microRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate (original) (raw)
Abstract
Hagfish and lampreys are the only living representatives of the jawless vertebrates (agnathans), and compared with jawed vertebrates (gnathostomes), they provide insight into the embryology, genomics, and body plan of the ancestral vertebrate. However, this insight has been obscured by controversy over their interrelationships. Morphological cladistic analyses have identified lampreys and gnathostomes as closest relatives, whereas molecular phylogenetic studies recover a monophyletic Cyclostomata (hagfish and lampreys as closest relatives). Here, we show through deep sequencing of small RNA libraries, coupled with genomic surveys, that Cyclostomata is monophyletic: hagfish and lampreys share 4 unique microRNA families, 15 unique paralogues of more primitive microRNA families, and 22 unique substitutions to the mature gene products. Reanalysis of morphological data reveals that support for cyclostome paraphyly was based largely on incorrect character coding, and a revised dataset is not decisive on the mono- vs. paraphyly of cyclostomes. Furthermore, we show fundamental conservation of microRNA expression patterns among lamprey, hagfish, and gnathostome organs, implying that the role of microRNAs within specific organs is coincident with their appearance within the genome and is conserved through time. Together, these data support the monophyly of cyclostomes and suggest that the last common ancestor of all living vertebrates was a more complex organism than conventionally accepted by comparative morphologists and developmental biologists.
Keywords: complexity, cyclostomata, evolution, organ, homology
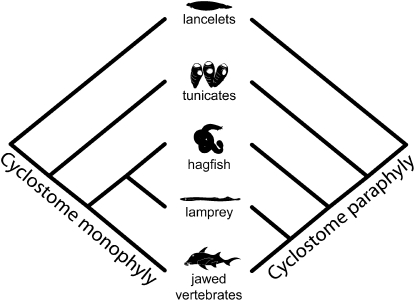
The origin and early evolution of vertebrates have been a focus of molecular and organismal evolutionary biology because of the fundamental events that attended this formative episode of our own evolutionary history over one-half billion years ago (1). However, attempts to integrate these perspectives have been stymied by the different phylogenetic perspectives afforded by molecular and morphological datasets. Molecular datasets, incorporating protein-coding genes, ribosomal RNA genes, and/or mitochondrial genes (2–21), invariably find that the jawless hagfish and lampreys constitute a clade, Cyclostomata (Fig. 1, on the left). In contrast, morphological datasets (22–36) have supported a closer relationship between lampreys and gnathostomes, rendering Cyclostomata paraphyletic (Fig. 1, on the right) and hagfish not vertebrates but mere craniates (33).
Fig. 1.
The two competing hypotheses. Either lampreys are more closely related to hagfish than they are to gnathostomes, making Cyclostomata monophyletic (on the left), or lampreys are more closely related to gnathostomes than they are to hagfish, making Cyclostomata paraphyletic (on the right).
Attempts have been made to reconcile these two views: a number of morphological characters have been identified that support the monophyly of cyclostomes (37, 38), but they have been overwhelmed by a seemingly far greater number of characters supporting cyclostome paraphyly (30, 31). Indeed, an analysis of combined morphological and molecular datasets has suggested that the signal of cyclostome paraphyly in morphological datasets is stronger than the signal for monophyly from molecular data (39). The interrelationships of hagfish, lampreys, and gnathostomes thus remain uncertain, and this has become a classic example of phylogenetic conflict between morphological and molecular data (7, 39). If morphological phylogenies are correct, hagfish provide an experimental model for investigating the evolutionary assembly of the vertebrate body plan shared by lampreys and gnathostomes. Alternatively, if the molecular phylogenies are correct, then it would indicate that the shared similarities of lampreys and gnathostomes are convergent or that these characters are absent through loss in the hagfish lineage. These would represent the most extraordinary examples of convergence or degeneracy, respectively, in vertebrate evolutionary history (18, 35).
