Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis (original) (raw)
. Author manuscript; available in PMC: 2010 Oct 29.
Summary
The enediyne antibiotic calicheamicin (CLM) γ1I is a prominent antitumor agent that is targeted to DNA by a novel aryltetrasaccharide comprised of an aromatic unit and four unusual carbohydrates. Herein we report the heterologous expression and the biochemical characterization of the two ‘internal’ glycosyltransferases CalG3 and CalG2 and the first structural elucidation of an enediyne glycosyltransferase (CalG3). In conjunction with the previous characterization of the ‘external’ CLM GTs CalG1 and CalG4, this study completes the functional assignment of all four CLM GTs, extends the utility of enediyne GT-catalyzed reaction reversibility, and presents conclusive evidence of a sequential glycosylation pathway in CLM biosynthesis. This work also reveals the common GT-B structural fold can now be extended to include enediyne GTs.
Keywords: enediyne, cancer, glycosyltransferase, carbohydrate, biosynthesis, crystallography
Introduction
Calicheamicin (CLM) γ1I (Fig. 1, 1) from Micromonospora echinospora spp. calichensis is a prominent member of the enediyne family due to its unprecedented molecular architecture, remarkable mechanism of action, and clinical utility (Galm et al., 2005; Thorson et al., 2000; Van Lanen and Shen, 2008). Structurally, CLM is a member of the 10-membered enediynes which all share a signature bicyclo[7.3.1]enediyne core.Like all enediynes, CLM-induced oxidative DNA strand scission is enabled by rapid enediyne cycloaromatization to form a highly reactive diradical species (Zein et al., 1989; Zein et al., 1988). This reactive intermediate is exquisitely positioned by the CLM aryltetrasaccharide (Fig. 1), the critical DNA docking element of CLM (Kumar et al., 1997; Walker et al., 1993). The incredible potency of CLM has also been harnessed for clinical use via conjugation to tumor-targeting antibodies, as exemplified by the γ-CD33 antibody conjugate (Mylotarg) approved by FDA in 2000 to treat acute myelogenous leukemia (AML) (Sievers and Linenberger, 2001). Similarly-appended CLM-antibody conjugates to treat other cancers are steadily progressing through clinical trials (Boghaert et al., 2004; DiJoseph et al., 2005; Hamann et al., 2005).
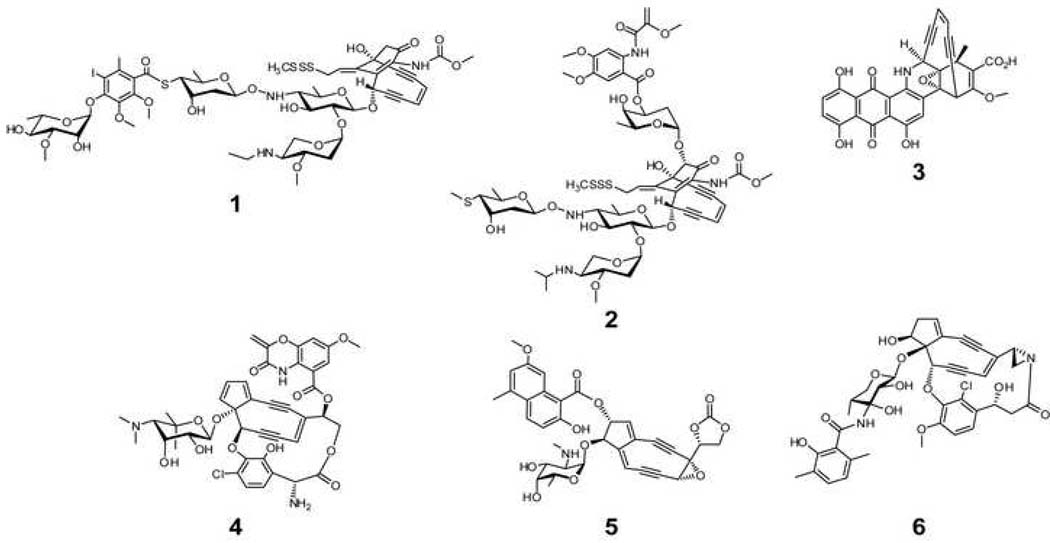
Fig. 1. Representative naturally occurring enediynes.
ncluding the 10-membered enediynes calicheamcin γ1I (1), esperamicin (2), dynemicin (3) and 9-membered chromoprotein enediynes C-1027 (4), neocarzinostatin (5), and maduropeptin (6). The CLM aryltetrasaccharide is highlighted in blue.
Consistent with their many novel structural and pharmacological features, enediyne biosynthetic pathways are also rich with unique enzyme-catalyzed biotransformations. Early metabolic labeling studies suggested the 9- and 10-membered enediynes to derive from distinct biosynthetic pathways (Hensens et al., 1989; Lam et al., 1993; Tokiwa et al., 1992). In contrast, the recent cloning and characterization of gene clusters encoding both 9-membered – including C-1027 (Fig. 1, 4) (Liu et al., 2002), neocarzinostatin (Fig. 1, 5) (Liu et al., 2005), and maduropeptin (Fig. 1, 6) (Van Lanen et al., 2007) - and 10-membered – including CLM (Ahlert et al., 2002), esperamicin (Fig. 1, 2) (Ahlert, Shepard and Thorson, unpublished, AY267372), and dynemicin (Fig. 1, 3) (Gao and Thorson, 2008; Zazopoulos et al., 2003) - enediynes revealed a unified, divergent polyketide paradigm for enediyne core biosynthesis (Ahlert et al., 2002; Liu et al., 2002; Zhang et al., 2008). Some enediyne-producing organisms also rely upon a novel ‘self-sacrifice’ resistance protein (as exemplified by the CLM protein, CalC) for enediyne self-resistance (Biggins et al., 2003; Singh et al., 2006). Shen and coworkers were the first to demonstrate the elegant application of pathway engineering to produce chromoprotein enediyne analogs with drastically differing activities (Kennedy et al., 2007a; Kennedy et al., 2007b; Liu et al., 2002) while ‘sugar exchange’ and ‘aglycon exchange’ reactions catalyzed by the CLM glycosyltransferases (GTs) CalG1 and CalG4 recently enabled the production of more than 70 differentially-glycosylated CLM variants (Zhang et al., 2006b). While this latter study also provided in vitro biochemical characterization of CalG1 and CalG4 as the CLM 3-_O_-methyl-rhamnosyltransferase and aminopentosyltransferase, respectively (Zhang et al., 2006b), the function of the remaining CLM glycosyltransferases CalG2 and CalG3 remain unresolved. In addition, while the structures for a variety of natural product-associated glycosyltransferase have emerged in recent years (Bolam et al., 2007; Mittler et al., 2007; Mulichak et al., 2003; Mulichak et al., 2001; Mulichak et al., 2004), enediyne GTs remain structurally uncharacterized (Van Lanen and Shen, 2008).
Herein we report the further study of the ‘internal’ stages of CLM glycosylation. Specifically, using a combination of GT reversibility and sugar nucleotide surrogates, CalG3 was verified as the requisite calicheamicinone 4,6-dideoxy-4-hydroxylamino-α-D-glucosyltransferase and demonstrated to accept a set of 10 alternative sugar nucleotide donors. The structural studies highlighted herein, also revealed, for the first time, that enediyne GTs such as CalG3 also adopt a GT-B fold common to natural product GTs. This structural study also illuminated key catalytic residues and snapshots of a dynamic loop anticipated to participate in NDP-binding. The application of a surrogate sugar nucleotide also enabled the confirmation of CalG2 as the remaining ‘internal’ GT – the 4-deoxy-thio-α-D-digitoxosyltransferase – and the first characterized hydroxylamino glycosidic bond-forming GT. In conjunction with our previous report (Zhang et al., 2006b), this study completes the functional assignment of all four CLM GTs, extends the concept of reversibility of enediyne GT-catalyzed reactions, and highlights the first crystal structure of an enediyne GT.
Results
Overexpression and purification of CalG3 and CalG2
Analysis of the CLM biosynthetic gene cluster from M. echinospora revealed four putative GT-encoded genes, calG1, calG2, calG3 and calG4 (Ahlert et al., 2002), implicating a distinct GT for each sugar of the CLM aryltetrasaccharide. Consistent with this, in vitro biochemical characterization confirmed CalG1 and CalG4 as the CLM 3-_O_-methyl-rhamnosyltransferase and aminopentosyltransferase, respectively (Zhang et al., 2006b). Analysis of CalG3 revealed highest homology to characterized GTs which operate upon aromatic acceptors, such as the nogalamycin SnogD (37% identity) (Torkkell et al., 1997) and elloramycin ElmGT (36% identity) (Blanco et al., 2001), while CalG2 more closely resembled GTs which act upon carbohydrate acceptors including the CLM CalG4 (50% identity) (Zhang et al., 2006b) and avermectin AveBI (42% identity) (Wohlert et al., 2001; Zhang et al., 2006a). Based upon this simple analysis, CalG3 was proposed as the putative calicheamicinone 4,6-dideoxy-4-hydroxylamino-α-D-glucosyltransferase and CalG2 postulated as the subsequent 4-deoxy-thio-α-D-digitoxosyltransferase (anticipated to form the signature CLM hydroxylamino glycosidic bond). To complete the CLM GT annotation, recombinant _N_-His10-CalG2 and _N_-His10-CalG3 fusion proteins were overproduced in E. coli and subsequently purified by nickel-affinity chromatography to >95% homogeneity (Fig. S1) with overall yields of 10–15 mg per liter of culture.
