Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods (original) (raw)
Abstract
A novel protein superfamily with over 600 members was discovered by iterative profile searches and analyzed with powerful bioinformatics and information visualization methods. Evidence exists that these proteins generate a radical species by reductive cleavage of _S_-adenosylmethionine (SAM) through an unusual Fe-S center. The superfamily (named here Radical SAM) provides evidence that radical-based catalysis is important in a number of previously well- studied but unresolved biochemical pathways and reflects an ancient conserved mechanistic approach to difficult chemistries. Radical SAM proteins catalyze diverse reactions, including unusual methylations, isomerization, sulfur insertion, ring formation, anaerobic oxidation and protein radical formation. They function in DNA precursor, vitamin, cofactor, antibiotic and herbicide biosynthesis and in biodegradation pathways. One eukaryotic member is interferon-inducible and is considered a candidate drug target for osteoporosis; another is observed to bind the neuronal Cdk5 activator protein. Five defining members not previously recognized as homologs are lysine 2,3-aminomutase, biotin synthase, lipoic acid synthase and the activating enzymes for pyruvate formate-lyase and anaerobic ribonucleotide reductase. Two functional predictions for unknown proteins are made based on integrating other data types such as motif, domain, operon and biochemical pathway into an organized view of similarity relationships.
INTRODUCTION
Sophisticated iterative profile methods have dramatically extended the power of sequence homology searches (1–3). These tools are useful for creating a larger context for database search results. Whereas a strong match in a BLAST search can be used to infer similar function, the weaker similarity detectable by an iterative profile method illuminates a more distant relationship and is evidence of a conserved fold in the protein structure (2). An anonymous sequence without significant pairwise similarity can often be linked in this way with proteins that have been characterized experimentally (4).
Iterative profile searches are easy to perform but can be difficult to interpret because the data sets returned are large. A query is linked to numerous sequences, each with multiple links to other data sources, creating a large information landscape that can be hard to navigate. As a result, when performing iterative profile searches on the most interesting and novel sequences, a scientist is likely to be overwhelmed with data presented simply as long linear lists, a sharply limited view of information that is inherently multi-dimensional.
We have applied powerful bioinformatics and information visualization techniques to overcome these obstacles in the analysis of an important new protein superfamily that we discovered using iterative profile searching. We call this new superfamily Radical _S_-adenosylmethionine (SAM) after the defining characteristics of its best-studied members. Radical SAM is an ancient and diverged group with 645 unique sequences from 126 species found to date from all three domains of life. At least half the proteins are of unknown activity. We use exploratory statistical methods to analyze the sequence similarity relationships and integrate these results with other data types (motif, domain, operon structure, biochemical pathway and the biomedical literature) for discovery efforts on previously uncharacterized sequences. Our results are part of a larger effort to scale up biological knowledge production using four accelerating factors: (i) information visualization; (ii) large computational resources; (iii) new mathematical strategies; (iv) collaborative problem solving environment technology.
MATERIALS AND METHODS
Detection of the superfamily
The reader can directly observe evidence for distant sequence similarity in the Radical SAM superfamily using the Web version of PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) at the National Center for Biotechnology Information (NCBI, Bethesda, MD). For example, enter any gi identifier from Figure 1 (such as 128228 for the Azotobacter vinelandii NifB protein) and iterate to convergence with the default threshold. In this work PSI-BLAST (2) searches were performed locally using command line searches against the non-redundant protein database downloaded from NCBI onto a Sun Ultra 60 workstation. Search results were analyzed and tested for the closure property with standard Unix tools. The 54 sequences directly tested include 30 proteins associated with at least some biochemical information as well as others chosen to represent the most distant members. False negatives for any individual search were measured against the set of unique and complete sequences already accepted as belonging to the superfamily from multiple other searches. False positives were measured against a list that also included redundant sequences and fragments. False positives ranged from 0 to 12%, with a median value of 0.2%. False negatives ranged from 0.7 to 16%, with a median value of 3%. There were seven sequences that required either an increased or decreased threshold [from the default Expect (E) value of 0.001] for convergence to occur. (Searches that fortuitously include significant numbers of unrelated sequences in a profile do not converge but rather ‘explode’ and can pull in the entire database.) With the 15 N-terminal/motif deletion sequences the median rate of false positives was unchanged, but false negatives increased to 5%. PROBE (3) is a powerful iterative profile search tool that was used in this case as a convenient method for extracting alignment blocks from the defined set of highly diverged Radical SAM proteins. These blocks were edited to show the strongest conserved regions.
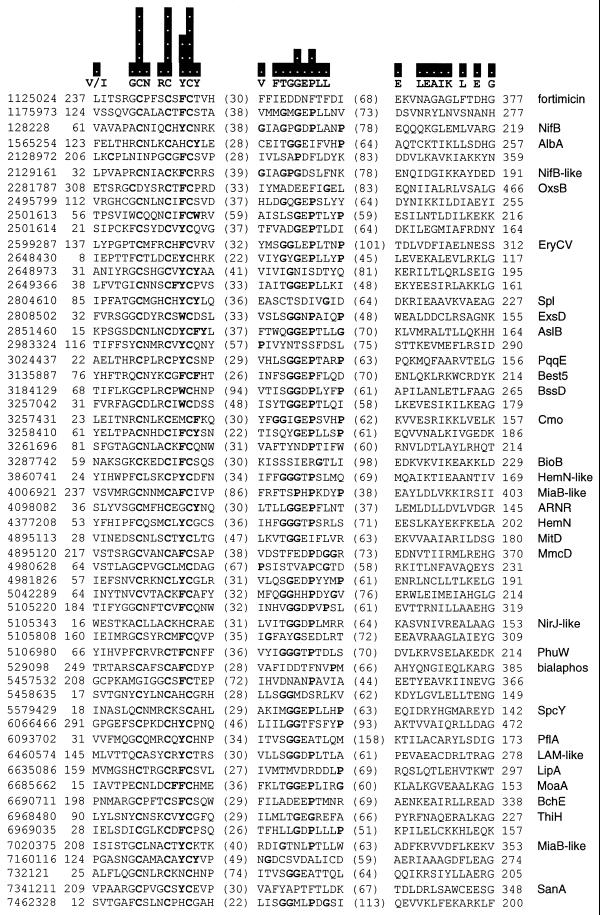
Figure 1.