We attempted to resolve the interrelationships of hagfish, lampreys, and gnathostomes through analysis of their microRNA (miRNA) repertoire. miRNAs are small, noncoding regulatory genes implicated in the control of cellular differentiation and homeostasis and as such, might be involved in the evolution of complexity (40–42). Because ancient miRNAs show a level of sequence conservation exceeding that of ribosomal DNA (43), it is possible to discern the evolutionary origins of miRNA families at even the deepest levels of animal phylogeny (43, 44). The rarity with which ancient miRNAs were lost within most evolutionary lineages, coupled with the continuous acquisition of miRNAs through geologic time in all metazoan lineages examined to date, makes miRNAs one of the most useful classes of characters in phylogenetics (45). Thus, miRNAs can be used to discern the interrelationships among the major vertebrate lineages and simultaneously, lend insight into the origin of vertebrate characteristics.
We constructed small RNA libraries from total RNA (Methods) from ammocoete larvae of the brook lamprey Lampetra planeri, from a single adult individual of the Atlantic hagfish Myxine glutinosa, from the catshark Scyliorhinus canicula, and for nine individually processed organs/regions (brain, gills, gut, heart, kidney, liver, mouth, muscle, and skin) from a single adult individual of the sea lamprey Petromyzon marinus. Using a combination of high-throughput 454 pyrosequencing and Illumina technology, we identified miRNAs from each library and found that shared gains of miRNAs support the monophyly of cyclostomes (lamprey and hagfish). We also revised, expanded, and reanalyzed an extensive morphological dataset previously found to support cyclostome paraphyly (23) and show that cyclostome monophyly is just as likely given these data. In addition, profiling the miRNA expression within nine organs of P. marinus shows conservation with known expression profiles in homologous organs across vertebrates. Our data suggest that the role of miRNAs within specific organs is coincident with their appearance within the genome, and thus, miRNAs may have played a role in the acquisition of organismal complexity in vertebrates.
Results and Discussion
miRNAs Shared Between Lampreys and Hagfish Support Cyclostome Monophyly.
Derivative cDNA libraries from the brook lamprey L. planeri, the sea lamprey P. marinus, and the Atlantic hagfish M. glutinosa were sequenced using high-throughput 454 pyrosequencing (Methods), yielding 422,122 (59,759 nonredundant) parsed high-quality reads. Additionally, we sequenced small RNAs from the catshark Scyliorhinus canicula using Illumina technology, yielding 333,294 (127,015 nonredundant) parsed high-quality reads. The resulting reads from all four taxa were then interrogated using miRMiner (43) to identify known and unknown miRNAs (Dataset S1).
Because the genome traces of the sea lamprey P. marinus are publicly available (http://www.ncbi.nlm.nih.gov/genomeprj?term=petromyzon), we first focused on elucidating the miRNA repertoire of this species. We identified 245 miRNA genes in P. marinus, including one family lost in gnathostomes (miR-315) and a second family lost in osteichthyans (mir-281) (Dataset S1). An additional 24 miRNA genes are inferred to be present in the genome of P. marinus, because, although the genes could not be located in the trace archives, reads of these phylogenetically conserved miRNAs were discovered in our libraries (e.g., miR-31, -34, -122, etc.) (Dataset S1). Of the 269 genes present in P. marinus, 202 are conserved in other animals, with 21 shared only with the brook lamprey, L. planeri (Dataset S1).
Lampreys lack the bilaterian miRNAs miR-71, miR-242, miR-252, and miR-278, as do urochordates and all other vertebrates examined to date. However, very few miRNA genes have been lost within the lamprey lineage itself: only a single miRNA family seems to have been lost in P. marinus (miR-214), because reads were detected in L. planeri (Dataset S1); however, reads were not detected in P. marinus, and the gene was not located in the trace archives. Conversely, we failed to detect transcripts of only two miRNA families in _L. planeri_—the lowly expressed miRNAs (Dataset S1) miR-202 and miR-875 (although we did not examine reads from an adult individual, and no genomic sequence for this species is currently available to confirm a true absence). Therefore, these two lamprey species share a miRNA complement of at least 200 genes and between them, have together lost no more than three miRNA families total since they last shared a common ancestor some time in the last 10–40 million y (10).