Preparation of the putative acceptor CLM T0
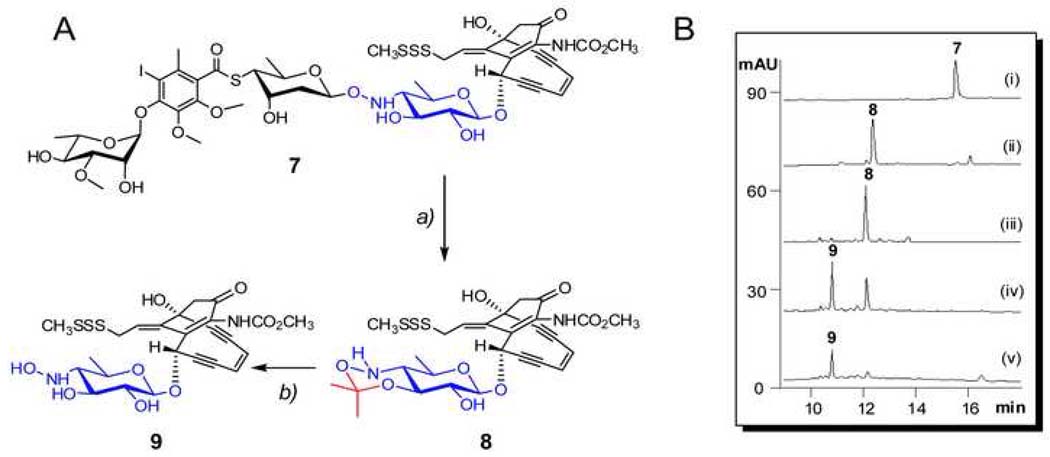
The availability of an appropriate acceptor was critical to the in vitro characterization of CalG2 and CalG3. To address this issue, the truncated CLM analogue CLM T0 (Fig. 2A, 9) was prepared via a slight modification of a literature procedure (Walker et al., 1992). Specifically, CLM α3I (Fig. 2A, 7 and Fig. 2B, i) was refluxed in wet acetone with pyridine _p_-toluenesulfonate and the reaction monitored via HPLC. After refluxing for 19 h, a product clearly distinct from starting material, but with the characteristic CLM core UV signature, was generated (Fig. 2B, ii). The product was purified (Fig. 2B, iii) and determined to have a mass of 625.1 [M+H] by APCI-MS analysis - 40 daltons greater than expected product 9. Subsequent 1H NMR analysis (Fig. S2) revealed the new product to be 8 (Fig. 2A), an isopropylidene adduct formed by the dehydration of acetone in the presence of tosylate. Reasoning that the adduct could be easily removed under mild acidic conditions, 8 was incubated with 0.2% TFA to provide 9 in 4 h (Fig. 2B, iv and v). The identity of 9 was verified by high resolution ESI-MS ([M+Na]+, m/z C24H28N2NaO9S3, calc. 607.0855, found 607.0866) and 1H NMR (Fig. S3).
Fig. 2. Preparation of CLM T0 (9) from CLM α3I (7).
(A) Schematic of the strategy – (a) refluxing in acetone, 65°C, 20 h; (b) incubated in 0.2% TFA, RT, 4 h. (B) HPLC analyses of the preparation - (i) the starting material 7; (ii) refluxed for 20 h; (iii) purified 8 from the reaction mixture of (ii); (iv) 8 incubated with 0.2% TFA at RT for 2 h; and (iv) 8 incubated with 0.2% TFA at RT for 4 h.
Reversibility of CalG3-catalyzed reaction
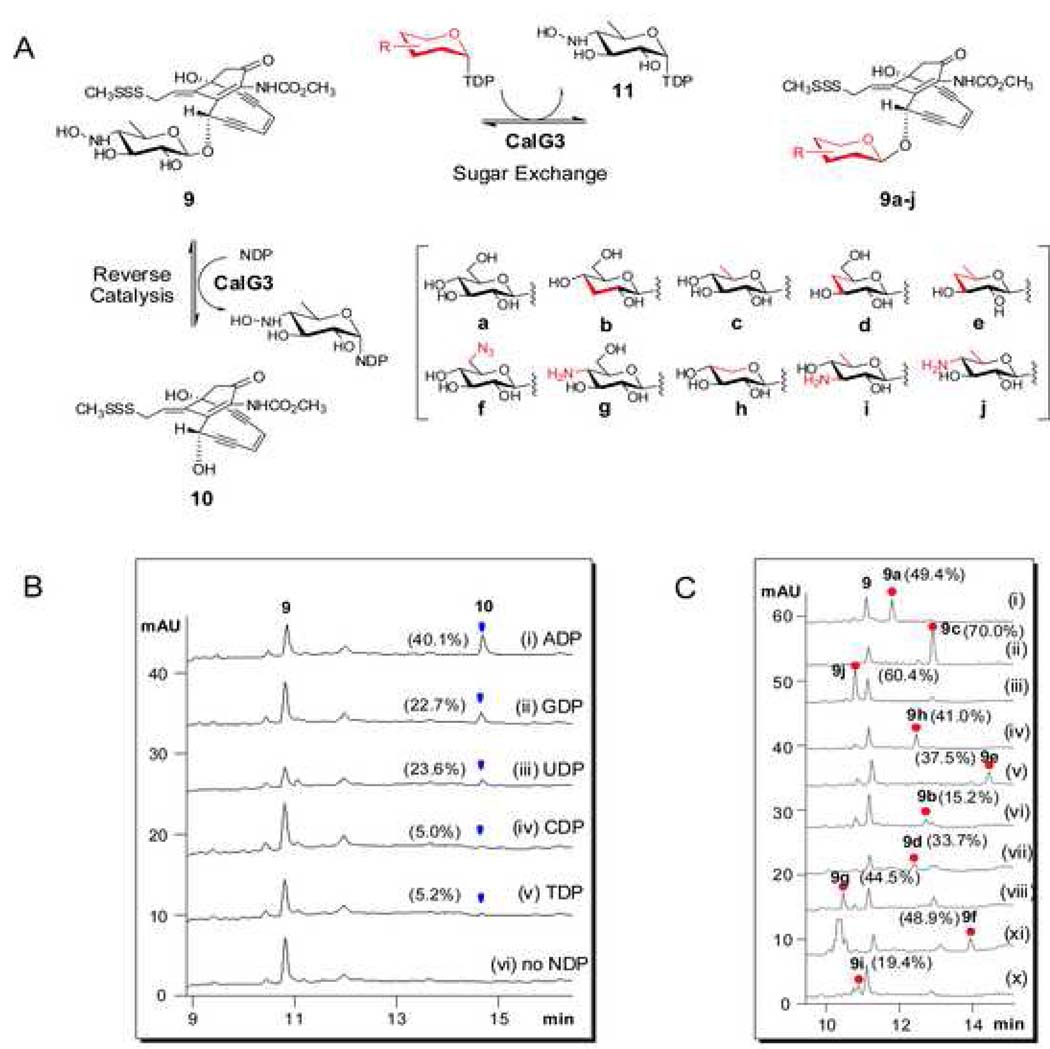
Following recently established protocols for GT reversibility (Minami et al., 2005; Zhang et al., 2006a; Zhang et al., 2006b; Zhang et al., 2007), CalG3 and CalG2 were first investigated for their ability to catalyze the excision of 4,6-dideoxy-4-hydroxylamino-α-D-glucosyl moiety from CLM T0 (9) (Fig. 3A). No reaction was observed with 9 (50 µM) and CalG2 (7.5 µM) in the presence of 2 mM NDP (ADP, CDP, GDP, UDP or TDP) in Tris-HCl (10 mM, pH 7.6). In contrast, the incubation of 9 (50 µM) with CalG3 (7.5 µM) in the presence of various NDPs (2 mM) led to new product after 2 h at 30 °C (Fig. 3B, i–v). This transformation was determined to be both CalG3- and NDP-dependent (Fig. 3B, vi) and the new product was subsequently identified as the deglycosylated calicheamicinone (10, Fig. 3A) by LC-MS (calcd. 423, found 446.0 [M+Na]+ and 422 [M-H]−) (Table S1). Unlike prior reports on the reversibility of GT-catalyzed reactions which indicated such transformations to be NDP-specific, CalG3 reversibility was surprisingly observed with ADP (conversion rate of 40%; Fig. 3B, i), GDP (23%; Fig. 3B, ii), UDP (24%; Fig. 3B, iii), TDP (trace; Fig. 3B, iv) and even CDP (trace; Fig. 3B, v). The apparent reversibility of this reaction was also enhanced at lower pH (Fig. S4A). Cumulatively, these studies highlight the clear reversibility of the CalG3-catalyzed reaction and are consistent with CalG3 as the requisite 4,6-dideoxy-4-hydroxylamino-α-D-glucosyltransferase involved in 1 biosynthesis.