PROBE alignment of Radical SAM superfamily members. The PROBE iterative profile tool is used to represent the distant sequence similarity between Radical SAM proteins in the form of alignment blocks. Only the strongest regions of the strongest blocks are displayed. Additional motifs emerge in sub-categories of proteins. The sequences included by PROBE in the alignment model represent every group in the BLAST distance dendrogram at the level of 29 clusters and 48 groups at the level of 50. The bars above the sequences show the information content of the PROBE model at each residue position, ranging from 0 to 5 bits. Above the sequences are shown individual residues with the highest information value in each position. Residues are marked in bold for positions with information content of 2 bits or greater. Also marked in bold are aromatic residues (Y, F and W) adjacent to the third cysteine in the first motif, as well as glycine and proline residues (G and P) in the second motif. Each protein is labeled on the left with a gi number, identifying the sequence in the GenBank database, and on the right with a protein name or pathway when known. The coordinates at the left of the alignment blocks suggest the size of independent N-terminal domains and, at the right, the size of C-terminal domains when compared against the sequence length.
BLAST E scores are reported in the computer style of standard scientific notation (e.g. 3e-20 represents 3 × 10–20).
Analysis and visualization of superfamily data
Standard Unix tools, S-PLUS (MathSoft, Cambridge, MA), the OmniViz Pro software package (OmniViz, Richland, WA) and custom Perl programs were used for superfamily analysis. At the time the analysis was initiated there were 533 unique and complete Radical SAM proteins in the database. The conserved core domains (estimated at ∼200 residues and starting at the conserved cysteine motif) were extracted from the Radical SAM sequences using an S-PLUS script. A Perl program was used to perform a complete BLAST comparison of the core domains to produce a matrix of BLAST E values with a high score cut-off of 1000 and then to transform the matrix (lowest score of 0 set to 1e-200, all missing scores to 10 000 and take logn). The transformed matrix was then imported into OmniViz Pro for hierarchical clustering (complete linkage with Euclidean distance). Data files produced by OmniViz Pro were imported into S-PLUS to produce a preliminary dendrogram representation. Cluster membership in the dendrogram was analyzed by making a Galaxy visualization in OmniViz Pro at each level of interest for the purpose of capturing the cluster membership list. These lists were examined individually and were also combined into a spreadsheet for a convenient view of the data. Inclusion of the location of the cysteine motif and the size of the proteins in the spreadsheet allowed patterns to be detected in the size of N- and C-terminal domains. The dendrogram visualization was created with an S-PLUS program that generated the colored blocks from the hierarchical clustering results and Adobe Illustrator (San Jose, CA).
The NCBI Web Entrez interface was used for access to MEDLINE and large sets of abstracts were downloaded using Network Entrez for analysis with the SPIRE technology (http://multimedia.pnl.gov:2080/infoviz/technologies.html), which provides an interactive topic map based on word frequency analysis.
RESULTS
Discovery of a novel superfamily
A small collection of proteins with diverse functions have been noted to share an unusual Fe-S cluster associated with generation of a free radical by reductive cleavage of SAM. This group consists of lysine 2,3-amino mutase (LAM), biotin synthase (BioB), lipoic acid synthase (LipA) and the activating proteins for pyruvate formate-lyase (PflA) and anaerobic ribonucleotide reductase (NrdG). These ‘deoxyadenosyl radical’ enzymes have been the focus of detailed experimental work, including UV-Vis, EPR, Mössbauer, resonance Raman, variable temperature magnetic circular dichroism and mutagenesis experiments (5–12). SAM has been described as equivalent to a ‘poor man’s coenzyme B12’ in the reaction catalyzed by LAM (13). Very recently, K edge X-ray absorption spectroscopy experiments have provided important mechanistic evidence for the direct role of the unusual Fe-S cluster in LAM in the reductive cleavage of SAM (14–16).
Despite the attention they have received, the deoxyadenosyl radical proteins have not been previously recognized as homologous sequences, although a characteristic cysteine motif has been noted (7). We applied sensitive bioinformatics methods that detected distant sequence similarity between these five protein groups. This observation is evidence for a shared ancestor and supports the prediction of a common fold for the core domain. Our results also link these enzymes to a larger collection of known and unknown functions, a list that includes proteins found at unresolved steps in familiar biosynthetic pathways, such as thiamin, heme, heme d1, bacteriochlorophyll, molybdopterin, nitrogenase cofactor, pyrroloquinoline quinone, desosamine and others in secondary metabolism.
We detected distant sequence conservation between the Radical SAM proteins with PSI-BLAST iterative profile searching (2). We observed that these proteins form a closed set with the following property. Each sequence detects the same hit list within a small margin of error after iteration to convergence with a conservative threshold (for details see Materials and Methods). Proteins classified as belonging to the superfamily were either directly tested for this closure property (54 sequences) or shown to be strongly similar to one that was. All of the 645 unique and complete sequences collected in this manner were observed to contain an unusual conserved cysteine motif, most often near the N-terminus or in some longer sequences in the middle. These include 592 proteins with an exact match to the consensus CxxxCxxC and 53 variants with a small increased distance between the first two cysteine residues.
We also tested 15 diverse Radical SAM proteins after removal of the N-terminus including the cysteine motif. This deletion had the effect of reducing the sensitivity of the searches, but not the specificity. Interestingly, the oxygen-sensing regulatory protein FNR has been described as containing an Fe-S cluster similar to those found in the deoxyadenosyl radical proteins both in the cysteine motif and in a reversible transition from [2Fe-2S]2+ to [4Fe-4S]2+ controlled by the presence of oxygen (16,17). However, FNR proteins were never detected in any of the Radical SAM searches. Therefore, the presence of the cysteine motif is not necessary or sufficient for inclusion in the superfamily by PSI-BLAST detection of distant sequence similarity.