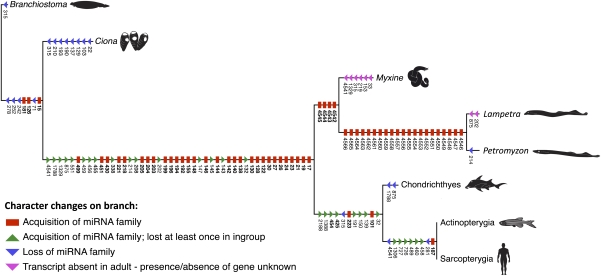
To determine the phylogenetic position of hagfish, we analyzed the conserved miRNA complement of M. glutinosa. Of the 46 vertebrate-specific miRNA families shared between lamprey and gnathostomes (Fig. 2), we detected all but two in our hagfish library: miR-1329 (which is expressed exclusively in the lamprey kidney) (Dataset S1) and miR-4541, an miRNA family found thus far only in the two sharks and the two lamprey species (Dataset S1). However, the hagfish shares four unique miRNA families with the lampreys that are not found or expressed in gnathostomes or in any other animal species investigated to date, miR-4542, miR-4543, miR-4544, and miR-4545 (Dataset S1 and Fig. S1), and a phylogenetic analysis based on the presence and absence of miRNA families (Dataset S2) supports the monophyly of the cyclostomes (Fig. 2 and Fig. S2).
Fig. 2.
Phylogenetic distribution of all miRNA families analyzed in chordates (see Dataset S2 for data matrix and Fig. S2 for complete phylogenetic analysis). Cyclostomes share four miRNA families not found in any other animal species investigated to date, and a maximum parsimony analysis supports the monophyly of Cyclostomata. Note that miRNA families specific to a single species are not indicated, but losses of more primitive families are indicated. Of particular interest is the number of miRNA families acquired in the stem lineage leading to the vertebrate crown group.
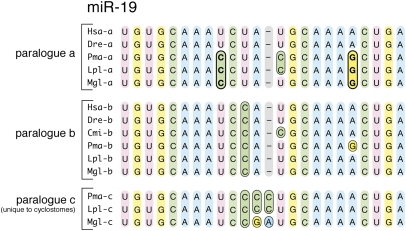
Further evidence of cyclostome monophyly is found in the paralogy group relations within miRNA families (46). Fifteen paralogues of previously described miRNA families (Fig. 3 and Dataset S1) are shared by the hagfish and lampreys to the exclusion of gnathostomes—we did not detect a single paralogue supporting cyclostome paraphyly. Finally, we examined the mature sequences of each miRNA to ask if polarizable nucleotide substitutions had occurred that supported either cyclostome monophyly or paraphyly (or some other set of relations). We did not find any nucleotide substitutions in the mature sequence of any vertebrate miRNA that is shared between gnathostomes and lampreys to the exclusion of hagfish (or between hagfish and gnathostomes to the exclusion of lamprey). However, we did find 22 derived nucleotide substitutions in the mature sequences of 18 miRNAs exclusive to the three cyclostome taxa investigated (Fig. 3 and Dataset S1). Thus, the acquisition of miRNA families, miRNA genes, and the nucleotide substitution patterns of conserved miRNA genes all support cyclostome monophyly.
Fig. 3.
The presence of paralogues of more primitive miRNA families and conserved nucleotide substitutions both support the monophyly of Cyclostomata. Shown is miR-19 as an example of a group of miRNAs that shows both conserved nucleotide substitutions (19a; Top, bold) with respect to the other paralogue(s) (19b and 19c; Middle and Bottom) and the possession of a paralogue (miR-19c) not present in any known gnathostome (Dataset S1 has the complete description of both paralogues and nucleotide substitutions supporting cyclostome monophyly). Cmi, Callorhinchus milii; Dre, Danio rerio; Hsa, Homo sapiens; Lpl, Lampetra planeri; Mgl, Myxine glutinosa; Pma, Petromyzon marinus.
Phenotypic Cladistic Data Do Not Distinguish Between Cyclostome Monophyly vs. Paraphyly.