Fig. 3. CalG3-catalyzed reverse reaction and ‘sugar exchange’ reaction.
(A) Schematic of the CalG3-catalyzed formation of 10 from 9 via reverse catalysis and the production of 10 novel CLM variants 9a–j via ‘sugar exchange’. (B) HPLC analyses of CalG3-catalyzed reverse reactions. In these reactions, 50 µM 9 was incubated with 7.5 µM CalG3 for 2 h at 30 °C in the presence of (i) 2 mM ADP; (ii) 2 mM GDP; (iii) 2 mM UDP; (iv) 2 mM CDP; (v) 2 mM TDP and (vi) no NDP. Percent conversions were indicated in the parentheses. (C) The production of 9a–j via CalG3-catalyzed ‘sugar exchange’. 50 µM 9 was incubated with 7.5 µM CalG3 at 30°C overnight in the presence of various TDP-sugars (300 µM, Fig. S4). Percent conversions were indicated in the parentheses.
CalG3-catalyzed ‘sugar exchange’
Given the lack of calicheamicinone availability (10, Fig. 3A), the CalG3 sugar nucleotide specificity was alternatively probed with 9 as an acceptor for putative GT-catalyzed ‘sugar-exchange’ reactions. In a GT-catalyzed sugar exchange reaction, first observed in the context of CalG1 catalysis (Zhang et al., 2006b), the native sugar of a natural glycoside can be substituted in situ by unnatural sugars supplied as NDP-sugar donors. Five commercially available NDP-glucoses, including ADP-, CDP-, GDP-, UDP- and TDP-Glc were examined in the CalG3 ‘sugar exchange’ reaction with 9. Remarkably, all 5 NDP-glucoses were established as CalG3 donor substrates, albeit with varying ‘sugar exchange’ efficiencies in the end point assay (50 µM 9, 2 mM NDP-Glc, 7.5 µM CalG3, 30 °C overnight). Interestingly, TDP-glucose exhibited the highest conversion rate of 9 to 9a (49.3%), followed by UDP-Glc (36.0%), CDP-Glc (25.3%), GDP-Glc (10.6%) and ADP-Glc (9.8%). In contrast to the influence of pH upon reaction reversibility, the CalG3-catalyzed sugar exchange reaction was enhanced at higher pH (Fig. S4B and Fig. S4C).
The sugar exchange promiscuity of CalG3 was subsequently probed directly with 9 and a small library of 22 TDP-sugars (Fig. S5) comprised of 20 TDP-D-sugars (including commercially available TDP-α-D-glucose and unnatural sugar nucleotides generated via chemoenzymatic synthesis with functionality variations such as deoxy, amino, and azido at the sugar C2, C3, C4, C5 or C6 positions) and two TDP-L-sugars (TDP-β-L-rhamnose and TDP-α-L-rhamnose). From this substrate specificity analysis, 10 sugar nucleotides were identified as CalG3 substrates (Fig. 3A & 3C, i–x) to ultimately provide 10 novel CLM variants 9a–j (Fig. 3A, Table S1) with ‘sugar exchange’ conversions ranging from 15 to 70% (Fig. 3C). Cumulatively, these studies revealed CalG3 to be a relatively promiscuous GT and further highlighted GT-catalyzed ‘sugar exchange’ as an expeditious method for natural product diversification.
CalG3 structure
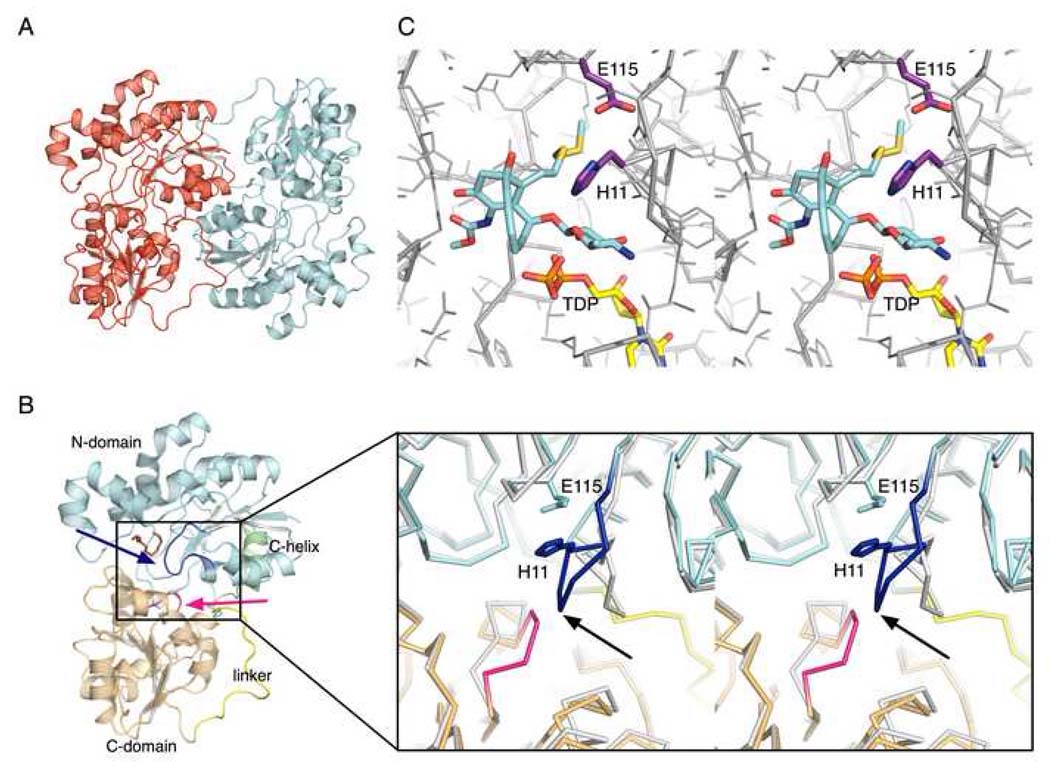
The structure of CalG3 was determined by single wavelength anomalous diffraction using a 2.8 Å dataset collected from SeMet-labeled crystals (from I222 crystal form) and refined against this dataset as well as a native dataset to a resolution of 1.9 Å (from P21 crystal form). Data collection, phasing and refinement statistics are summarized in Table I. Consistent with gel filtration, asymmetric units of both crystal forms revealed two molecules of CalG3 arranged into dimers with C2 symmetry (Fig. 4A). Analysis of the binding interface of these dimers by the PISA server (Krissinel and Henrick, 2007) revealed the total buried surface area for the complex of 3300–3600 Å2. The dimers of both crystal forms align with an all-atom rmsd of 0.76 Å, while all observed monomer conformers from both crystal forms align with rmsd ~0.48 Å. These results confirm that CalG3 forms dimers with a closely similar quaternary arrangement in both crystal forms and this form of protein thus may be relevant in vivo. A detailed backbone comparison of the CalG3 molecules found in the two crystal forms revealed that several secondary structure elements undergo slight shifts; these include surface exposed segments containing residues 54–76, 205–212, and 219–226. In addition, a tetraglycine loop spanning residues 285–288 (Fig. 4B, pink) undergoes a conformational change.
Table I.
Crystal parameters, X-ray data collection, phasing, and refinement statistics.
| SeMet | Native | |
|---|---|---|
| Crystal parameters | ||
| Space group | 1222 | P21 |
| Unit-cell parameters (Å, °) | a = 106.7, b = 119.3, c = 156.0 | a = 57.4, b = 97.7,c = 63.0 |
| β = 90.6 | ||
| Data collection statistics | ||
| Wavelength (Å) | 0.97918 | 0.97918 |
| Energy (eV) | 12,662 | 12,662 |
| Resolution range (Å) | 26.75–2.80 (2.90–2.80) | 38.76–1.68 (1.74–1.68) |
| No. of reflections (measured/unique)a | 180890/24883 | 482455/68513 |
| Completeness (%) | 99.2 (94.9) | 86.3 (43.6) |
| R_merge_b | 0.121 (0.426) | 0.051 (0.496) |
| Redundancy | 7.3 (6.3) | 7.0 (3.9) |
| Mean l/sigma(l) | 9.8 (4.0) | 25.1 (2.4) |
| Phasing statisticsc | ||
| Mean FOM (centric/acentric) | 0.344/0.090 | |
| Phasing Power (isomorphous/anomalous) | 0.0/1.17 | |
| Cullis R-factor (isomorphous/anomalous) | 0.0/0.79 | |
| Refinement and model statistics | ||
| Resolution range | 26.75–2.79 (2.86– 2.79) | 38.76–1.90 (1.95–1.90)d |
| No. of reflections (work/test) | 23613/1270 | 50954/2745 |
| R_cryst_e | 0.187 (0.311) | 0.160 (0.186) |
| R_free_f | 0.243 (0.378) | 0.212 (0.275) |
| Rmsd bonds (Å) | 0.011 | 0.014 |
| Rmsd angles (°) | 1.336 | 1.417 |
| ESU from Rfree(Å)g | 0.342 | 0.146 |
| B factor–Wilson plot (Å2) | 50.3 | 24.3 |
| B factor – monomer A/B/waters (Å2)h | 31.4/33.0/23.3 | 31.4/30.2/36.4 |
| No. of protein molecules/all atoms | 2/5783 | 2/6378 |
| No. of waters | 45 | 579 |
| No. of auxiliary molecules | 2 MOPS | 1 PEG |
| Ramachandran Plot by MOLPROBITY (%) | ||
| Favoured regions | 97.5 | 98.3 |
| Additional allowed regions | 2.5 | 1.6 |
| Outliers | 0.0 | 0.1 |
| PBD code |
Fig. 4. Structure of CalG3.