We used the PROBE (3) software against the diverged set of Radical SAM sequences to extract alignment blocks and show the strongest sections of these in Figure 1. The cysteine motif in the first block has the highest information density (in units of bits). A conserved aromatic residue (Y, F or W) adjacent to the third cysteine may function to lower the midpoint potential of the cluster by limiting solvent exposure (16). The second block contains a glycine-rich sequence resembling the SAM-binding site in methyltransferases (18) and could play a role in binding this molecule for reductive cleavage.
Protein sequences evolve more quickly than the corresponding three-dimensional structures and, as our results illustrate, proteins with a common fold may only show faint sequence conservation that approaches the limit of detection. However, these patterns can be extracted with sensitive bioinformatics approaches and the information they contain has quantitative value, as exemplified by recent successes in ‘threading’ protein fold prediction programs that include PSI-BLAST results as a term in the calculations (19).
Organizing and visualizing superfamily relationships
The Radical SAM classification places 645 proteins into a single conceptual box but does not illuminate any details of how the members are organized. Although a phylogenetic tree is a useful way to analyze a sequence family, it is difficult to create a multiple alignment for this purpose with highly diverged proteins (20). We applied clustering, a well-known approach in exploratory statistics for extracting groups, to characterize the sequence similarity relationships between the Radical SAM core domains and generate a dendrogram (21). In this approach we used a tree representation not to represent phylogeny but rather to display sequence similarity relationships between superfamily homologs. We first generated a feature matrix based on complete BLAST comparisons between the conserved core domain in each Radical SAM member. We then used hierarchical clustering (complete linkage with Euclidean distance) on the BLAST E score feature matrix to organize the sequences and produce the dendrogram. This algorithm results in a hierarchical tree with the property that variance within each cluster is minimized. We found this preliminary dendrogram to be useful in many ways, for example in identifying misnamed sequences, classifying unknown sequences and in supporting the definition of unique features that characterize sub-groups. However, navigating the raw dendrogram with its associated lists of groups was a difficult and rate-limiting step in the analysis. We therefore created a visual prototype for an automated and interactive solution.
Visual display of information is considered a ‘broad bandwidth’ pathway to the human brain. Powerful visual problem solving approaches have been applied to the analysis of complex hierarchical data (22–28). We used aspects of this research to create a new visual representation for our data. We applied a measure of cluster cohesion that we have named the maximum BLAST E (mBE) score to the clusters in the dendrogram as the basis for color coding groups of closely related proteins that may share a common function (Fig. 2). This mBE cluster cohesion metric is defined as the maximum of the BLAST E values in the subset of the original feature matrix defined by the proteins in the group under consideration. Therefore, our measure of the ‘tightness’ of any cluster is directly based on the largest of all the BLAST comparisons for the proteins in a group. Strong relationships in the dendrogram are depicted with colored blocks that appear when the mBE value is <1. Further divisions within groups are represented by the color scheme, with cool colors representing looser groups and warm colors tighter ones. This visual encoding essentially creates a level of abstraction on distracting details and facilitates interpretation of the results.
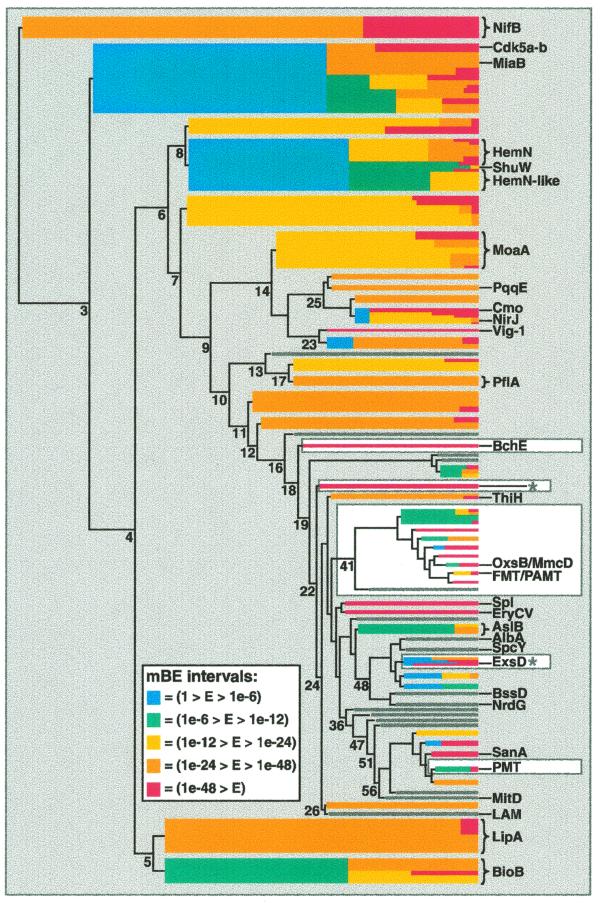
Figure 2.
Dendrogram visualization of Radical SAM core domains. The sequence similarity relationships in the superfamily are visualized using the mBE measure of cluster cohesion to filter a dendrogram produced by hierarchical clustering of a distance matrix. On the left is the line drawing of the original dendrogram, which conveys a representation of between cluster distances of the basic groups. On the right, with increasing cluster number, colored bars are superimposed over the most complex region of the original line drawing using an mBE threshold value applied to the groups. A colored bar appears when a group of sequences appears at a node with an mBE value of <1.0. Further divisions within groups are represented by the color scheme, with cool colors representing looser groups and warm colors tighter ones. Many nodes of the dendrogram are labeled with the number of clusters found at that level. The right edge of the dendrogram is at 200 clusters. Groups discussed in the domain and operon analyses are highlighted with a white background. The ExsD and the _Pyrococcus furiosus_ protein groups from the operon discussion are labeled with asterisks. The color scale for mBE intervals is as follows: blue (1 > E > 1e-6); green (1e-6 > E > 1e-12); yellow (1e-12 > E > 1e-24); orange (1e-24 > E > 1e-48); red (1e-48 > E). Many small branches of less interest have been ‘closed’ (represented by grey bars) to place distracting details in the background.