The phylogenetic distribution of vertebrate miRNAs corroborates molecular sequence data in supporting cyclostome monophyly (2–21), contradicting what has been considered an equally strong signal from phenotypic datasets supporting cyclostome paraphyly (22–36). To determine the source of this discordance, we augmented a phenotypic dataset based on the nervous system (23), with characters representative of other organ systems recoded from observations and the primary literature rather than recycled from previous analyses (SI Text and Dataset S3). In so doing, we considered all characters that have been marshaled previously in support of cyclostome monophyly or paraphyly. We find that, although the revised dataset (SI Text) marginally favors cyclostome paraphyly (monophyly is one step longer in a tree of 237 steps) (Fig. S3), Templeton (47), Kishino–Hasegawa (48), and approximate two-tailed Shimodaira–Hasegawa (49) tests reveal that the dataset is indecisive on this question (Templeton: P = 0.8415; K–H: P = 0.8421; approximate S–H is one-half P of K–H) (49). This is because much of the evidence traditionally interpreted as supporting cyclostome paraphyly has been based on spurious character design. For example, many of the characters are inapplicable to the outgroup, making it impossible to discriminate between the primary or secondary absence in hagfish of characters otherwise found only in lampreys and gnathostomes (e.g., the proximity of the atrium and ventricle of the heart, radial muscles, and retinal synaptic ribbons). In addition, some characters have been coded as absent in hagfish when data have merely been lacking (e.g., heart response to catecholamines, pituitary control of gametogenesis, and sexual dimorphism). Finally, the uncritical recycling of characters and their codings between generations of analyses has resulted in the repeated use of obsolete data (50). For instance, similarities in the immune system of lampreys and gnathostomes have been exploited to draw a distinction from hagfish (30–35, 51). However, it has been long established that lampreys and hagfish share a distinct type of adaptive immune system based on variable lymphocyte receptors, rather than the Ig-based T and B antigen receptors that characterize the lymphocytes of jawed vertebrates (52), and thus, similarities in the immune system of lampreys and jawed vertebrates are convergent.
miRNA Expression Profiles Are Conserved Across Vertebrates.
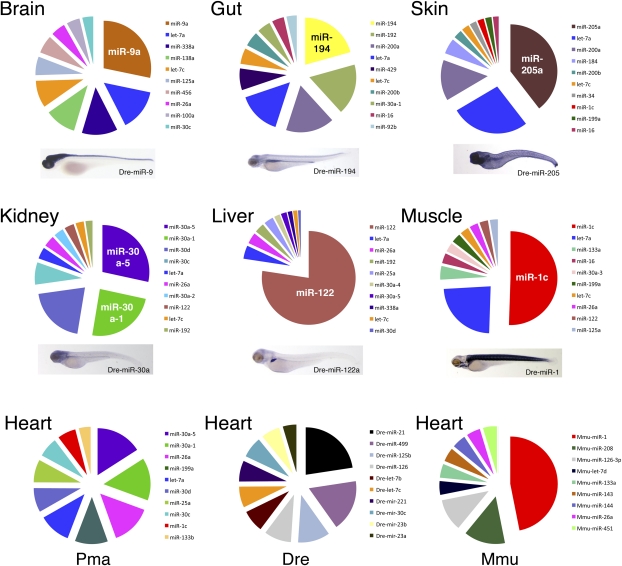
The origin of vertebrates occurred in association with a very high rate of miRNA family innovation, and it has been proposed that this is a causal association, because where expression data are available, vertebrate miRNAs are often expressed in tissues and organs that are unique to vertebrates (41). This hypothesis predicts that the organ-specific expression of vertebrate-specific miRNAs is highly conserved, such that data from the zebrafish (Danio) and the mouse (Mus) are representative not only of osteichthyans (the clade that they circumscribe) but of vertebrates more generally. Our phylogenetic results indicate that a comparison of existing data with lampreys will provide an adequate test of the hypothesis, because together, these taxa circumscribe the clade of all living vertebrates (Fig. 2). Expression data for seven different P. marinus organs are shown in Fig. 4. Similar to Danio (53, 54) and Mus (55), each lamprey organ expresses a specific suite of miRNAs that gives the organ a unique miRNA expression profile. For example, ignoring the ubiquitously expressed let-7, the four highest expressed miRNA genes in the lamprey brain are miR-9a, miR-338a, miR-138a, and miR-125a, whereas the four highest expressed miRNA genes in the lamprey gut are miR-194, miR-192, miR-200a, and miR-429 (Fig. 4 and Dataset S4). Furthermore, similar to mouse (56), the lamprey brain is the most complex of the organs queried, and the gut and liver are the least, at least in terms of the number of different miRNAs expressed (Dataset S4). With just one exception (the heart), the miRNA with the highest expression in each of the lamprey organs is also expressed in that same organ in both Danio (Fig. 4 Insets) and hagfish (Fig. S4). Thus, homologous organs in vertebrates more often than not (57) express homologous miRNAs, consistent with the hypothesis that miRNAs (e.g., miR-30 and miR-122) were instrumental in the evolutionary origin of vertebrate-specific organs (e.g., kidney and liver, respectively) (41).
