A phosphotyrosine displacement mechanism for activation of Src by PTPα (original) (raw)
Abstract
Protein tyrosine phosphatase α (PTPα) is believed to dephosphorylate physiologically the Src proto-oncogene at phosphotyrosine (pTyr)527, a critical negative-regulatory residue. It thereby activates Src, and PTPα overexpression neoplastically transforms NIH 3T3 cells. pTyr789 in PTPα is constitutively phosphorylated and binds Grb2, an interaction that may inhibit PTPα activity. We show here that this phosphorylation also specifically enables PTPα to dephosphorylate pTyr527. Tyr789→Phe mutation abrogates PTPα–Src binding, dephosphorylation of pTyr527 (although not of other substrates), and neoplastic transformation by overexpressed PTPα in vivo. We suggest that pTyr789 enables pTyr527 dephosphorylation by a pilot binding with the Src SH2 domain that displaces the intramolecular pTyr527–SH2 binding. Consistent with model predictions, we find that excess SH2 domains can disrupt PTPα–Src binding and can block PTPα-mediated dephosphorylation and activation in proportion to their affinity for pTyr789. Moreover, we show that, as predicted by the model, catalytically defective PTPα has reduced Src binding in vivo. The displacement mechanism provides another potential control point for physiological regulation of Src-family signal transduction pathways.
Keywords: dephosphorylation/mitosis/protein tyrosine phosphatase α/SH2 domain/Src
Introduction
The Src protein tyrosine kinase is involved in multiple signal transduction events; it and closely related Src family members are also involved in many cellular functions including mitogenesis, cell cycle progression and neuronal differentiation (for review, see Thomas and Brugge, 1997). Src is ubiquitously expressed, but is found at highest levels in brain, platelets and osteoclasts (for review, see Brown and Cooper, 1996). In fibroblasts, Src is bound to perinuclear membranes, endosomes and secretory vesicles, and at the cytoplasmic face of the plasma membrane where it can interact with a variety of growth factor and fibronectin receptors (for review, see Brown and Cooper, 1996). Src kinase activity is negatively regulated during most of the cell cycle, but is transiently activated by growth-factor- and cell-cycle-dependent phosphorylations and dephosphorylations at multiple sites within the protein.
The most important regulatory phosphorylation site in Src is Tyr527 (chicken Src numbering), six residues from the C-terminus. When phosphorylated, as in ∼90–95% of Src in normally growing fibroblasts (Shenoy et al., 1992), phosphotyrosine (pTyr)527 binds to the Src SH2 domain (for review, see Thomas and Brugge, 1997). This intramolecular association stabilizes a compact, inactive form of Src (Sicheri et al., 1997; Xu et al., 1997; for review, see Shalloway and Taylor, 1997) in which neither the SH2 domain, pTyr527, nor the Src SH3 domain is accessible for binding. The Tyr527→Phe mutant Src(Y527F) is a constitutively active kinase that induces focus formation and anchorage-independent growth in vitro and tumors in vivo. This mutant stably autophosphorylates at Tyr416, which is located within the Src catalytic domain (Cartwright et al., 1987; Kmiecik and Shalloway, 1987).
Tyr527 phosphorylation is reduced during activation of Src following thrombin stimulation of platelets (Clark and Brugge, 1993), attachment of fibroblasts to fibronectin-coated plates (Kaplan et al., 1995) and during mitosis (Bagrodia et al., 1991; Kaech et al., 1991). In principle, reduced phosphorylation could result from either decreased Tyr527-directed kinase activity or increased Tyr527directed phosphatase activity. At least in mitosis, it is most likely that activation results from increased phosphatase activity, since (i) mitosis-specific Cdc2-mediated phosphorylation of Src weakens the SH2–pTyr527 association yielding increased SH2 accessibility and increased availability of pTyr527 to phosphatases (Shenoy et al., 1989, 1992; Bagrodia et al., 1994; Stover et al., 1994), and (ii) vanadate, a tyrosine phosphatase inhibitor, blocks the mitotic activation of Src (Bagrodia et al., 1991). Since pTyr527 dephosphorylation requires Src membrane localization (Bagrodia et al., 1993), it seems likely that a membrane-localized protein tyrosine phosphatase (PTP) is involved.