For example, at the level of eight clusters in the dendrogram a group of 40 HemN-related sequences appear with an mBE value of 0.028 (Fig. 2 and Table 1). This group, shown in blue (a cool color reflecting lower cohesion) divides again at 37 clusters, producing 19 HemN sequences (mBE 3e-24, yellow) and 21 HemN-like sequences (mBE 1e-8, green). Examining the sequences, we observed that the HemN proteins all contain an extra cysteine at the end of the conserved motif (CxxxCxxCxC) but the HemN-like proteins, such as in Bacillus subtilus (29), and the PhuW, HutW and ShuW virulence proteins (30) do not. Thus, our visual algorithm presents these two related but distinct groups of proteins in an intuitive fashion and facilitates integration of this motif information into the analysis.
Table 1. Radical SAM superfamily.
| Proteina | Sequenceb | Function | Clusterc | mBEd | Reference |
|---|---|---|---|---|---|
| NifB | 18 | Nitrogenase cofactor (FeMoCo) | 2 | 9e-25 | (33) |
| MiaB branch | 53 | (Two known functions) | 3 | 0.001 | |
| MiaB | 17 | Methylthiolation of tRNA | 45 | 8e-42 | (34) |
| Nclk-binding | 6 | Cdk5 activator binding | 45 | 3e-59 | (36) |
| BioB/LipA | 46 | Biotin and lipoic acid synthases | 4 | >1 | |
| BioB | 20 | Sulfur transfer to dethiobiotin | 5 | 3e-8 | (7) |
| LipA | 26 | Sulfur transfer to octanoate | 5 | 9e-29 | (9) |
| HemN branch | 40 | Heme biosynthesis | 8 | 0.028 | |
| HemN | 19 | Coproporphyrinogen III oxidase | 37 | 3e-24 | (38) |
| HemN-like | 21 | Coproporphyrinogen III oxidase | 37 | 1e-8 | (29) |
| PhuW | 6 | Associated with virulence | 85 | 1e-8 | (30) |
| MoaA | 28 | Pterin formation in molybdopterin | 14 | 8e-19 | (65) |
| PflA | 7 | Pyruvate formate-lyase activation | 17 | 7e-42 | (11) |
| BchE | 3 | Bacteriochlorophyll | 18 | 2e-91 | (39) |
| Best5 | 3 | Eukaryotic interferon-inducible | 23 | 1e-101 | (46) |
| PqqE | 6 | Pyrroloquinoline quinone | 25 | 1e-44 | (44) |
| ThiH | 4 | Thiazole ring formation in thiamin | 27 | 4e-31 | (32) |
| SplB | 2 | Spore photoproduct lyase | 34 | 1e-127 | (59) |
| DesII | 3 | Desosamine moiety in antibiotics | 34 | 1e-84 | (56) |
| AtsB | 7 | Formylglycine in sulfatases | 43 | 9e-12 | (42) |
| Cmo | 3 | Putative cofactor modification | 46 | 1e-79 | (67) |
| NirJ | 10 | Heme d1 biosynthesis | 46 | 5e-16 | (41) |
| BssD | 8 | Benzylsuccinate synthase | 53 | 3e-5 | (58) |
| ExsD | 12 | Succinoglycan production | 60 | 5e-6 | (62) |
| NrdG | 6 | Anaerobic ribonucleotide reductase | 61 | 0.0001 | (10) |
| SpcY | 2 | Spectinomycin biosynthesis | 82 | 7e-14 | (49) |
| AlbA | 6 | Subtilosin biosynthesis | 83 | 0.05 | (50) |
| SanA | 1 | Nikkomycin biosynthesis | 93 | – | (51) |
| P-methylase | 2 | Bialaphos biosynthesis | 101 | 0.013 | (53) |
| MitD | 1 | Mitomycin C biosynthesis | 102 | – | (45) |
| MmcD | 3 | Mitomycin C biosynthesis | 108 | 1e-11 | (45) |
| OxsB | 3 | Oxetanocin biosynthesis | 108 | 1e-11 | (52) |
| Methylases | 2 | Fortimicin, fosfomycin | 130 | 1e-129 | (53) |
| LAM | 2 | Lysine 2,3-aminomutase | 155 | 2e-7 | (13) |
Interactive visualization strategies are beginning to be part of the analyst’s basic toolkit in working with large-scale information. We rely on the SPIRE technology (31) to explore large sets of biomedical records through interactive topographical maps based on word frequency statistics (http://multimedia.pnl.gov:2080/infoviz/technologies.html). In a similar way, we envision an automated and interactive version of our dendrogram visualization as a data mining tool that supports biological problem solving by creating a map of superfamily sequences and providing a framework for the integration of diverse data types in the analysis of unknown proteins.
Radical SAM superfamily proteins
We explored the organized view of the Radical SAM proteins to find 30 distinct groups associated with at least some biochemical data (Table 1). Interestingly, the most distantly related clusters (diverging first in the hierarchical tree) seem to share an involvement with sulfur transfer; these include the NifB, MiaB, BioB and LipA proteins (7–9,32–35). A mechanism for sulfur transfer from the Fe-S cluster in BioB has been proposed (32). Like biotin synthase, the NifB proteins act as reagents and not catalysts in existing in vitro assays (7,33). A group of 53 sequences, including MiaB (appearing at the level of three clusters with an mBE value of 0.001), also contains a novel human Cdk5 activator-binding protein that binds the neuronal Cdc2-like kinase (Nclk) involved in regulation of neuronal differentiation and neuro-cytoskeletal dynamics (36).
Other Radical SAM proteins, such as ThiH of thiamin and MoaA of molybdopterin biosynthesis, are found in pathways with sulfur transfer, but most likely do not act in this role directly. Interestingly, however, the MoaA proteins contain the residues GG at the C-terminus, a motif that is adenylated for sulfur transfer in the MoaD proteins, as in ubiquitin (37). In thiazole biosynthesis, sulfur is mobilized from cysteine in a manner similar to the molybdopterin pathway, with adenylation/thiocarboxylate formation at a C-terminal GG motif in ThiS (32).