Fig. 4.
miRNA expression profile of seven different lamprey organs. Only the top 10 highest expressed miRNAs (Dataset S4) are shown, and each specific miRNA is given a distinct color for all pie charts. Below each pie chart is the expression pattern of the highest expressed gene in the lamprey library in the zebrafish (54)—note the concordance between the lamprey and zebrafish for all organs queried except for the heart (Bottom). Pma, Petromyzon marinus; Dre, Danio rerio; Mmu, Mus musculus.
Conclusions
Hinging on debate over the interrelationships of living jawless and jawed vertebrates has been the nature of the ancestral vertebrate and the pattern and sequence of organismal and genomic evolution, on which hypotheses of developmental evolution are based. We conclude that cyclostomes are monophyletic, and thus, characters reconstructed as lamprey and gnathostome synapomorphies are actually shared primitive characters of all vertebrates, with hagfish anatomy having degenerated to a remarkable degree (18, 36). Cyclostome paraphyly (22) and a hierarchical distinction between craniates and vertebrates (33) afforded insight into the assembly of vertebrate characters (58). With the recognition of cyclostome monophyly, however, that taxonomic distinction and evolutionary insight are lost. Evidently, the crown ancestor of vertebrates was more complex, phenotypically and developmentally, than has been perceived hitherto (58), making attempts to explain mechanistically the distinction between vertebrates and invertebrates even more formidable. Nonetheless, in reconciling phylogenies grounded in genotype and phenotype, we provide a holistic framework for uncovering the formative events in the evolutionary emergence of vertebrates. We predict that the renaissance in hagfish embryology (59) will further show the loss of vertebrate characters, but with the recognition of cyclostome monophyly, attempts to dissect the assembly of the vertebrate body plan can be focused on comparative analysis of lamprey development and genomics. The prolific origin of miRNA families in the vertebrate stem-lineage and their expression in vertebrate-specific tissues and organs supports the idea that miRNAs played a pivotal role, as part of a broader gene regulatory landscape, in the assembly of the vertebrate body plan (41).
Methods
Total RNA Extraction, Northern Analysis, and Small RNA Library Construction.
Embryonic brook lampreys (L. planeri) were collected from Highland Water, upstream of Millyford Bridge, New Forest National Park (United Kingdom) and allowed to develop in captivity at 16 °C in filtered river water until hatching. Adult sea lamprey (P. marinus) were collected from Lake Champlain (Vermont), and a single individual was dissected to isolate the brain, gut, gills, heart, kidney, liver, mouth and tongue, muscle, and skin. Atlantic hagfish (M. glutinosa) were collected at Kristineberg Marine Station, Gulmarsfjord, Sweden and purchased from Gulf of Maine Inc. (Pembroke, ME). RNA was extracted from 20 combined larval L. planeri, from each dissected tissue and organ derived from a single adult P. marinus, and from a single adult M. glutinosa. From these animals, small RNA libraries were constructed individually and sequenced with a unique barcode using 454 DNA pyrosequencing (Branford, CT) as described previously (43). The resulting reads were then analyzed with miRMiner to identify known and unknown miRNAs (43), with additional filters for transfer and ribosomal RNAs written with custom shell scripts.
RNA was also extracted from the brain, gut, heart, kidney, liver, muscle, and skin derived from a single adult M. glutinosa, and northern analyses using Starfire probes (IDT) designed against the mature miRNA sequence (sequences available on request) were performed as previously described (43). Catshark (S. canicula) embryos were obtained from commercial sources, and RNA was extracted from five embryos near hatching. S. canicula RNA was sequenced for small RNAs using the Illumina sequencing platform and analyzed using miRMiner as described (43). All genomic inquiries for miRNAs in P. marinus and Callorhinchus milii (elephant shark) were made through National Center for Biotechnology Information using the available genomic traces. All alignments and sequence analyses were performed using MacVector (v. 10.0.2). Secondary structures of precursor miRNA transcripts were predicted using mFold (60).