(A) A ribbon diagram of the CalG3 dimer with monomers color-coded in red and cyan. (B) The CalG3 monomer is formed by closely opposed N-terminal- (cyan) and the C-terminal-domains (khaki). These distinct domains are connected by a linker (yellow) and their interaction is stabilized by the C-terminal helix (green). The blue arrow indicates the putative catalytic loop, the magenta arrow points to a pyrophosphate-binding tetraglycine loop spanning residues 285–288. An ordered portion of a polyethylene glycol molecule (brown) has been found in the cavity formed by the N-domain. The inset highlights Cot-trace of CalG3 (cyan, yellow, khaki) in the active site with putative catalytic diad residues H11 and E115 highlighted. The gray Cα-trace is that of a docked model, which incorporates experimentally-observed conformational changes in the pyrophosphate binding loop (magenta) and modeled changes of the "catalytic loop" (blue, black arrow). (C) Manually docked model of the CalG3 with CLM T0 (carbon-cyan, oxygen-red, sulfur-yellow, nitrogen-blue) and a dinucleotide TDP (carbon-yellow, oxygen-red, nitrogen-blue, phosphorus-orange) in the active site.
The structure of the CalG3 monomer revealed that this protein consists of two closely opposed globular domains, the N-terminal domain (residues 1–193, cyan in Fig. 4B) and the C–terminal domain (residues 209–360, khaki in Fig. 4B) connected with a linker (residues 194–208, yellow in Fig. 4B) and stabilized by interaction of the C-terminal helix (residues 362–375, green in Fig. 4B) and the N-terminal domain. Both the N- and C-terminal domain adopt a fold with Rossmann topology and a 3-layer α-β-α sandwich architecture that shows homology to glycogen phoshorylase B. The structurally homologous cores of these domains can be aligned with a rmsd of 3.1 Å for 103 residues. However, the domains show dramatic variations in size and conformation of several loop regions as well as topology of their C-termini. Full length CalG3 belongs to the GT-B clan of GTs and adopts a UDP-glycosyltransferase/glycogen phosphorylase fold found in many GTs. Based on the VAST server (Madej et al., 1995), the closest overall structural homologs of CalG3 include Streptomyces fradiae dTDP-D-olivose-transferase UrdGT2 (which structurally aligns with CalG3 for 369 residues with 24% identity and rmsd of 3.1 Å, PDB ID 2p6p) (Mittler et al., 2007) and Amycolatopsis orientalis TDP-_epi_-vancosaminyltransferase GtfA (363 residues with 20% identity and rmsd of 4.0 Å, PDB ID 1pnv, 1pn3, 1rrv) (Mulichak et al., 2003).
In vitro characterization of CalG2
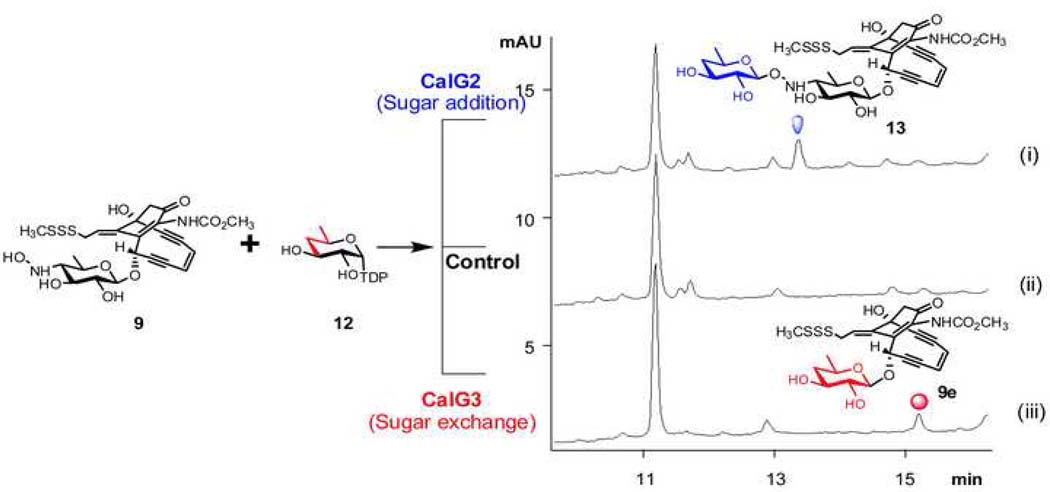
Given the lack of availability of the natural substrate NDP-4-deoxy-4-thio-α-D-digitoxose, the biochemical function of CalG2 was examined with a small library of 22 surrogate TDP-sugar substrates (Fig. S5). Of this set, only one, TDP-4,6-dideoxy-α-D-glucose (12, Fig. 5) led to the CalG2-catalyzed transformation of 9 to a new product with a mass (calcd. 714, found 737 [M+Na]+) (Table S1) consistent with 13 (Fig. 5, i). Notably, this new product was clearly distinct from the CalG3-catalyzed ‘sugar exchange’ product 9e (Fig. 5, iii), derived from the same set of starting materials 9 and 12 (Fig. 5), and its production was enzyme-dependent (Fig. 5, ii). The incubation of 8 with 12 and CalG2 under identical conditions led to no change, consistent with the 8 isopropylidene as masking the CalG2 acceptor nucleophile. Cumulatively, these in vitro studies confirmed that CalG2 was capable of adding a sugar to 9, consistent with CalG2 as the requisite CLM 4-deoxy-4-thio-α-D-digitoxosyltransferase.
Fig. 5. Differential reactions with 9 and 12 in the presence of CalG2 and CalG3.
(i) 50 µM 9, 300 µM 12 in the presence of 7.5 µM CalG2 at 30 °C overnight; (ii) 50 µM 9, 300 µM 12 in the absence of enzymes at 30 °C overnight; (iii) 50 µM 9, 300 µM 12 in the presence of 7.5 µM CalG3 at 30 °C overnight.
Discussion
CLM GTs and CLM biosynthesis
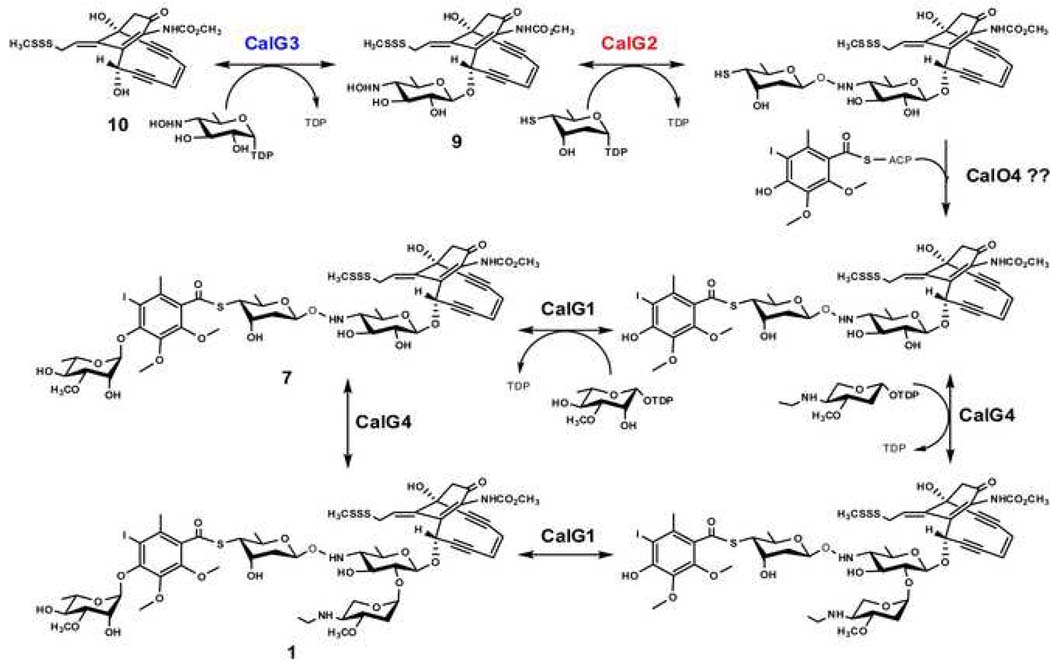
Calicheamicin (1) has two distinct structural regions: the enediyne aglycon, or ‘warhead’, which consists of a highly functionalized bicyclo[7.3.1.]tridecadiyne core structure with an allylic trisulfide serving as the initial trigger for warhead cycloaromatization; the aryltetrasaccharide, which is composed of a set of unusual carbohydrate and aromatic units and docks the metabolite specifically into the minor groove of DNA (Kumar et al., 1997; Walker et al., 1994). Two possible scenarios for the final stages of CLM biosynthesis have been put forth – sequential GT-catalyzed glycosylation of the calicheamicinone core (as highlighted in Fig. 6) or coupling an intact aryltetrasaccharide unit to the calicheamicinone core (Rothstein and Love, 1991; Thorson et al., 1999). The first of these was based upon the conventional routes to secondary metabolite glycosylation while the second derived from the structural similarities between the CLM aryltetrasaccharide and the orthosomycins avilamycin or evernimicin (Hosted et al., 2001; Weitnauer et al., 2001). Consistent with either putative NDP-sugar or aryltetrasaccharide intermediates, random chemical mutagenesis of the CLM producing strain M. echinospora led to undefined water soluble intermediates that could be transformed to CLM by other blocked mutant strains (Rothstein and Love, 1991). Characterization of the CLM biosynthetic gene cluster (Ahlert et al., 2002) revealed four putative GT-encoding genes (calG1, calG2, calG3 and calG4) which enabled the initial biochemical characterization of the CLM GTs CalG1 and CalG4 (Zhang et al., 2006b), providing the first support for the pathway highlighted in Fig. 6. Specifically, this prior work revealed CalG1 and CalG4 as the 3-_O_-rhamonsyltransferase and aminopentosyltransferase, respectively, the order of which appeared indiscriminate. The current study extends this work by confirming CalG3 and CalG2 as the ‘internal’ sequential 4,6-dideoxy-4-hydroxylamino-α-D-glucosyltransferaseand 4-deoxy-thio-α-D-digitoxosyltransferase, respectively. Thus, this study completes the functional assignment of the four CLM GTs and provides further support for a sequential glycosylation pathway in CLM biosynthesis.