Radical SAM proteins often provide an anaerobic or oxygen-independent mechanism that is found as an aerobic reaction in other proteins, for example HemN (38), BchE (39) and, possibly, ThiH (32). The HemN protein catalyzes an oxygen-independent oxidation in anaerobic heme biosynthesis and has been shown to require NADH and either SAM or ATP and methionine for in vitro activity (40), which Thauer is reported to have explained with the hypothesis of deoxyadenosyl radical chemistry (38). In heme d1 biosynthesis the anaerobic production of oxo groups at positions C3 and C8 is of special interest as a possible role for the NirJ protein (41). The oxidation of serine or cysteine to formylglycine is catalyzed by sulfatase activation proteins similar to AslB; the activation of arylsulfatase has been observed under both aerobic and anaerobic growth conditions (42,43).
Radical SAM proteins are associated with several ring-forming reactions. ThiH functions in thiazole ring formation from tyrosine, cysteine and 1-deoxy-d-xylulose-5-phosphate (32), PqqE in cyclization of the tyrosine amino acid backbone with glutamate addition to form the cofactor PQQ (44) and BchE in isocyclic ring formation in bacteriochlorophyll (39). The MitD protein in the mitomycin C gene cluster may catalyze mitosane ring formation from the condensation of 3-amino-5-hydroxybenzoic acid, d-glucosamine and carbamoyl phosphate (45).
Three eukaryotic interferon-inducible members are found, including a rat gene (best5) expressed during osteoblast differentiation and bone formation and a candidate drug target for osteoporosis (46), a human gene (cig5) induced during cytomegalovirus infection (47) and a trout gene (vig1) induced during rhabdovirus infection (48).
Many examples from secondary metabolism pathways, such as antibiotic and herbicide biosynthesis, are found, including spectinomycin (49), subtilosin (50), nikkomycin (51), mitomycin C (45), oxetanocin (52), fortimicin, fosfomycin and bialaphos biosynthesis (53,54) and the desosamine moiety of erythromycin (55), oleandomycin (56), methymycin, neomethymycin, narbomycin and pikromycin (57). Biodegradation is represented by BssD in toluene catabolism (58) and DNA repair by spore photoproduct lyase (59).
Two functional predictions for unknown proteins
Problem solving in the genomics era increasingly depends upon traversing complex data landscapes with computational and visualization approaches. We present two examples of functional prediction for unknown proteins in the Radical SAM superfamily based on a multi-dimensional approach to data mining. These analyses were performed in a semi-manual fashion as a preliminary effort in the large-scale automation of the approach.
Although a dendrogram for the Radical SAM core domains is a useful tool for classification and annotation, it essentially provides only a one-dimensional analysis of the superfamily proteins based on the single data type of sequence similarity. Features such as motif, domain, operon, biosynthetic pathway, chemical structure and properties described in the biochemical literature can all provide important functional clues to the biologist when viewed in an organized context. We use the similarity dendrogram as a framework for the integration of multiple data types for the purpose of gaining leverage in the functional prediction of unknown proteins.
Example 1. Many Radical SAM sequences have independent N- or C-terminal domains (Fig. 1). We correlated the sizes of these independent domains with cluster membership in the dendrogram. For example, at the level of 41 clusters in the dendrogram (Fig. 2) a group of 26 proteins appears that shows poor cohesion even after multiple divisions, a feature that often suggests divergent functions. However, 25 of these proteins possess a long N-terminal extension of ∼200 residues. PSI-BLAST searches show that these N-terminal sequences have distant sequence similarity (Fig. 3). The proteins in this group, linked by both cluster membership and a shared N-terminal domain, include the fortimicin (FMT) and fosfomycin (PAMT) methyltransferases (53,54), OxsB (52) (oxetanocin) and MmcD (45) (mitomycin C). Also sharing the N-terminal domain but located in different clusters are the BchE proteins (38,39) and the bialaphos P-methylase (PMT) (53,54).
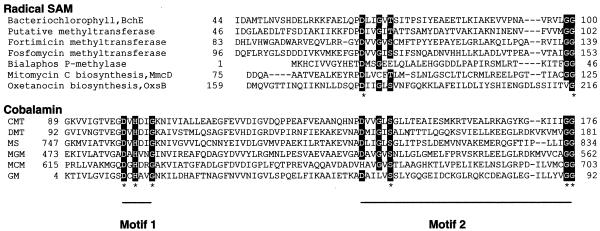
Figure 3.
Multiple alignment of the Radical SAM N-terminus and cobalamin-binding domains. A multiple alignment between cobalamin-binding proteins and a subset of Radical SAM N-terminal domains shows that Motif 2 but not Motif 1 is conserved between the two groups. Motif 1 is the ‘base-off’ consensus that supplies a histidine residue to displace the dimethylbenzimidazole moiety from cobalamin. The conserved Motif 2 residues are known to play a role in binding cobalamin in these corrinoid proteins. The Radical SAM proteins are a bacteriochlorophyll biosynthesis protein BchE (gi|114858), a putative methyltransferase (gi|6686119), a fortimicin methyltransferase (gi|1125024), a fosfomycin methyltransferase (gi|2144248), a bialaphos P-methylase (gi|529098), the mitomycin C biosynthesis protein MmcD (gi|4895120) and the oxetanocin biosynthesis protein OxsB (gi|7474372). The corrinoid-binding proteins are CMT (corrinoid methyltransferase, gi|7483270), DMT (dimethylamine corrinoid protein, gi|4262424), MS (methionine synthase, gi|400244), MGM (2-methyleneglutarate mutase, gi|543481), MCM (methylmalonyl-CoA mutase, gi|1942488) and GM (glutamate mutase, gi|7245512).