A candidate for a physiological pTyr527 phosphatase is PTPα, an ∼130 kDa membrane-spanning PTP that is widely expressed but, like Src, is particularly abundant in the brain (Kaplan et al., 1990; Krueger et al., 1990). Although it contains a short extracellular domain and is often called a receptor PTP, no extracellular ligand is known (for reviews, see Neel and Tonks, 1997; Schaapveld et al., 1997). It contains two cytoplasmic catalytic domains, D1 and D2, of which only D1 is significantly active in vitro and in vivo (Wang and Pallen, 1991; den Hertog et al., 1993; Wu et al., 1997; Harder et al., 1998). Zheng et al. (1992) showed that overexpression of PTPα in rat embryo fibroblasts activates Src and causes neoplastic transformation, and den Hertog et al. (1993) showed that overexpression of PTPα in P19 embryonal carcinoma cells causes Tyr527 dephosphorylation, activation of Src and neuronal differentiation (similar to that observed following introduction of activated Src into P19 cells; Alema et al., 1985). Moreover, PTPα mRNA levels are increased during neuronal differentiation (den Hertog et al., 1993) at the same time that Src expression and kinase activity are increased (Sorge et al., 1984; Lynch et al., 1986; Boulter and Wagner, 1988; Bjelfman et al., 1990). PTPα also dephosphorylates Fyn (Bhandari et al., 1998), and Ponniah et al. (1999) and Su et al. (1999) recently showed that Src and Fyn have increased Tyr527 phosphorylation and decreased activity in knockout PTPα–/– mice. The PTPα–/– cells have a reduced rate of spreading on fibronectin substrates and deficiencies in integrin-mediated signaling responses (Su et al., 1999) that are similar to those observed in knockout mice lacking Src, Yes and Fyn (Klinghoffer et al., 1999). These results indicate that PTPα is an important Src activator in vivo.
PTPα can be phosphorylated by protein kinase C at Ser180 and 204 (Tracy et al., 1995), resulting in stimulation of its activity (den Hertog et al., 1995). Tyr789, a residue four amino acids away from the C-terminus, is also constitutively phosphorylated (den Hertog et al., 1994; Su et al., 1994). Its phosphorylation level is normally ∼20%, but this can be increased by transient overexpression of Src, suggesting that Src may phosphorylate Tyr789 in vivo (den Hertog, 1994). The sequence downstream from Tyr789 (…Y789ANF) fits the consensus binding site for the SH2 domain of the signal-transducing adapter protein Grb2 (Songyang et al., 1993), and wild-type (wt) PTPα but not the Tyr789→Phe mutant PTPα(Y789F) binds Grb2 in vitro and in vivo (den Hertog et al., 1994; Su et al., 1994). These findings suggest that Tyr789-phosphorylated PTPα might modulate Grb2-mediated signaling (den Hertog et al., 1994). However, Sos, the Ras exchange factor that is recruited to growth factor receptors by Grb2 (for review, see Pawson, 1995), is not detected in PTPα immunoprecipitates (den Hertog et al., 1994).
An alternative role for Tyr789 phosphorylation might be to regulate dephosphorylation of Src by PTPα. Evidence to date has suggested that this phosphorylation has either a negative or no effect on dephosphorylation: tyrosine phosphorylation of bacterially expressed PTPα has been reported to decrease PTPα activity (den Hertog et al., 1994) and a Tyr789→Phe PTPα mutant also induces neuronal differentiation of P19 cells (den Hertog and Hunter, 1996), which has been suggested to proceed via activation of Src (den Hertog et al., 1993). In contrast, here we show that Tyr789→Phe mutation abrogates the ability of PTPα to dephosphorylate and activate Src in vitro and in vivo, and to transform NIH 3T3 cells neoplastically. To explain this, we propose and test a model in which phosphorylated Tyr789 displaces pTyr527 from the Src SH2 domain in a manner that specifically enables PTPα to dephosphorylate pTyr527.
Results
Generation and characterization of PTPα inducible cell lines
Polyclonal antibodies were made in rabbits against recombinant glutathione _S_-transferase (GST)–PTPα(B20) fusion protein, which contains the PTPα cytoplasmic domain. These immunoprecipitated a single protein band of molecular weight ∼140 kDa from NIH 3T3 cells. Excess GST–PTPα(B20), but not GST, was able to inhibit band detection completely, thus verifying antibody specificity (data not shown).