Morphological Analysis.
The phenotypic dataset was coded directly from the primary literature and from direct observation of anatomy (SI Text). We designed and coded characters using a contingent coding strategy, because it is the only approach that is theoretically and operationally valid in instances, as here, where many of the characters are inapplicable to the outgroup (61). We restricted our analyses to a parsimony-based approach, because phenotypic support for hagfish–lamprey–gnathostome relationships has always been debated using this method of phylogenetic inference. The cladistic parsimony analyses, Bremer support index calculations, and Templeton and Kishino–Hasegawa tests were performed in PAUP*4.0b10 running on Mac OS9 within a Sheepshaver 2.3 emulator on an Intel MacBook.
Supplementary Material
Corrected Supporting Information
Acknowledgments
We thank E. Marsden, S. Shimeld, and M. Thorndyke for access to materials and J. Mallatt, E. Sperling, D. Pisani, and M. Schubert for comments on a previous draft. P.C.J.D. is supported by the Biotechnology and Biological Sciences Research Council, European Commission Seventh Framework Programme EU FP7, The Leverhulme Trust, Natural Environment Research Council, and National Endowment for Science Technology and the Arts (NESTA); K.J.P. is supported by the National Aeronautics and Space Administration/Ames and National Science Foundation. A.M.H. was supported by Award Number T32GM008704 from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in miRBase, www.mirbase.org.
See Commentary on page 19137.
*This Direct Submission article had a prearranged editor.
References
- 1.Shimeld SM, Holland PWH. Vertebrate innovations. Proc Natl Acad Sci USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock DW, Whitt GS. Evidence from 18S ribosomal RNA sequences that lampreys and hagfishes form a natural group. Science. 1992;257:787–789. doi: 10.1126/science.1496398. [DOI] [PubMed] [Google Scholar]
- 3.Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- 4.Lartillot N, Philippe H. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos Trans R Soc Lond B Biol Sci. 2008;363:1463–1472. doi: 10.1098/rstb.2007.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 6.Yu SY, et al. Phylogenetic analysis of 48 gene families revealing relationships between hagfishes, lampreys, and Gnathostomata. J Genet Genomics. 2008;35:285–290. doi: 10.1016/S1673-8527(08)60041-7. [DOI] [PubMed] [Google Scholar]
- 7.Goodman M, Miyamoto MM, Czelusniak J. Pattern and processes in vertebrate phylogeny revealed by coevolution of molecules and morphologies. In: Patterson C, editor. Molecules and Morphology in Evolution: Conflict or Compromise? Cambridge, UK: Cambridge University Press; 1987. pp. 141–176. [Google Scholar]
- 8.Delarbre C, et al. The complete nucleotide sequence of the mitochondrial DNA of the agnathan Lampetra fluviatilis: Bearings on the phylogeny of cyclostomes. Mol Biol Evol. 2000;17:519–529. doi: 10.1093/oxfordjournals.molbev.a026332. [DOI] [PubMed] [Google Scholar]
- 9.Kuraku S, Hoshiyama D, Katoh K, Suga H, Miyata T. Monophyly of lampreys and hagfishes supported by nuclear DNA-coded genes. J Mol Evol. 1999;49:729–735. doi: 10.1007/pl00006595. [DOI] [PubMed] [Google Scholar]
- 10.Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoolog Sci. 2006;23:1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- 11.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 12.Lanfranchi G, Pallavicini A, Laveder P, Valle G. Ancestral hemoglobin switching in lampreys. Dev Biol. 1994;164:402–408. doi: 10.1006/dbio.1994.1210. [DOI] [PubMed] [Google Scholar]