With further PSI-BLAST iterations of the N-terminal domain (with BchE for example) proteins outside the Radical SAM superfamily are detected, which share the property of binding cobalamin (60; Fig. 3). This result is interesting in the light of experimental evidence that the fortimicin, fosfomycin and bialaphos methyltransferases (53,54) and, recently, BchE (61) utilize a cobalamin cofactor. Methylation reactions commonly occur through electrophilic attack of a methyl cation. However, the fortimicin, fosfomycin and bialaphos biosynthesis proteins each transfer a methyl group to an electrophilic site (54). Taken together, these data (dendrogram, domain and biochemical) reinforce each other and suggest that the candidate methyltransferase proteins found at cluster level 41 in our analysis are likely to share an unusual chemical mechanism even as they have diverged in sequence as a result of acting on distinct substrates and pathways.
Example 2. Operon data can be powerfully integrated with clustering results and biochemical data in a similar way. Little is known about the Radical SAM member ExsD (62) except that proteins in this operon impact on succinoglycan biosynthesis. We used the neighboring ExsC protein to search an operon database (made by extracting protein links from Radical SAM nucleotide records) as a means of finding other superfamily members with this linkage. ExsC homologs are found adjacent to nine Radical SAM proteins, located in two clusters (Fig. 2), one containing ExsD and another with a Pyrococcus furiosus protein (63). Therefore, it appears likely that the location of ExsD next to ExsC is not fortuitous. These results suggest an interesting hypothesis. ExsC is strongly related to 6-pyruvoyl tetrahydrobiopterin synthase, the second step in tetrahydrobiopterin biosynthesis (64). The first step in tetrahydrobiopterin biosynthesis is the production of a pterin ring by GTP cyclooxidase I. Interestingly, the MoaA proteins provide a unique mechanism for production of a pterin ring from GTP in molybdopterin biosynthesis (65). Therefore, by analogy, the ExsD Radical SAM protein and its neighbor in the operon ExsC could be the first two steps of an unusual pterin synthesis pathway.
DISCUSSION
With over 600 unique sequences, 30 known functions and many additional unknowns, the existing biochemical and genetic data on the Radical SAM proteins easily represent over 1 000 000 person-hours of experimental work in the laboratory. With identification of the superfamily this knowledge base becomes a resource supporting the laboratory efforts of a newly defined community of experimental scientists. All the Radical SAM proteins can now be evaluated for radical chemistry as well as other properties. The usefulness of the classification is illustrated by experiments performed by Nicholson and co-workers based on the observation that spore photoproduct lyase contains the characteristic cysteine motif of the deoxyadenosyl radical proteins (59). They modified an assay for anaerobic ribonucleotide reductase and successfully measured spore photoproduct lyase activity in vitro for the first time.
Radical SAM represents a mechanistic solution for the catalysis of difficult chemical reactions. Robert H. Abeles, who uncovered many unusual enzymatic reactions, is reported to have said ‘if you can formulate on paper a mechanism in two-electron steps, then there is no radical involved’, and this comment is still a practical one (66). However, the many two-electron mechanisms proposed for proteins in this new superfamily can now be seen as too conservative and can be reasonably made more radical.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Gus J.Calapristi and Jim J.Thomas for helpful discussions and Wanda F.Mar and Leigh K.Williams for technical assistance. This work was performed at the Pacific Northwest National Laboratory (PNNL), which is operated by Battelle for the US Department of Energy. Funding support was provided by PNNL Laboratory Directed Research and Development and by Battelle.
References
- 1.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuwald A.F., Liu,J.S., Lipman,D.J. and Lawrence,C.E. (1997) Extracting protein alignment models from the sequence database. Nucleic Acids Res., 25, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional, and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 5.Wu W., Booker,S., Lieder,K.W., Bandarian,V., Reed,G.H. and Frey,P.A. (2000) Lysine 2,3-aminomutase and _trans_-4,5-dehydrolysine: characterization of an allylic analogue of a substrate-based radical in the catalytic mechanism. Biochemistry, 39, 9561–9570. [DOI] [PubMed] [Google Scholar]
- 6.Frey P.A. and Moss,M.L. (1987) _S_-adenosylmethionine and the mechanism of hydrogen transfer in the lysine 2,3-aminomutase reaction. Cold Spring Harbor Symp. Quant. Biol., 52, 571–577. [DOI] [PubMed] [Google Scholar]
- 7.Duin E.C., Lafferty,M.E., Crouse,B.R., Allen,R.M., Sanyal,I., Flint,D.H. and Johnson,M.K. (1997) [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry, 36, 11811–11820. [DOI] [PubMed] [Google Scholar]
- 8.Ollagnier-de Choudens S., Sanakis,Y., Hewitson,K.S., Roach,P., Baldwin,J.E., Münck,E. and Fontecave,M. (2000) Iron-sulfur center of biotin synthase and lipoate synthase. Biochemistry, 39, 4165–4173. [DOI] [PubMed] [Google Scholar]
- 9.Miller J.R., Busby,R.W., Jordan,S.W., Cheek,J., Henshaw,T.F., Ashley,G.W., Broderick,J.B., Cronan,J.E.,Jr and Marletta,M.A. (2000) Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry, 39, 15166–15178. [DOI] [PubMed] [Google Scholar]
- 10.Ollagnier S., Meier,C., Mulliez,E., Gaillard,J., Schuenemann,V., Trautwein,A., Mattioli,T., Lutz,M. and Fontecave,M. (1999) Assembly of 2Fe-2S and 4Fe-4S clusters in the anaerobic ribonucleotide reductase from Escherichia coli. J. Am. Chem. Soc., 121, 6344–6350. [Google Scholar]
- 11.Külzer R., Pils,T., Kappl,R., Hüttermann,J. and Knappe,J. (1998) Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. J. Biol. Chem., 273, 4897–4903. [DOI] [PubMed] [Google Scholar]
- 12.Knappe J., Neugebauer,F.A., Blaschkowski,H.P. and Ganzler,M. (1984) Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc. Natl Acad. Sci. USA, 81, 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey P.A., Ballinger,M.D. and Reed,G.H. (1998) _S_-adenosylmethionine: a ‘poor man’s coenzyme B12’ in the reaction of lysine 2,3-aminomutase. Biochem. Soc. Trans, 26, 304–310. [DOI] [PubMed] [Google Scholar]
- 14.Cosper N.J., Booker,S.J., Ruzicka,F., Frey,P.A. and Scott,R.A. (2000) Direct FeS cluster involvement in generation of a radical in lysine 2,3-aminomutase. Biochemistry, 39, 15668–15673. [DOI] [PubMed] [Google Scholar]
- 15.Frey P.A. and Reed,G.H. (1993) Lysine 2,3-aminomutase and the mechanism of the interconversion of lysine and β-lysine. Adv. Enzymol. Relat. Areas Mol. Biol., 66, 1–39. [DOI] [PubMed] [Google Scholar]
- 16.Johnson M.K. (1998) Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol., 2, 173–181. [DOI] [PubMed] [Google Scholar]
- 17.Kiley P.J. and Beinert,H. (1999) Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev., 22, 341–352. [DOI] [PubMed] [Google Scholar]
- 18.Niewmierzycka A. and Clarke,S. (1999) _S_-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae: identification of a novel protein arginine methyltransferase. J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- 19.Panchenko A.R., Marchler-Bauer,A. and Bryant,S.H. (2000) Combination of threading potentials and sequence profiles improves fold recognition. J. Mol. Biol., 296, 1319–1331. [DOI] [PubMed] [Google Scholar]
- 20.Hershkovitz M.A. and Leipe,D.D. (1998) Phylogenetic analysis. In Baxevanis,A.D. and Ouellette,B.F.F. (eds), Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. John Wiley and Sons, New York, NY, pp. 189–230.