Fig. 6. The proposed CLM glycosylation pathway.
CalG3 reaction reversibility and sugar exchange
The reversibility of GT-catalyzed reactions has enabled GT biochemical characterization, the syntheses of exotic sugar nucleotides, and the differential glycosylation of various complex natural products (Zhang et al., 2006a; Zhang et al., 2006b; Zhang et al., 2007). As described in the present study, a similar strategy facilitated the functional assignment of CalG3 as the requisite CLM 4,6-dideoxy-4-hydroxylamino-α-D-glucosyltransferase and allowed for the generation a small set of novel CLM analogs via CalG3-catalyzed ‘sugar exchange’. While this study revealed CalG3 to be among the growing list of inherently promiscuous GTs (Salas and Mendez, 2007; Thibodeaux et al., 2007), a remarkable distinction of the CalG3 reaction from previously studied examples (Minami et al., 2005; Zhang et al., 2006a; Zhang et al., 2006b; Zhang et al., 2007) is the apparent reversibility with ‘non-native’ NDPs. Specifically, biochemical characterization of the CLM sugar nucleotide pathways reveals all four aryltetrasaccharide sugars to derive from TDP/UDP-sugar precursors (Bililign et al., 2002; Johnson et al., unpublished) while the present study reveals purine-based nucleotides to be optimal substrates in the reverse direction. Although this highlights the use of caution when employing reversibility as a means to determine GT substrate specificity, the nucleotide specificity of CalG3 sugar exchange reactions with the NDP-Glc series is consistent with a TDP/UDP-sugar–dependent process. Simulation of GT-catalysis has revealed the equilibrium constant (_K_eq) to be the single most critical factor governing reaction efficiency (Melancon et al., 2006). The observed modulation of the CalG3 equilibrium via nucleotides (Fig. 3 and Fig. 3B) and/or pH (Fig. S4) is also consistent with a thermodynamically-controlled process.
CalG3 structure
While the sequence homology of GalG3 and its closest structural homologs is low (<25% identity), the modular design and the structural similarity between GT-B glycosyltransferases implicate the C-terminal domain as involved in sugar nucleotide-binding. Specifically, the tetraglycine loop 285–288 (Fig 4B, inset), which adopted different conformations within the distinct CalG3 crystal forms, most likely interacts with the sugar nucleotide pyrophosphate while an adjacent large cavity within the C-terminal domain of CalG3 is anticipated to accommodate the corresponding nucleoside. The N-terminal domain also contains a large cavity, which in our high-resolution model binds an extended chemical entity (likely an ordered fragment of polyethylene glycol used during crystallization, Fig. 4B, brown). Based on structural homology of CalG3 to other natural product GTs, this cavity likely binds calicheamicinone.
Using this information as a guide, the products of the reaction, CLM T0 and dTDP, were manually docked into the CalG3 model (Fig. 4C). Specifically, the dinucleotide positioning was initially guided by the high structural conservation of C-terminal domains of CalG3 and oleandomycin glycosytransferase OleI in complex with UDP (PDB ID 2iya) (Bolam et al., 2007). Subsequent CLM T0 orientation in the corresponding N-terminal cavity was defined by the surface of the cavity and the geometric constraints of a putative SN2 reaction between the acceptor hydroxyl group and the sugar nucleotide anomeric carbon. Based upon this model, the acceptor hydroxyl group is located near a putative catalytic diad (His11 and Glu115). The first residue of the diad, His11, is highly conserved as His or Asp among GT homologs (based upon a multiple sequence alignment of 1070 sequences from NR85S database by FFA03 (Jaroszewski et al., 2005) and a structure-based multiple sequence alignment (Fig. S6). Consistent with this, mutation of the His11 equivalent in the oleadromycin GT OleI (His25 to Ala) led to complete loss of catalytic activity (Bolam et al., 2007). Moreover, the CalG3 His11-containing loop (N1 loop, residues 5–12, Fig. 4B, inset, black arrow) has to undergo conformational changes during the substrate binding and catalysis. In the CalG3 model, this dynamic loop resides between the calicheamicinone binding cavity and an internal cavity filled with a cluster of well ordered water molecules, the removal of which allows for an expansion of the calicheamicinone binding cavity to accommodate the requisite hexose.
The second member of the CalG3 putative catalytic diad (Glu115), while less conserved in general, is most often found to be Asp or Glu in GT sequence homologs (Fig. S6). The CalG3-substrate complex model also predicts the access to the calicheamicinone hydroxyl nucleophile to be sterically hindered, and thus, activation of this hydroxyl may require a water-mediated process. Similar water-containing catalytic ‘triads’ have been described in variety of enzymes, including phospholipases (Scott et al., 1990). In some flavonoid GTs, a neighboring serine within the same loop has been implicated (along with the His/Asp-Asp/Glu diad) as part of a catalytic ‘triad’ (Offen et al. 2006). Interestingly, CalG3 also contains a Ser within the N1 loop but in a different position, the function of which remains to be elucidated. In addition, consistent with other GTs, the CalG3-substrate model is consistent with the conserved Gln311 – sugar, His284 – pyrophosphate and Trp268 – base stacking components of the sugar nucleotide (Fig. S6). In comparison to other UDP/dTDP-sugar nucleotide-utilizing GTs, CalG3 Leu271 is topologically equivalent to the TDP-sugar-utilizing GT GtfA Leu280, an amino acid noted as a potential selectivity filter for 2’-deoxy nucleotides. In contrast, the TDP-sugar-utilizing Gt OleI has a Gln315 at the same position which hydrogen bonds the nucleotide 2’-OH. Finally, this model also implicates the cavity formed by the residues Leu14, Pro15, Gln137, and Arg135 may accommodate the CLM trisulfide-SSSMe trigger.
Significance
In conjunction with our previous report (Zhang et al., 2006b), this study completes the functional assignment of all four CLM GTs, extends the concept of reversibility of enediyne GT-catalyzed reactions, and highlights the first crystal structure of an enediyne GT. From a structural biology perspective, this work reveals the common GT-B structural fold can now be extended to include enediyne GTs. Given the notable architectural distinctions of enediynes from other natural products for which glycosyltransferase structures have been elucidated (including glycopeptides, aromatic polyketides and macrolides), this work adds to the structural blueprints for engineering and/or evolving novel glycosylation catalysts (Williams et al., 2007; Williams et al., 2008a; Williams et al. 2008b). From a biosynthetic perspective, this work also completes the functional annotation of the four calicheamicin GTs, highlights the first characterization of a hydroxylamino glycosidic bond-forming GT, and presents conclusive evidence of a sequential glycosylation pathway in CLM biosynthesis. Furthermore, this work highlights the utility of the reversibility of GT-catalyzed reactions and sugar nucleotide surrogate substrates for both the elucidation of enzyme function and the diversification of therapeutically important natural products.
Materials and methods
Materials
E coli DH5α and BL21(DE3) competent cells were purchased from Invitrogen (Carlsbad, CA). The pET-16b E coli expression vector was purchased from Novagen (Madison, WI). Primers were purchased from Intergrated DNA Technology (Coralville, IA). Pfu DNA polymerase was purchased from Stratagene (La Jolla, CA). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). All other chemicals were reagent grade or better and purchased from Sigma (St. Louis, MO). Calicheamicinα3I (7, Fig. 2) was provided by Wyeth Research (Pearl River, New York). Analytical HPLC was run on a Varian Prostar 210/216 system connected to a Prostar 330 photodiode array detector (Varian, Walnut Creek, CA). Mass spectra (MS) were obtained by using electrospray ionization on an Agilent 1100 HPLC-MSD SL quadrupole mass spectrometer (Agilent Technologies, Palo Alto, CA) connected with a UV/Vis diode array detector.