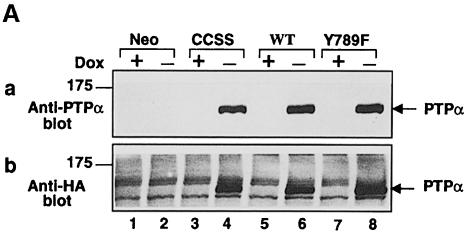
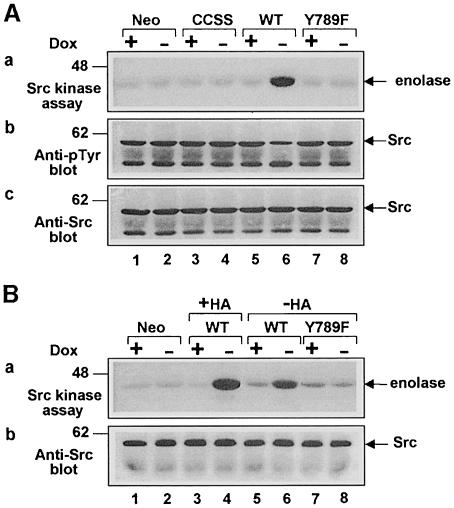
As described in Materials and methods, we created cell lines for _tet_-off inducible expression (Gossen and Bujard, 1992; Shockett et al., 1995) of hemagglutinin (HA) epitope-tagged human wt PTPα, the Tyr789→Phe mutant PTPα(Y789F), or the Cys 433→Ser/Cys 723→Ser double mutant PTPα(C433S/C723S), which is phosphatase defective because of the mutations in the catalytic domains (Streuli et al., 1990). Immunoblotting with anti-PTPα polyclonal antibody showed that PTPα levels increased to half-maximum by 6–8 h after removal of doxycycline (data not shown) and reached a maximum by 16–18 h (Figure 1A, panel a). The maximal expression levels were at least 15 times the endogenous level (Figure 1B, cf. odd-numbered lanes). Negligible protein was detected in the presence of doxycycline or in control cells that had been cotransfected with the empty expression vector (Figure 1A, panel a). Detection of the overexpressed proteins in the induced cell lines was blocked by competition with GST–PTPα(B20) fusion protein, and similar results were obtained using a second polyclonal antibody (data not shown). As expected, bands of the same molecular weight (∼140 kDa) were detected in the induced PTPα-expressor cells by the anti-HA epitope antibody (Figure 1A, panel b).
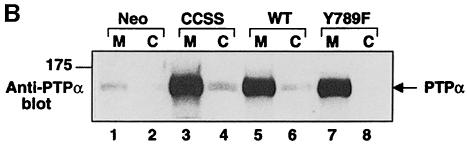
Fig. 1. PTPα inducible expression and subcellular localization. Cell line names are explained in Table I. (A) Lysates containing 20 μg of total cell protein were prepared from control cells (lanes 1 and 2) and from HA-tagged wt (lanes 5 and 6) and mutant (lanes 3–4 and 7–8) PTPα overexpressor cells that had been grown in the absence (induction of expression, even-numbered lanes) or presence (suppression of expression, odd-numbered lanes) of 5 ng/ml doxycycline (Dox). Lysates were immunoblotted with either anti-PTPα polyclonal (panel a) or anti-HA monoclonal (panel b) antibodies. (B) Membrane (M) and cytosolic (C) subcellular fractions from induced cells were purified, and aliquots containing 25 μg of total protein were immunoblotted with anti-PTPα polyclonal antibody 7–091. (C) As in (A), except that lysates were immunoblotted with anti-pTyr antibody 4G10. The positions of molecular weight markers (in kDa) are indicated.
Comparisons of anti-pTyr immunoblots of immunoprecipitated endogenous and overexpressed PTPα proteins indicated that the overexpressed proteins were only tyrosine phosphorylated at ∼40% the level of endogenous PTPα (data not shown). Since ∼20% of the endogenous PTPα in NIH 3T3 cells is tyrosine phosphorylated (den Hertog et al., 1994), we estimate that ∼8% of the overexpressed PTPα was phosphorylated at Tyr789.
Anti-pTyr immunoblots of unfractionated whole-cell lysates (Figure 1C) revealed a band at ∼140 kDa that was detectable in the induced wt PTPα and PTPα(C433S/C723S) lanes (4 and 6), but not in the control, uninduced or PTPα(Y789F) lanes. This is probably tyrosine-phosphorylated PTPα (cf. Figure 4, panel b, below). As expected, tyrosine phosphorylations of multiple proteins were increased in the induced wt PTPα overexpressor cells (Figure 1C, lane 6). This probably reflects increased tyrosine kinase activity of PTPα-activated Src family members.