- 21.Everitt B.S. (1993) Cluster Analysis, 3rd Edn. Edward Arnold, London, UK.
- 22.Robertson G.G., Mackinlay,J.D. and Card,S.K. (1991) Cone trees: animated 3D visualizations of hierarchical information. In Proceedings of CHI ACM Conference on Human Factors in Computing Systems. New York, NY, pp. 189–194.
- 23.Lamping J. and Rao,R. (1996) The hyperbolic browser: a focus + context technique for visualizing large hierarchies. J. Vis. Lang. Comput., 7, 35–55. [Google Scholar]
- 24.Jeong C. and Pang,A. (1998) Reconfigurable disc trees for visualizing large hierarchical information space. In Proceedings of IEEE Information Visualization. IEEE Computer Society, Los Alamitos, CA, pp. 19–25.
- 25.Munzner T. (1997) H3: laying out large directed graphs in 3D hyperbolic space. In Proceedings of IEEE Information Visualization. IEEE Computer Society, Los Alamitos, CA, pp. 2–10.
- 26.Johnson B. and Shneiderman,B. (1991) Tree-maps: a space filling approach to the visualization of hierarchical information structures. In Proceedings of IEEE Information Visualization. IEEE Computer Society, Los Alamitos, CA, pp. 284–291.
- 27.Wills G. (1998) An interactive view for hierarchical clustering. In Proceedings of IEEE Information Visualization. IEEE Computer Society, Los Alamitos, CA, pp. 26–31.
- 28.Stasko J., Guzdial,M. and McDonald,K. (1999) Evaluating space-filling visualizations for hierarchical structures. In Proceedings of Late Breaking Hot Topics/IEEE Information Visualization Symposium. IEEE Computer Society, Los Alamitos, CA, pp 35–38.
- 29.Hippler B., Homuth,G., Hoffmann,T., Hungerer,C., Schumann,W. and Jahn,D. (1997) Characterization of Bacillus subtilis hemN. J. Bacteriol., 179, 7181–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyckoff E.E., Duncan,D., Torres,A.G., Mills,M., Maase,K. and Payne,S.M. (1998) Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol., 28, 1139–1152. [DOI] [PubMed] [Google Scholar]
- 31.Wise J.A., Thomas,J.J., Pennock,K., Lantrip,D., Pottier,M., Schur,A. and Crow,V. (1995) Visualizing the non-visual: spatial analysis and interaction with information from text documents. In Proceedings of IEEE Information Visualization. IEEE Computer Society, Los Alamitos, CA, pp. 51–58.
- 32.Begley T.P., Xi,J., Kinsland,C., Taylor,S. and McLafferty,F. (1999) The enzymology of sulfur activation during thiamin and biotin biosynthesis. Curr. Opin. Chem. Biol., 3, 623–629. [DOI] [PubMed] [Google Scholar]
- 33.Allen R.M., Chatterjee,R., Ludden,P.W. and Shah,V.K. (1995) Incorporation of iron and sulfur from NifB cofactor into the iron-molybdenum cofactor of dinitrogenase. J. Biol. Chem., 270, 26890–26896. [DOI] [PubMed] [Google Scholar]
- 34.Esberg B, Leung,H.-C.E., Tsui,H.-C.T., Björk,G.R. and Winkler,M.E. (1999) Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol., 181, 7256–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed K.E., Morris,T.W. and Cronan,J.E.,Jr (1994) Mutants of Escherichia coli K-12 that are resistant to a selenium analog of lipoic acid identify unknown genes in lipoate metabolism. Proc. Natl Acad. Sci. USA, 91, 3720–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ching Y.P., Qi,Z. and Wang,J.H. (2000) Cloning of three neuronal Cdk5 activator binding proteins. Gene, 242, 285–294. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan K.V. (1997) Biosynthesis and processing of the molybdenum cofactors. Biochem. Soc. Trans, 25, 757–761. [DOI] [PubMed] [Google Scholar]
- 38.Akhtar M. (1994) The modification of acetate and propionate side chains during the biosynthesis of haem and chlorophylls: mechanistic and stereochemical studies. Ciba Found. Symp., 180, 131–155. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki J.Y., Bollivar,D.W. and Bauer,C.E. (1997) Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet., 31, 61–89. [DOI] [PubMed] [Google Scholar]
- 40.Tait G.H. (1972) Coproporphyrinogenase activities in extracts of Rhodopseudomonas spheroides and Chromatium strain D. Biochem. J., 128, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zumft W.G. (1997) Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev., 61, 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szameit C., Miech,C., Balleininger,M., Schmidt,B., von Figura,K. and Dierks,T. (1999) The iron sulfur protein AtsB is required for posttranslational formation of formylglycine in the Klebsiella sulfatase. J. Bacteriol., 274, 15375–15381. [DOI] [PubMed] [Google Scholar]
- 43.Kertesz M.A. (2000) Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev., 24, 135–175. [DOI] [PubMed] [Google Scholar]
- 44.Goodwin P.M. and Anthony,C. (1998) The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv. Microbiol. Physiol., 40, 1–80. [DOI] [PubMed] [Google Scholar]
- 45.Mao Y., Varoglu,M. and Sherman,D.H. (1999) Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem. Biol., 6, 251–263. [DOI] [PubMed] [Google Scholar]