Chemoenzymatic synthesis of TDP-sugars
The set of TDP-sugars employed were generated chemoenzymatically as previously described (Barton et al., 2002; Jiang et al., 2001; Jiang J. et al., 2000). Specifically, the RmlA (Ep, glucose-1-phosphate thymidylyltransferase) reaction was carried out in Tris-HCl buffer (50 mM, pH 8.0) containing 5 mM MgCl2, 1 U inorganic pyrophosphatase, 10 µM of purified Ep, 8 mM sugar-1-phosphate and 6 mM TTP, and incubated at 37°C for 2 hrs. The formation of sugar nucleotides (5 – 24, Fig. 2) was analyzed by HPLC using a reverse phase column Luna C18, 5 µm, 250 × 4.6 mm with UV detection at 254 nm. A phosphate buffer A (30 mM potassium phosphate pH 6.0, 5 mM tetrabutylammonium hydrogensulfate, 2 % acetonitrile) was used as mobile phase and the HPLC was run with a 0 – 50 % gradient of buffer B (acetonitrile) over 30 min.
Preparation of CLM T0 (9) from calicheamicin α3I (7)
CLM α3I (7, 20 mg, 0.017 mmol) was dissolved in wet acetone (20 mL) followed by the addition of pyridine _p_-toluenesulfonate (0.4 mg, 0.002 mmol). The reaction was heated to reflux (65 oC) and monitored by TLC (9:1 CHCl3:MeOH) and reverse-phase HPLC (Phenomenex Luna C18, 4.6 × 250 mm column) (Fig. 2). After 19 h, full conversion of 7 to a product with a UV spectrum characteristic to the CLM core structure had occurred. Purification was performed by silica gel column chromatography (9:1 CHCl3:MeOH) to yield the isopropylidenylated analog 8 as a light brown oil (10 mg, 97%, TLC Rf = 0.35 9:1 CHCl3:MeOH), which was characterized by 1H NMR on a Varian UNITYInova 400 MHz instrument (Fig. S3) and by APCI-MS using an Agilent 1100 Series LC/MSD. Compound 8 (10 mg, 0.016 mmol) was hydrolyzed by dissolving in 1 mL of a 1:1 MeOH:H2O solution in the presence of TFA (12 µL) and agitating for 2 d, using TLC and reverse-phase HPLC to observe the reaction (Fig. 2). The solvent was removed in vacuo and the crude oil purified by preparatory reverse-phase HPLC (Supelco Discovery BIO 10 × 250 mm, 5 µm column) using a gradient of 10–100% acetonitrile in water over 20 min. at a rate of 10 mL/min with UV detection at 280 nm. Lyophilization yielded 9 as a white powder (5.8 mg, 62%, TLC Rf = 0.32 9:1 CHCl3:MeOH). Characterization was performed on a Varian UNITYInova 500 MHz NMR with a capillary probe (Fig. S2) and by high-resolution MS using a Waters LCT time-of-flight mass spectrometer. Compund 8 -1H NMR (_d_6-acetone, 400 MHz)δ 7.86 (s, 1 H), 6.46 (dd, _J_= 10.3, 5.0 Hz, 1 H), 6.15 (d, J = 1.5 Hz, 1 H), 6.04 (d, J = 9.4 Hz, 1 H), 5.98 (dd, J = 9.4, 1.5 Hz, 1 H), 4.75 (d, J = 7.9 Hz, 1 H), 4.23 (t, _J_= 9.1 Hz, 1 H), 4.19-4.13 (m, 2 H), 4.03 (t, J = 9.2 Hz, 1 H), 3.94 (dd, J = 14.9, 5.0 Hz, 1 H), 3.65 (s, 3 H), 3.48 (t, _J_= 8.4 Hz, 1 H), 3.05 (d, _J_= 17.2 Hz, 1 H), 2.73 (d, _J_= 17.2 Hz, 1 H), 2.54 (s, 3 H), 2.28 (s, 3 H), 2.13 (s, 3 H), 1.18 (d, _J_= 6.2 Hz, 3 H); MS (APCI) m/z C27H33N2O9S3 ([M+H]+) 625.1, calc. 625.1. Compund 9 - 1H NMR (_d_6-acetone, 500 MHz) δ 6.46 (dd, _J_= 10.3, 4.9 Hz, 1 H), 6.14 (s, 1 H), 6.03 (d, _J_= 9.3 Hz, 1 H), 5.97 (d, _J_= 9.3 Hz, 1 H), 4.72 (d, _J_= 7.7 Hz, 1 H), 4.23 (t, _J_= 9.2 Hz, 1 H), 4.18-4.11 (m, 2 H), 4.00 (t, J = 9.3 Hz, 1 H), 3.93 (dd, J = 14.9, 4.9 Hz, 1 H), 3.65 (s, 3 H), 3.49 (dd, _J_= 9.2, 7.7 Hz, 1 H), 3.05 (d, J = 17.0 Hz, 1 H), 2.73 (d, _J_= 17.0 Hz, 1 H), 2.53 (s, 3 H), 1.17 (d, J = 6.0 Hz, 3 H); HRMS (ESI) m/z C24H28N2NaO9S3 ([M+Na]+) 607.0866, calc. 607.0855.
Cloning, expression and purification of GTs
The calG2 and calG3 genes from the calicheamicin producer,Micromonospora echinospora LL6600, were amplified from genomic DNA by using primer pairs: 5’- cacggacggagtcgcatatggcccacctc - 3’ (forward, _Nde_I) and 5’- gccggtggatccgcggggcg −3’(reverse, BamHI) for calG2; 5’-gaagggctcccatatgcgcgtgctgttc-3’(forward,_Nde_I)and5’- gggcgacgagatctgctcaacccgagatg - 3’ (reverse, _Bgl_II) for calG3, using Pfu DNA polymerase. PCR products were digested with _NdeI/Bam_HI (calG2) or _Nde_I/_Bgl_II (calG3) and ligated into the pET16b expression vector (_NdeI/Bam_HI - to generate the _N_-terminal MGHHHHHHHHHH fusion) to give plasmids pCAM3.2 (CalG2) and pCAM11.2 (CalG3), respectively.
For CalG3 expression, a single transformant of E. coli BL21(DE3)/pCAM11.2 was inoculated into 4 ml LB medium supplemented with 100 µg/ml of ampicilin and grown at 37 °C overnight. The starter cultures were inoculated into 1 L LB medium with 100 µg/ml of ampicilin and was initially grown at 28 °C to an OD600 value of 0.5 – 0.7. Expression was induced with the addition of 0.4 mM of isopropyl-β-D-thiogalactopyranoside (IPTG) followed by continued growth with shaking for 16 hrs. The cells obtained from 1 L of culture were washed twice with buffer A (20 mM NaH2PO4, pH 7.5, 500 mM NaCl, 10 mM imidazole) and resuspended in 30 ml of buffer A supplemented with 1 mg/ml of lysozyme. After 10 min incubation on ice, the cells were lysed by 3 rounds of French-press (1,200 psi, Thermo IEC) and the insoluble material was removed by centrifugation at 30,000 g for 1 hr (4°C). The supernatant was loaded onto the HisTrap HT column (1 ml, GE Healthcare) and the N-(His)10-tagged CalG3 was eluted with a linear gradient of imidazole (10 – 500 mM) in buffer A using a FPLC-AKTA system (GE Healthcare). The purified protein was desalted through PD-10 column (GE Healthcare) and stored in the buffer containing 10 mM Tris-HCl (pH 8.0), 100 mM NaCl and 10 % glycerol until use. Protein concentration was determined by Bradford assay. _N_-(His)10-tagged CalG2 was expressed and purified following the same protocol from E. coli overexpression strain BL21(DE3)/pCAM3.2.
CalG3 assays
Generally, CalG3 assays were performed in a total volume of 100 µl containing 50 µM of CLM T0 (9) and 2 mM NDP (Fig. 3B) or 300 µM of TDP-sugars (Fig. S4) with incubation at 30°C for 2 h (reverse reaction) or overnight (sugar exchange) in the presence of 7.5 µM of CalG3, in Tris-HCl buffer (10 mM, pH 7.6) containing 1 mM of MgCl2. The assay mixtures lacking CalG3 served as controls. The reactions were subsequently quenched by the addition of 100 µl methanol and were centrifuged to remove proteins. The reactions were monitored by HPLC (Phenomenex Luna C18, 5µm, 250 × 4,6 mm; 10% CH3CN to 100% CH3CN over 20 min, 1 mL/min, 280 nm). The conversion rate was calculated by dividing the integrated area of glycosylated product with the sum of integrated area of product and the remaining substrate. All newly-formed products were also analyzed by LC-MS (ESI) with both positive (+) and negative (−) modes.