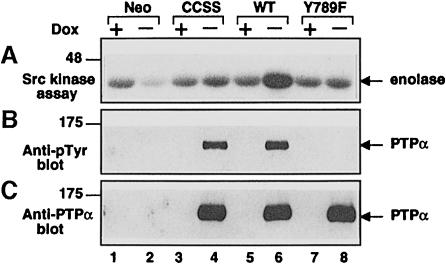
Fig. 4. In vitro dephosphorylation and activation of Src by PTPα. Eluates containing anti-HA immunopurified wt and mutant PTPα and equivalent volumes of mock-purified eluates were prepared from induced (–Dox) and uninduced (+Dox) PTPα overexpressor and control cells. (A) Five percent of each eluate was pre-incubated in dephosphorylation buffer with wt Src (bound to GammaBind Sepharose beads) that had been immunoprecipitated from Src overexpressor cells. The Src was then washed and subjected to in vitro kinase assay with [γ-32P]ATP and acid-denatured enolase as in Figure 3. (B) Twenty percent of each eluate was immunoblotted with anti-pTyr antibody. (C) Twenty percent of each eluate was immunoblotted with anti-PTPα antibody. The positions of molecular weight markers (in kDa) are indicated.
Membrane and cytosolic fractions prepared from induced wt and mutant PTPα overexpressor cells were immunoblotted (Figure 1B). In all cases, >80% of the PTPα proteins were in the membrane fraction, indicating that neither the mutations nor the high expression levels grossly affected subcellular localization.
Cell lines that inducibly expressed wt PTPα and PTPα(Y789F) without the HA tag were similarly constructed for use as controls in some experiments where the epitope tag was not required for technical reasons. However, unless otherwise noted, expressor cells for the epitope-tagged proteins were used.
Tyr789→Phe mutation blocks transformation by PTPα
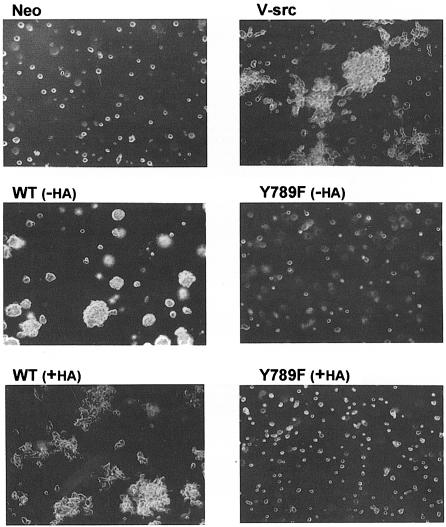
Induced control cells and HA-tagged and -untagged wt and mutant PTPα overexpressor cells were tested for their abilities to induce foci in monolayer culture when mixed with normal NIH 3T3 cells, or to form colonies when suspended in semi-solid media containing 0.3% soft agarose without doxycycline (Table I and Figure 2). As expected, after 14–16 days no foci or colonies were formed by either the control cells or cells that overexpressed phosphatase-defective PTPα(C433S/C723S), while both HA-tagged and -untagged wt PTPα overexpressor cells formed foci and colonies with moderate efficiency. These were similar to those formed by a v-_src_-transformed NIH 3T3 cell line. Notably, the Tyr789→Phe mutation completely abolished the ability of both tagged and untagged PTPα to form foci or induce growth in soft agarose.
Table I. Biological activities of wild-type and mutant PTPα.