- 46.Grewal T.S., Genever,P.G., Brabbs,A.C., Birch,M. and Skerry,T.M. (2000) Best5: a novel interferon-inducible gene expressed during bone formation. FASEB J., 14, 523–531. [DOI] [PubMed] [Google Scholar]
- 47.Zhu H., Cong,J.-P. and Shenk,T. (1997) Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl Acad. Sci. USA, 94, 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boudinot P., Massin,P., Blanco,M., Riffault,S. and Benmansour,A. (1999) vig-1, a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family. J. Virol., 73, 1846–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyutzkanova D., Distler,J. and Altenbuchner,J. (1997) A spectinomycin resistance determinant from the spectinomycin producer _Streptomyces flavopersicus._Microbiology, 143, 2135–2143. [DOI] [PubMed] [Google Scholar]
- 50.Zheng G., Yan,L.Z., Vederas,J.C. and Zuber,P. (1999) Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol., 181, 7346–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Möhrle V., Roos,U. and Bormann,C. (1995) Identification of cellular proteins involved in nikkomycin production in Streptomyces tendae Tü901. Mol. Microbiol., 15, 561–571. [DOI] [PubMed] [Google Scholar]
- 52.Morita M., Tomita,K., Ishizawa,M., Takagi,K., Kawamura,F., Takahashi,H. and Morino,T. (1999) Cloning of oxetanocin A biosynthetic and resistance genes that reside on a plasmid of Bacillus megaterium strain NK84-0128. Biosci. Biotechnol. Biochem., 63, 563–566. [DOI] [PubMed] [Google Scholar]
- 53.Kuzuyama T., Seki,T., Dairi,T., Hidaka,T. and Seto,H. (1995) Nucleotide sequence of fortimicin KL1 methyltransferase gene isolated from Micromonospora olivasterospora, and comparison of its deduced amino acid sequence with those of methyltransferases involved in the biosynthesis of bialaphos and fosfomycin. J. Antibiot., 48, 1191–1193. [DOI] [PubMed] [Google Scholar]
- 54.Seto H. and Kuzuyama,T. (1999) Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep., 16, 589–596. [DOI] [PubMed] [Google Scholar]
- 55.Summers R.G., Donadio,S., Staver,M.J., Wendt-Pienkowski,E., Hutchinson,C.R. and Katz,L. (1997) Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology, 143, 3251–3262. [DOI] [PubMed] [Google Scholar]
- 56.Trefzer A., Salas,J.A. and Bechthold,A. (1999) Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Nat. Prod. Rep., 16, 283–299. [DOI] [PubMed] [Google Scholar]
- 57.Xue Y., Zhao,L., Liu,H.-W. and Sherman,D.H. (1998) A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl Acad. Sci. USA, 95, 12111–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heider J., Spormann,A.M., Beller,H.R. and Widdel,F. (1999) Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev., 22, 459–473. [Google Scholar]
- 59.Rebeil R., Sun,Y., Chooback,L., Pedraza-Reyes,M., Kinsland,C., Begley,T.P. and Nicholson,W.L. (1998) Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J. Bacteriol., 180, 4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ludwig M.L. and Matthews,R.G. (1997) Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem., 66, 269–313. [DOI] [PubMed] [Google Scholar]
- 61.Gough S.P., Petersen,B.O. and Duus,J.O. (2000) Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc. Natl Acad. Sci. USA, 97, 6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Becker A., Küster,H., Niehaus,K. and Pühler,A. (1995) Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol. Gen. Genet., 249, 487–497. [DOI] [PubMed] [Google Scholar]
- 63.Jenney F.E.,Jr, Verhagen,M.F.J.M., Cui,X. and Adams,M.W.W. (1999) Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science, 286, 306–309. [DOI] [PubMed] [Google Scholar]
- 64.Bracher A., Eisenreich,W., Schramek,N., Ritz,H., Götze,E., Herrmann,A., Gütlich,M. and Bacher,A. (1998) Biosynthesis of pteridines: NMR studies on the reaction mechanisms of GTP cyclohydrolase I, pyruvoyltetrahydropterin synthase, and sepiapterin reductase. J. Biol. Chem., 273, 28132–28141. [DOI] [PubMed] [Google Scholar]
- 65.Rieder C., Eisenreich,W., O’Brien,J., Richter,G., Götze,E., Boyle,P., Blanchard,S., Bacher,A. and Simon,H. (1998) Rearrangement reactions in the biosynthesis of molybdopterin: an NMR study with multiply 13C/15N labelled precursors. Eur. J. Biochem., 255, 24–36. [DOI] [PubMed] [Google Scholar]
- 66.Buckel W. and Golding,B.T. (1999) Radical species in the catalytic pathways of enzymes from anaerobes. FEMS Microbiol. Rev., 22, 523–541. [Google Scholar]
- 67.Kletzin A., Mukund,S., Kelley-Crouse,T.L., Chan,M.K., Rees,D.C. and Adams,M.W.W. (1995) Molecular characterization of the genes encoding the tungsten-containing aldehyde ferredoxin oxidoreductase from Pyrococcus furiosus and formaldehyde ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol., 177, 4817–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]