- 13.Turbeville JM, Schulz JR, Raff RA. Deuterostome phylogeny and the sister group of the chordates: Evidence from molecules and morphology. Mol Biol Evol. 1994;11:648–655. doi: 10.1093/oxfordjournals.molbev.a040143. [DOI] [PubMed] [Google Scholar]
- 14.Lipscomb DL, Farris JS, Källersjo M, Tehler A. Support, ribosomal sequences and the phylogeny of the eukaryotes. Cladistics. 1998;14:303–338. doi: 10.1111/j.1096-0031.1998.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 15.Winchell CJ, Sullivan J, Cameron CB, Swalla BJ, Mallatt J. Evaluating hypotheses of deuterostome phylogeny and chordate evolution with new LSU and SSU ribosomal DNA data. Mol Biol Evol. 2002;19:762–776. doi: 10.1093/oxfordjournals.molbev.a004134. [DOI] [PubMed] [Google Scholar]
- 16.Mallatt J, Sullivan J, Winchell CJ. The relationship of lampreys to hagfishes: A spectral analysis of ribosomal DNA sequences. In: Ahlberg PE, editor. Major Events in Early Vertebrate Evolution: Palaeontology, Phylogeny, Genetics and Development. London: Taylor & Francis; 2001. pp. 106–118. [Google Scholar]
- 17.Mallatt J, Winchell CJ. Ribosomal RNA genes and deuterostome phylogeny revisited: More cyclostomes, elasmobranchs, reptiles, and a brittle star. Mol Phylogenet Evol. 2007;43:1005–1022. doi: 10.1016/j.ympev.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Delarbre C, Gallut C, Barriel V, Janvier P, Gachelin G. Complete mitochondrial DNA of the hagfish, Eptatretus burgeri: The comparative analysis of mitochondrial DNA sequences strongly supports the cyclostome monophyly. Mol Phylogenet Evol. 2002;22:184–192. doi: 10.1006/mpev.2001.1045. [DOI] [PubMed] [Google Scholar]
- 19.Mallatt J, Sullivan J. 28S and 18S rDNA sequences support the monophyly of lampreys and hagfishes. Mol Biol Evol. 1998;15:1706–1718. doi: 10.1093/oxfordjournals.molbev.a025897. [DOI] [PubMed] [Google Scholar]
- 20.Furlong RF, Holland PWH. Bayesian phylogenetic analysis supports monophyly of ambulacraria and of cyclostomes. Zoolog Sci. 2002;19:593–599. doi: 10.2108/zsj.19.593. [DOI] [PubMed] [Google Scholar]
- 21.Takezaki N, Figueroa F, Zaleska-Rutczynska Z, Klein J. Molecular phylogeny of early vertebrates: Monophyly of the agnathans as revealed by sequences of 35 genes. Mol Biol Evol. 2003;20:287–292. doi: 10.1093/molbev/msg040. [DOI] [PubMed] [Google Scholar]
- 22.Løvtrup S. The Phylogeny of the Vertebrata. New York: Wiley; 1977. p. 330. [Google Scholar]
- 23.Khonsari RH, Li B, Vernier P, Northcutt RG, Janvier P. Agnathan brain anatomy and craniate phylogeny. Acta Zool. 2009;90:52–68. [Google Scholar]
- 24.Jefferies RPS. The Ancestry of the Vertebrates. London: British Museum Natural History; 1986. [Google Scholar]
- 25.Forey PL, Janvier P. Agnathans and the origin of jawed vertebrates. Nature. 1993;361:129–134. [Google Scholar]
- 26.Forey PL. Yet more reflections on agnathan-gnathostome relationships. J Vertebr Paleontol. 1984;4:330–343. [Google Scholar]
- 27.Janvier P. The dawn of the vertebrates: Characters versus common ascent in the rise of current vertebrate phylogenies. Palaeontology. 1996;39:259–287. [Google Scholar]
- 28.Janvier P. Early Vertebrates. Oxford: Oxford University Press; 1996. [Google Scholar]
- 29.Donoghue PCJ, Smith MP. The anatomy of Turinia pagei (Powrie) and the phylogenetic status of the Thelodonti. Trans R Soc Edinb Earth Sci. 2001;92:15–37. [Google Scholar]
- 30.Donoghue PCJ, Forey PL, Aldridge RJ. Conodont affinity and chordate phylogeny. Biol Rev Camb Philos Soc. 2000;75:191–251. doi: 10.1017/s0006323199005472. [DOI] [PubMed] [Google Scholar]
- 31.Gess RW, Coates MI, Rubidge BS. A lamprey from the Devonian period of South Africa. Nature. 2006;443:981–984. doi: 10.1038/nature05150. [DOI] [PubMed] [Google Scholar]
- 32.Forey PL. Agnathans recent and fossil, and the origin of jawed vertebrates. Rev Fish Biol Fish. 1995;5:267–303. [Google Scholar]