| Protein expressed | HA tag | Expressiona plasmid | Cell lineb | Nicknamec | % colony formationd in soft agarose | % foci formede in cell mixing |
|---|---|---|---|---|---|---|
| – | pTet-SPLICE | NIH(pTet-SPLICE/COS)1 | Neo | ⩽0.2 | ⩽0.2 | |
| NIH(pTet-SPLICE/COS)2 | ⩽0.2 | ⩽0.2 | ||||
| Wild-type PTPα | + | pTPTPα | NIH(pTPTPα/COS)1 | wt | 12 ± 0.7 | 14 ± 1.7 |
| NIH(pTPTPα/COS)2 | 16 ± 0.3 | 17 ± 1.8 | ||||
| NIH(pTPTPα/COS)3 | 17 ± 0.4 | 20 ± 2.6 | ||||
| − | pNTPTPα | NIH(pNTPTPα/COS)1 | 26 ± 0.7 | nt | ||
| PTPα(C433S/C723S) | + | pTPTPα(CCSS) | NIH(pTPTPαCCSS/COS)1 | CCSS | ⩽0.2 | ⩽0.2 |
| NIH(pTPTPαCCSS/COS)2 | ⩽0.2 | ⩽0.2 | ||||
| PTPα(Y789F) | + | pTPTPα(Y789F) | NIH(pTPTPαY789F/COS)1 | Y789F | ⩽0.2 | ⩽0.2 |
| NIH(pTPTPαY789F/COS)2 | ⩽0.2 | ⩽0.2 | ||||
| NIH(pTPTPαY789F/COS)3 | ⩽0.2 | ⩽0.2 | ||||
| − | pNTPTPα(Y789F) | NIH(pNTPTPαY789F/COS)1 | ⩽0.2 | nt | ||
| NIH(pNTPTPαY789F/COS)2 | ⩽0.2 | nt | ||||
| v-Src | − | pMvsrc | NIH(pMvsrc/foc/ep)A1 | 39 ± 1.4 | 44 ± 1 |
Fig. 2. Colony formation in soft agarose by PTPα overexpressor cells. Control (Neo), HA-tagged (+HA) and -untagged (-HA) wt and Tyr789→Phe mutant PTPα-expressor cells, and a previously characterized v-_src_-transformed cell line (NIH[pMvsrc/foc/ep]A1; Kmiecik and Shalloway, 1997) were cultured in suspension in media containing 0.3% agarose and no doxycycline. Colonies were photographed after 14 days. Cells that overexpressed PTPα(CCSS) formed no colonies and looked identical to the Neo cells (not shown). Similar results were obtained in four independent experiments.
To check that this inability to transform cells did not simply reflect loss of protein overexpression during the culture assays, cells from the cell-mixing experiment were reselected using G418, and wt PTPα and PTPα(Y789F) levels in several resistant subclones were analyzed by immunoblotting. This showed that both the wt and Y789F proteins were overexpressed at levels similar to those shown in Figure 1 (data not shown). Thus, these data indicate that pTyr789 is required for significant PTPα-transforming ability.
Tyr789→Phe mutation blocks dephosphorylation and activation of Src by PTPα in vivo and in vitro
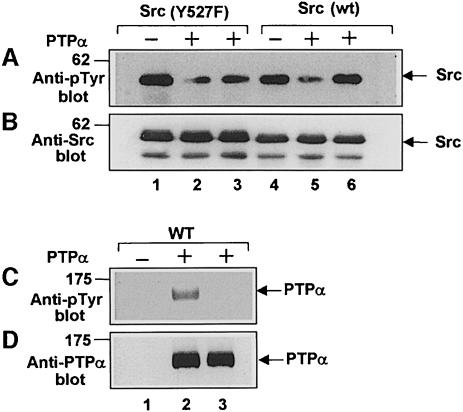
The simplest explanation for the requirement of pTyr789 for transformation is that PTPα(Y789F) fails to dephosphorylate Tyr527, and hence to activate Src. To test this, the specific kinase activity of endogenous Src immunoprecipitated from the uninduced and induced epitope-tagged PTPα overexpressor lines was measured (Figure 3). Induction had no effect on Src expression levels (Figure 3A, panel c). As expected, induction of wt PTPα overexpression caused an ∼30% decrease in the level of Src tyrosine phosphorylation in vivo (Figure 3A, panel b) and an ∼3-fold increase in Src specific kinase activity; the phosphatase-defective PTPα(C433S/723S) had no effect (Figure 3A, panel a). Importantly, PTPα(Y789F) also had no effect on either Src phosphotyrosine level or kinase activity (Figure 3, lanes 7 and 8), indicating that this mutant does not act on Src in vivo.
Fig. 3. Effect of PTPα overexpression on Src in vivo tyrosine phosphorylation and kinase activity. Cell line names are explained in Table I. (A) Src was immunoprecipitated from lysates containing 1 mg of total cell protein prepared from control cells (lanes 1 and 2), wt PTPα (lanes 5 and 6) and mutant PTPα (lanes 3–4 and 7–8) expressor cells that had been grown in the presence (odd-numbered lanes) or absence (even-numbered lanes) of doxycycline (Dox). Each immunoprecipitate was divided into three fractions (containing 10, 45 and 45%, respectively, of the total), which were either: (a) subjected to kinase assay in buffer containing [γ-32P]ATP and acid-denatured enolase followed by 9% SDS–PAGE and autoradiography of the reaction products; (b) immunoblotted using anti-pTyr mAb 4G10; or (c) immunoblotted using anti-Src mAb 327. The positions of molecular weight markers (in kDa) are indicated. (B) As in (A) except that Src was immunoprecipitated from lysates from cells overexpressing untagged (–HA) wt PTPα and PTPα(Y789F) (lanes 5–8). Epitope-tagged (+HA) wt PTPα (lanes 3 and 4) was included as a positive control.