To assess the pH range for CalG3 catalysis, potassium phosphate buffers (50 mM, pH 6.0, pH 7.0, pH 7.6 and pH 8.0) were used (9 was instable > pH 8.0). The reaction mixtures contained 50 µM 9, 2 mM NDP (or 2 mM NDP-glucose) and 7.5 µM CalG3 and were incubated at 30 °C for 2 h (reverse reaction) or overnight (sugar exchange).
CalG2 assays
Generally, CalG2 assays were performed in a total volume of 100 µl containing 50 µM of CLM T0 (9) and 2 mM of NDP-glucoses (Fig. 3B) or 300 µM of TDP-sugars (Fig. S4) with incubation at 30°C overnight in the presence of 7.5 µM CalG2, in Tris-HCl buffer (10 mM, pH 7.6) containing 1 mM of MgCl2. The assay mixtures without addition of CalG2 served as controls. The reactions were quenched by the addition of 100 µl methanol and were centrifuged to remove proteins. The formation of new CLM products was monitored by HPLC analysis as described above for CalG3 assays.
CalG3 crystallization
Crystals of selenomethionine-labeled (SeMet) CalG3 were grown at 277 K by the hanging drop method from a 10 mg/ml protein solution in a protein buffer (50 mM NaCl, 10 mM TRIS pH 7.5) mixed with an equal amount of the well solution (16% (w/v) polyethylene glycol 4000, 200 mM triammonium citrate, 100mM MOPS pH 7.0). Rod-shaped crystals with dimensions up to 400 µm × 20 µm × 20 µm grew fused in parallel clusters, only occasionally as usable single needles. Crystals were cryoprotected at 277 K by soaking in the well solution containing 0%, 10%, and 20% (v/v) ethylene glycol and were flash frozen in a stream of cryogenic nitrogen gas at 100 K. The native crystals of CalG3 that lead to a high resolution dataset were obtained by sitting drop method from a 10 mg/ml protein solution in the protein buffer mixed with an equal amount of the well solution [25% (w/v) polyethylene glycol 1500; condition D1 of Hampton IndexHT screen]. Crystals were discovered 8 months after the initial set-up and were never again successfully reproduced. Crystals were cryoprotected at 277 K in Fomblin MW2500 (Aldrich) and were flash frozen in a stream of cryogenic nitrogen gas at 100 K. The diffraction quality of crystals was evaluated using a laboratory X-ray diffraction instrument equipped with a Bruker AXS Proteum R CCD detector and a Microstar rotating anode generator using copper Ka radiation (Bruker, Madison, WI). All attempts to prepare crystals of complexes of CalG3 with its substrates, via co-crystallization with ligands or soaking crystals with ligands, were unsuccessful.
CalG3 structure determination
X-ray diffraction data for both SeMet-labeled and native CalG3 were collected at the General Medicine and Cancer Institute Collaborative Access Team (GM/CA-CAT) 23-ID-D beamline at the Advanced Photon Source at Argonne National Laboratory. Each of the 290 diffraction images for the native crystal was collected at the crystal-to-detector distance of 220 mm and exposed for 2 s with 100-fold attenuation of the incident beam. The data was collected in a single pass with 1.25 deg oscillation per frame. Each of the 180 diffraction images for the selenomethionine-labeled crystal was collected at wavelength of 0.97918 Å at the crystal-to-detector distance of 300 mm and exposed for 6 s with 100-fold attenuation of the incident beam. The data was collected in a single pass with 1 deg oscillation per frame. The diffraction images were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997). The native crystals belong to the space group P21 with unit cell parameters a = 57.4 Å, b = 97.7 Å, c = 63.0 Å, β = 90.6°. The selenomethionine-labeled crystals belong to the space group I222 with unit cell parameters a = 106.7 Å, b = 119.3 Å, c = 155.9 Å.
The selenomethionine substructure of the SeMet-labeled crystals of CalG3 was determined using HySS (Adams et al., 2002; Uson and Sheldrick, 1999). These programs identified 13 consensus anomalous sites. The structure was automatically phased using autoSHARP (de la Fortelle and Bricogne, 1997) with the help of auxiliary programs from the CCP4 (Collaborative Computational Project Number 4, 1994) suite. The initial phase information was significantly improved by two fold averaging during the density modification as implemented in CNS (Brunger et al., 1998). Resulting map at 2.8 Å resolution was of a high quality and allowed for partial building of the model in ARP/wARP (Perrakis et al., 1999). This model was improved by manual building in COOT (Emsley and Cowtan, 2004) and used as a search model in molecular replacement trials in MOLREP (Vagin and Teplyakov, 1997) against the 1.9 Å native diffraction data. The high resolution model of CalG3 was next built in ARP/wARP using the phase information derived from the successfully placed low resolution model. The automatically build model contained 614 residues of which 591 had side-chains assigned and had R = 25.3 % (Rfree = 31.0%). The structure was completed in multiple cycles of manual building in COOT and refinement in REFMAC5 (Murshudov et al., 1997). Final refinement protocol included TLS refinement with 5 TLS-groups per monomer based on the TLSMD server (Painter and Merritt, 2006) analysis. The final refined model has R = 16.0% (Rfree = 21.2%). In addition to residues −2–375 of monomer A and residues −6–374 of monomer B, the final model contains 579 waters and a molecule of polyethylene glycol. Finally, we used the final high-resolution model of CalG3 (in P21) as a starting point for refinement of the lower-resolution model of the SeMet-labeled CalG3 (in I222).
PDB depositions
The CalG3 1.9 Å native and 2.8 Å Se-Met structures have been deposited under the pdb accession numbers 3DOR and 3DOQ, respectively.
Supplementary Material
01
Acknowledgements
We thank the University of Wisconsin–Madison School of Pharmacy Analytical Facility for analytical support, Dr. Philip R. Hamann (Wyeth Research) for providing CLM α3I and Dr. Gavin J. Williams for helpful discussion. B.R.G. is a postdoctoral fellow of the American Cancer Society (PF-05-016-01-CDD) and J.S.T is a UW HI Romnes Fellow. This research was supported in part by National Institutes of Health Grants CA84374 and National Cooperative Drug Discovery Group Grant U19 CA113297 from the National Cancer Institute. GM/CA-CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- Barton WA, Biggins JB, Jiang J, Thorson JS, Nikolov DB. Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering. Proc. Natl. Acad. Sci. USA. 2002;99:13397–13402. doi: 10.1073/pnas.192468299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins JB, Onwueme KC, Thorson JS. Resistance to enediyne antitumor antibiotics by CalC self-sacrifice. Science. 2003;301:1537–1541. doi: 10.1126/science.1086695. [DOI] [PubMed] [Google Scholar]
- Bililign T, Shepard EM, Ahlert J, Thorson JS. On the origin of deoxypentoses: evidence to support a glucose progenitor in the biosynthesis of calicheamicin. ChemBioChem. 2002;3:1143–1146. doi: 10.1002/1439-7633(20021104)3:11<1143::AID-CBIC1143>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Blanco G, Patallo EP, Brana AF, Trefzer A, Bechthold A, Rohr J, Mendez C, Salas JA. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem. Biol. 2001;8:253–263. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- Boghaert ER, Sridharan L, Armellino DC, Khandke KM, DiJoseph JF, Kunz A, Dougher MM, Jiang F, Kalyandrug LB, Hamann PR, et al. Antibody-targeted chemotherapy with the calicheamicin conjugate hu3S193-N-Acetyl gamma calicheamicin dimethyl hydrazide targets lewisy and eliminates Lewis(y)-positive human carcinoma cells and xenografts. Clin. Cancer Res. 2004;10:4538–4549. doi: 10.1158/1078-0432.CCR-04-0037. [DOI] [PubMed] [Google Scholar]
- Bolam DN, Roberts S, Proctor MR, Turkenburg JP, Dodson EJ, Martinez-Fleites C, Yang M, Davis BG, Davies GJ, Gilbert HJ. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc. Natl. Acad. Sci. USA. 2007;104:5336–5341. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiJoseph JF, Popplewell A, Tickle S, Ladyman H, Lawson A, Kunz A, Khandke K, Armellino DC, Boghaert ER, Hamann PR, et al. Antibody-targeted chemotherapy of B-cell lymphoma using calicheamicin conjugated to murine or humanized antibody against CD22. Cancer Immunol. Immun. 2005;54:11–24. doi: 10.1007/s00262-004-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Method Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- DiJoseph JF, Popplewell A, Tickle S, Ladyman H, Lawson A, Kunz A, Khandke K, Armellino DC, Boghaert ER, Hamann PR, et al. Antibody-targeted chemotherapy of B-cell lymphoma using calicheamicin conjugated to murine or humanized antibody against CD22. Cancer Immunol Immun. 2005;54:11–24. doi: 10.1007/s00262-004-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem. Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- Gao Q, Thorson JS. The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710. FEMS Microbiol Lett. 2008;282:105–114. doi: 10.1111/j.1574-6968.2008.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann PR, Hinman LM, Beyer CF, Lindh D, Upeslacis J, Shochat D, Mountain A. A calicheamicin conjugate with a fully humanized anti-MUC1 antibody shows potent antitumor effects in breast and ovarian tumor xenografts. Bioconjugate Chem. 2005;16:354–360. doi: 10.1021/bc049794n. [DOI] [PubMed] [Google Scholar]