- 33.Janvier P. The phylogeny of the Craniata, with particular reference to the significance of fossil “agnathans”. J Vertebr Paleontol. 1981;12:121–159. [Google Scholar]
- 34.Hardisty MW. The Biology of Cyclostomes. London: Chapman & Hall; 1979. [Google Scholar]
- 35.Hardisty MW. Lampreys and Hagfishes: Analysis of Cyclostome Relationships. The Biology of Lampreys. London: Academic; 1982. [Google Scholar]
- 36.Hardisty MW. Lampreys: Life Without Jaws. Forrest Text, Tresaith-Ceredigion-SA43 2JG-UK; 2006. [Google Scholar]
- 37.Yalden DW. Feeding mechanisms as evidence of cyclostome monophyly. Zool J Linn Soc. 1985;84:291–300. [Google Scholar]
- 38.Mallatt J. Crossing a major morphological boundary: The origin of jaws in vertebrates. Zoology. 1997;100:128–140. [Google Scholar]
- 39.Near TJ. Conflict and resolution between phylogenies inferred from molecular and phenotypic data sets for hagfish, lampreys, and gnathostomes. J Exp Zool B Mol Dev Evol. 2009;312:749–761. doi: 10.1002/jez.b.21293. [DOI] [PubMed] [Google Scholar]
- 40.Christodoulou F, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heimberg AM, Sempere LF, Moy VN, Donoghue PCJ, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci USA. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 43.Wheeler BM, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 44.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperling EA, Peterson KJ. microRNAs and metazoan phylogeny: Big trees from little genes. In: Telford MJ, Littlewood DTJ, editors. Animal Evolution–Genomes, Trees and Fossils. Oxford: Oxford University Press; 2009. pp. 157–170. [Google Scholar]
- 46.Goodman M, Czelusniak J, Moore GW, Romero-Herrera AE, Matsuda G. Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst Zool. 1979;28:132–163. [Google Scholar]
- 47.Templeton A. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution. 1983;37:221–224. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 48.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 49.Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 50.Jenner RA. Bilaterian phylogeny and uncritical recycling of morphological data sets. Syst Biol. 2001;50:730–742. doi: 10.1080/106351501753328857. [DOI] [PubMed] [Google Scholar]
- 51.Nicholls H. Evolution: Mouth to mouth. Nature. 2009;461:164–166. doi: 10.1038/461164a. [DOI] [PubMed] [Google Scholar]
- 52.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 53.Soares AR, et al. Parallel DNA pyrosequencing unveils new zebrafish microRNAs. BMC Genomics. 2009;10:195. doi: 10.1186/1471-2164-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 55.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strauss WM, Chen C, Lee C-T, Ridzon D. Nonrestrictive developmental regulation of microRNA gene expression. Mamm Genome. 2006;17:833–840. doi: 10.1007/s00335-006-0025-7. [DOI] [PubMed] [Google Scholar]
- 57.Ason B, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janvier P. Ostracoderms and the shaping of the gnathostome characters. In: Ahlberg PE, editor. Major Events in Early Vertebrate Evolution: Palaeontology, Phylogeny, Genetics and Development. London: Taylor and Francis; 2001. pp. 172–186. [Google Scholar]
- 59.Ota KG, Kuraku S, Kuratani S. Hagfish embryology with reference to the evolution of the neural crest. Nature. 2007;446:672–675. doi: 10.1038/nature05633. [DOI] [PubMed] [Google Scholar]
- 60.Néron B, et al. Mobyle: A new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strong EE, Lipscomb D. Character coding and inapplicable data. Cladistics. 1999;15:363–371. doi: 10.1111/j.1096-0031.1999.tb00272.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Corrected Supporting Information