To exclude the possibility that the effect of the Tyr789 →Phe mutation was influenced by the presence of an HA tag, similar experiments were conducted using cells that overexpressed untagged wt PTPα and PTPα(Y789F) (Figure 3B). The results were equivalent to those observed with the HA-tagged variants: overexpression of wt PTPα, but not of PTPα(Y789F), increased Src kinase activity. [The somewhat lower level of Src activation by untagged wt PTPα relative to HA-tagged PTPα (cf. Figure 3B, panel a, lanes 4 and 6) reflects the 2.5-fold lower PTPα overexpression levels in the untagged cell line (6× endogenous) relative to the level in the tagged cell line. The expression level in the untagged PTPα(Y789F) line was 6.5× endogenous.]
To see whether these differences reflected changes in the direct action of PTPα on Src, HA-tagged wt and mutant PTPα were immunopurified with anti-HA monoclonal antibody (mAb) from induced overexpressor cells and were pre-incubated in dephosphorylation buffer with Src immunoprecipitated from Src overexpressor cells. Anti-pTyr immunoblots (Figure 4B) showed that, as expected, immunopurified PTPα and PTPα(C433S/C723S) were tyrosine phosphorylated, while PTPα(Y789F) was not. [Although PTPα can autodephosphorylate (den Hertog et al., 1994), time-course studies showed that there was no noticeable tyrosine dephosphorylation of wt PTPα during the 30 min incubation period (data not shown).] The immunoprecipitated Src was then assayed for kinase activity (Figure 4A). Pre-incubation with wt PTPα increased Src kinase activity ∼3-fold, while pre-incubation with equal amounts (Figure 4C) of PTPα(C433S/C723S) or PTPα(Y789F) had no significant effect (Figure 4A). Correspondingly, decreased Src tyrosine phosphorylation was observed only after pre-incubation with wt PTPα (data not shown, but see Figure 3). Eluates from induced control cells (Figure 4, lane 2) and from uninduced cells (odd-numbered lanes) contained no anti-HA-precipitable PTPα (Figure 4C) and caused no increase in Src activity. These data indicate that the dephosphorylation-mediated activation of Src by PTPα is greatly enhanced by Tyr789 phosphorylation.
Regulation of substrate specificity by Tyr789 phosphorylation
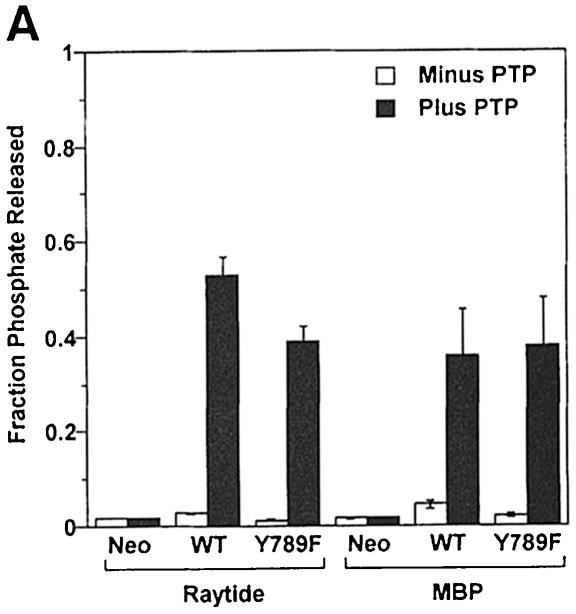
The previous results indicated that pTyr789 plays an essential role in the dephosphorylation and activation of Src by PTPα, both in vitro and in vivo. Because Tyr789 is located far from the active D1 catalytic domain, its mutation would not be expected to alter intrinsic catalytic activity, but it could play an important role in substrate specificity. To test this, wt and mutant PTPα were immunoprecipitated from uninduced and induced overexpressor cells with anti-HA mAb and tested for in vitro phosphatase activity using either the synthetic peptide Raytide or myelin basic protein (MBP) as substrates. [These two non-specific substrates have been used previously to measure the activity of a variety of phosphatases (Krueger et al., 1990).] Wild-type PTPα had a slightly higher dephosphorylation rate than PTPα(Y789F) on Raytide; rates were almost identical on MBP (Figure 5A). The immunoprecipitates from the control and uninduced cells had essentially no phosphatase activity, indicating that the activity being measured was indeed from the over– expressed PTPα. We conclude that the Tyr789→Phe mutation does not affect the intrinsic phosphatase activity of PTPα or its ability to dephosphorylate non-specific substrates.