- Hensens OD, Giner JL, Goldberg IH. Biosynthesis of Ncs chrom-a, the chromophore of the antitumor antibiotic neocarzinostatin. Journal of the American Chemical Society. 1989;111:3295–3299. [Google Scholar]
- Hosted TJ, Wang TX, Alexander DC, Horan AC. Characterization of the biosynthetic gene cluster for the oligosaccharide antibiotic, evernimicin, in Micromonospora carbonacea var. africana ATCC 39149. J. Ind. Microbiol. Biotechnol. 2001;27:386–392. doi: 10.1038/sj.jim.7000189. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Biggins JB, Thorson JS. Expanding the pyrimidine diphosphosugar repertoire: The chemoenzymatic synthesis of amino- and acetamidoglucopyranosyl derivatives. Angew. Chem. Int. Ed. Engl. 2001;40:1502–1505. doi: 10.1002/1521-3773(20010417)40:8<1502::AID-ANIE1502>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Jiang J, Biggins JB, T JS. A general enzymatic method for the synthesis of natural and "unnatural" UDP- and TDP-nucleotide sugars. J. Am. Chem. Soc. 2000;122:6803–6804. [Google Scholar]
- Kennedy DR, Gawron LS, Ju J, Liu W, Shen B, Beerman TA. Single chemical modifications of the C-1027 enediyne core, a radiomimetic antitumor drug, affect both drug potency and the role of ataxia-telangiectasia mutated in cellular responses to DNA double-strand breaks. Cancer Res. 2007a;67:773–781. doi: 10.1158/0008-5472.CAN-06-2893. [DOI] [PubMed] [Google Scholar]
- Kennedy DR, Ju J, Shen B, Beerman TA. Single modifications of an enediyne chromophore can confer an ability to induce DNA double strand breaks, interstrand crosslinks or both with concominant changes in PIKK regulation of DNA damage response. Proc. Natl. Acad. Sci. USA. 2007b;104:17632–17637. doi: 10.1073/pnas.0708274104. 104:17632–17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kumar RA, Ikemoto N, Patel DJ. Solution structure of the calicheamicin gamma1I-DNA complex. J. Mol. Biol. 1997;265:187–201. doi: 10.1006/jmbi.1996.0718. [DOI] [PubMed] [Google Scholar]
- Lam KS, Veitch JA, Golik J, Krishnan B, Klohr SE, Volk KJ, Forenza S, Doyle TW. Biosynthesis of esperamicin-a(1), an enediyne antitumor antibiotic. Journal of the American Chemical Society. 1993;115:12340–12345. [Google Scholar]
- Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem. Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- Melancon CE, Thibodeaux CJ, 3rd, Liu HW. Glyco-stripping and glyco-swapping. ACS Chem Biol. 2006;1:499–504. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]
- Minami A, Kakinuma K, Eguchi T. Aglycon switch approach toward unnatural glycosides from natural glycoside with glycosyltransferase VinC. Tetrahedron Lett. 2005;46:6187–6190. doi: 10.1021/ja042848j. [DOI] [PubMed] [Google Scholar]
- Mittler M, Bechthold A, Schulz GE. Structure and action of the C-C bond-forming glycosyltransferase UrdGT2 involved in the biosynthesis of the antibiotic urdamycin. J. Mol. Biol. 2007;372:67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc. Natl. Acad. Sci. USA. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9:547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43:5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. Embo J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Rothstein DM, Love SF. Isolation of Mutants Blocked in Calicheamicin Biosynthesis. J. Bacteriol. 1991;173:7716–7718. doi: 10.1128/jb.173.23.7716-7718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas JA, Mendez C. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Sievers EL, Linenberger M. Mylotarg: antibody-targeted chemotherapy comes of age. Curr. Opin. Oncol. 2001;13:522–527. doi: 10.1097/00001622-200111000-00016. [DOI] [PubMed] [Google Scholar]
- Scott DL, White SP, Otwinowski Z, Yuan W, Gelb MH, Sigler PB. “Interfacial catalysis: the mechanism of phospholipase A2.”. Science. 1990;250:1541–1546. doi: 10.1126/science.2274785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Hager MH, Zhang CS, Griffith BR, Lee MS, Hallenga K, Markley JL, Thorson JS. Structural insight into the self-sacrifice mechanism of enediyne resistance. ACS Chemical Biology. 2006;1:451–460. doi: 10.1021/cb6002898. [DOI] [PubMed] [Google Scholar]
- Thibodeaux CJ, Melancon CE, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- Thorson JS, Shen B, Whitwam RE, Liu W, Li Y, Ahlert J. Enediyne biosynthesis and self-resistance: A progress report. Bioorg. Chem. 1999;27:172–188. [Google Scholar]
- Thorson JS, Sievers EL, Ahlert J, Shepard E, Whitwam RE, Onwueme KC, Ruppen M. Understanding and exploiting nature's chemical arsenal: the past, present and future of calicheamicin research. Curr. Pharm. Des. 2000;6:1841–1879. doi: 10.2174/1381612003398564. [DOI] [PubMed] [Google Scholar]
- Tokiwa Y, Miyoshisaitoh M, Kobayashi H, Sunaga R, Konishi M, Oki T, Iwasaki S. Biosynthesis of dynemicin-a, a 3-ene-1,5-diyne antitumor antibiotic. Journal of the American Chemical Society. 1992;114:4107–4110. [Google Scholar]
- Torkkell S, Ylihonko K, Hakala J, Skurnik M, Mantsala P. Characterization of Streptomyces nogalater genes encoding enzymes involved in glycosylation steps in nogalamycin biosynthesis. Mol. Gen. Genet. 1997;256:203–209. doi: 10.1007/s004380050562. [DOI] [PubMed] [Google Scholar]
- Van Lanen SG, Oh T-J, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J. Am. Chem. Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr Opin Struct Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Van Lanen SG, Shen B. Biosynthesis of enediyne antitumor antibiotics. Curr. Topics Med. Chem. 2008;8 doi: 10.2174/156802608783955656. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Landovitz R, Ding WD, Ellestad GA, Kahne D. Cleavage behavior of calicheamicin gamma1I and calicheamicin T. Proc. Natl. Acad. Sci. USA. 1992;89:4608–4612. doi: 10.1073/pnas.89.10.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Murnick J, Kahne D. Structural characterization of a calichemicin-DNA complex by NMR. J. Am. Chem. Soc. 1993;115:7954–7961. [Google Scholar]
- Walker SL, Andreotti AH, Kahne DE. NMR characterization of calicheamicin gamma1I bound to DNA. Tetrahedron. 1994;50:1351–1360. [Google Scholar]
- Williams GJ, Zhang C, Thorson JS. Directed evolution of a natural product glycosyltransferase. Nat. Chem. Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- Williams GJ, Thorson JS. A high-throughput fluorescence-based glycosyltransferase screen and its application in glycosyltransferase directed-evolution. Nat. Protocols. 2008a;3:357–362. doi: 10.1038/nprot.2007.538. [DOI] [PubMed] [Google Scholar]
- Williams GJ, Goff RD, Zhang C, Thorson JS. Optimizing glycosyltransferase specificity via ‘hot spot’ saturation mutagenesis presents a new catalyst for novobiocin glycorandomization. Chem. Biol. 2008b doi: 10.1016/j.chembiol.2008.02.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Biosynthesis of the orthosomycin antibiotic avilamycin A: Deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tü57 and production of new antibiotics. Chem. Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- Wohlert S, Lomovskaya N, Kulowski K, Fonstein L, Occi JL, Gewain KM, MacNeil DJ, Hutchinson CR. Insights about the biosynthesis of the avermectin deoxysugar L-oleandrose through heterologous expression of Streptomyces avermitilis deoxysugar genes in Streptomyces lividans. Chem. Biol. 2001;8:681–700. doi: 10.1016/s1074-5521(01)00043-6. [DOI] [PubMed] [Google Scholar]
- Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- Zein N, Poncin M, Nilakantan R, Ellestad GA. Calicheamicin gamma1I and DNA: Molecular recognition process responsible for site-specificity. Science. 1989;244:697–699. doi: 10.1126/science.2717946. [DOI] [PubMed] [Google Scholar]
- Zein N, Sinha AM, McGahren WJ, Ellestad GA. Calicheamicin gamma1I: An antitumor antibiotic that cleaves double-stranded DNA site specifically. Science. 1988;240:1198–1201. doi: 10.1126/science.3240341. [DOI] [PubMed] [Google Scholar]
- Zhang C, Albermann C, Fu X, Thorson JS. The in vitro characterization of the iterative avermectin glycosyltransferase AveBI reveals reaction reversibility and sugar nucleotide flexibility. J. Am. Chem. Soc. 2006a;128:16420–16421. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]
- Zhang C, Fu Q, Albermann C, Li L, Thorson JS. The in vitro characterization of the erythronolide mycarosyltransferase EryBV and its utility in macrolide diversification. ChemBioChem. 2007;8:385–390. doi: 10.1002/cbic.200600509. [DOI] [PubMed] [Google Scholar]
- Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006b;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc. Natl. Acad. Sci. USA. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01