert, L. Borozdina, R. Bonesteel, C. Aston, E. Shaw, Q. Chen, C. Martinez, V. Koka, J. Lee, E. Chi, M. P. de Bethune, and K. Hertogs. 2009. The impact of individual human immunodeficiency virus type 1 protease mutations on drug susceptibility is highly influenced by complex interactions with the background protease sequence. J. Virol. 83**:**9512-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vergani, B., M. L. Cicero, O. Vigano, F. Sirianni, S. Ferramosca, P. Vitiello, P. Di Vincenzo, M. Pia De Pasquale, M. Galli, and S. Rusconi. 2008. Evolution of the HIV-1 protease region in heavily pretreated HIV-1 infected patients receiving Atazanavir. J. Clin. Virol. 41**:**154-159. [DOI] [PubMed] [Google Scholar]
- 46.Vermeiren, H., E. Van Craenenbroeck, P. Alen, L. Bacheler, G. Picchio, and P. Lecocq. 2007. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods 145**:**47-55. [DOI] [PubMed] [Google Scholar]
- 47.Watkins, T., W. Resch, D. Irlbeck, and R. Swanstrom. 2003. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 47**:**759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinheimer, S., L. Discotto, J. Friborg, H. Yang, and R. Colonno. 2005. Atazanavir signature I50L resistance substitution accounts for unique phenotype of increased susceptibility to other protease inhibitors in a variety of human immunodeficiency virus type 1 genetic backbones. Antimicrob. Agents Chemother. 49**:**3816-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72**:**5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziermann, R., K. Limoli, K. Das, E. Arnold, C. J. Petropoulos, and N. T. Parkin. 2000. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to amprenavir. J. Virol. 74**:**4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zolopa, A., W. Towner, D. Butcher, S. Wang, J. Maa, and D. Seekins. 2007. Resistance profile after viral rebound on atazanavir-containing therapy: focus on protease inhibitor-naive subjects in the IMPACT study (BMS AI424-128) [abstract 77]. Antivir. Ther. 12**:**S86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]