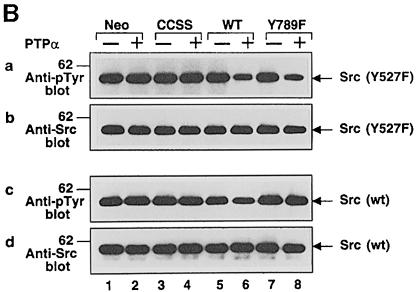
Fig. 5. In vitro dephosphorylation of non-specific and specific substrates by wt and mutant PTPα. (A) Raytide peptide and MBP were tyrosine phosphorylated in vitro using v-Src and [γ-32P]ATP. The radioactive substrates (Raytide: 3.5 × 105 c.p.m./lane, 2 μg/lane; MBP: 1.5 × 105 c.p.m./lane, 6 μg/lane) were then incubated in phosphatase buffer with anti-HA immunoprecipitates from lysates (containing 500 μg of total cell protein) prepared from uninduced and induced control and PTPα-expressor cells for 30 min (i.e. within the linear reaction/time regime) at 30°C. Reactions were terminated and the amount of [32P]phosphate released was measured by scintillation counting. Results are averages of two independent experiments; ranges are indicated. (B) Src was immunoprecipitated from either Src(Y527F) (panels a and b) or wt Src (panels c and d) overexpressor cells (see Table I for full names) and subjected to in vitro dephosphorylation with wt or mutant PTPα purified from induced overexpressor cells (lanes 4, 6 and 8). Mock-purified eluates from non-overexpressor cells (lanes 1 and 2) and uninduced PTPα expressor cells (lanes 3, 5 and 7) were used as controls. Reactions were terminated and divided into two equal aliquots, which were immunoblotted for phosphotyrosine content using anti-pTyr mAb 4G10 (panels a and c) or for Src levels using anti-Src mAb 327 (panels b and d). The positions of molecular weight markers (in kDa) are indicated.
We then incubated immunopurified wt and mutant PTPα in phosphatase buffer with immunoprecipitated wt Src, which is tyrosine phosphorylated almost exclusively at Tyr527, and with mutant Src(Y527F), which as a result of its Tyr527→Phe mutation is tyrosine phosphorylated almost exclusively at the autophosphorylation site Tyr416 (Kmiecik and Shalloway, 1987). Wild-type PTPα was able to dephosphorylate both pTyr527 (∼35% dephosphorylation) and pTyr416 (∼64%) (Figure 5B, panels a and c, lane 6). In contrast, PTPα(Y789F) was unable to dephosphorylate Tyr527 (Figure 5B, panel c, lane 8), but was able to dephosphorylate Tyr416 (∼65%) (panel a, lane 8) to the same extent as wt PTPα. The difference is remarkable since both these tyrosines are potential physiological PTPα substrates. We conclude that pTyr789 participates selectively in dephosphorylation of pTyr527 in Src.
Specific effect of dephosphorylation of PTPα on its ability to dephosphorylate pTyr527
The interpretation of the experiments described above rests on the assumption that the Tyr789→Phe mutation is equivalent to tyrosine dephosphorylation of PTPα. To verify this directly, we compared the dephosphorylating activities of tyrosine-phosphorylated and -dephosphorylated wt PTPα. As mentioned above, PTPα did not dephosphorylate itself in vitro under our experimental conditions. Instead, we used a modification of a method previously used to dephosphorylate Src (Bagrodia et al., 1993; Taylor et al., 1995): wt PTPα overexpressor cells were incubated in phosphate-buffered saline (PBS) containing 5 mM EDTA to leach out Mg2+ and other divalent cations that are required for endogenous kinase activity, so that the (divalent cation-independent) endogenous phosphatases could dephosphorylate the cellular proteins. PTPα was immunopurified with anti-HA antibody from EDTA-treated and -untreated cells and used to dephosphorylate Src(Y527F) and wt Src as in the previous experiment.
The EDTA treatment (Figure 6, lanes 3 and 6) removed all detectable phosphotyrosine in PTPα (Figure 6C, cf. lanes 2 and 3) without changing the amount of PTPα in the overexpressor cells (Figure 6D, cf. lanes 2 and 3). In agreement with the results above, we found that tyrosine-phosphorylated PTPα dephosphorylated pTyr527 (in wt Src) and pTyr416 [in Src(Y527F)] to roughly the same extent, and that prior dephosphorylation abrogated PTPα's ability to dephosphorylate pTyr527 while having little or no effect on its ability to dephosphorylate pTyr416 (Figure 6A, lanes 2, 3 and 5, 6).
Fig. 6. Effect of dephosphorylation of PTPα on its dephosphorylating activity. (A and B) Src was immunoprecipitated from either Src(Y527F) (lanes 1–3) or wt Src overexpressor cells (lanes 4–6) and incubated with (anti-HA) immunopurified wt PTPα (lanes 2 and 5), dephosphorylated immunopurified wt PTPα (lanes 3 and 6) or mock-purified eluates from uninduced PTPα overexpressor cells (lanes 1 and 4) as in Figure 5. Reactions were terminated and divided into two equal aliquots, which were immunoblotted with either anti-pTyr mAb 4G10 (A) or anti-Src mAb 327 (B). (The band in lane 2 was unevenly blotted because of a bubble.) (C and D) Aliquots of the purified (or mock-purified) PTPα used in the Src dephosphorylation reactions were immunoblotted with either anti-pTyr mAb 4G10 (C) or with anti-PTPα 7-091 polyclonal antibody (D). Lanes 1–3 in these panels correspond to lanes 1–3 and lanes 4–6 in the panels above. The positions of molecular weight markers (in kDa) are indicated.
A pTyr displacement model
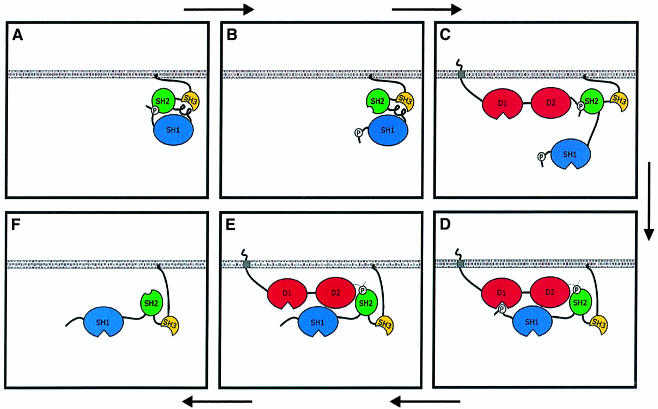
One mechanism by which pTyr789 could modulate substrate specificity is shown schematically in Figure 7. In this model, pTyr789 binding to the Src SH2 domain serves two important functions: (i) pTyr527 is prevented from binding the SH2 domain, thereby rendering it accessible to the D1 catalytic domain; and (ii) the relatively tight association of pTyr789 and the SH2 domain provides extended time for the D1 catalytic domain to bind and dephosphorylate pTyr527. Beyond explaining the results described above, this model gives rise to additional predictions that were tested in experiments described below: (i) wt PTPα will bind Src to a much greater extent than PTPα(Y789F). (ii) This binding will be stabilized once pTyr527 has been dephosphorylated and can not compete with pTyr789 for binding to the SH2 domain (i.e. as in Figure 7E); thus, the phosphatase-inactivating Cys433→Ser/Cys723→Ser double mutation will also reduce PTPα–Src binding. (iii) PTPα–Src binding will be inhibited by addition of free SH2 domain that binds pTyr789. (iv) Dephosphorylation and activation of Src will be inhibited by free SH2 domain that binds pTyr789.
Fig. 7. Phosphotyrosine displacement model for PTPα substrate specificity. Src pTyr527 is usually bound to the Src SH2 domain (A), but this binding is transiently dissociated by thermal fluctuations (B), providing an opportunity for PTPα pTyr789 binding instead (C). The transient binding provides an extended time period for rearrangement to a head-to-tail conformation (D) that will facilitate Tyr527 dephosphorylation by the D1 catalytic domain. Because there is no further competition from pTyr527, the pTyr789–SH2 binding will persist for a longer time, enhancing the association between PTPα and Src (E), prior to dissociation (F).
Association of wt and mutant PTPα and Src in vivo
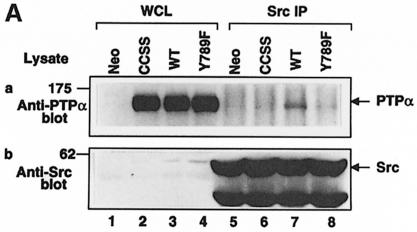
Anti-Src polyclonal antibody immunoprecipitates from induced wt and mutant PTPα overexpressor and control cells were immunoblotted with anti-PTPα and anti-Src antibodies (Figure 8A). Although the wt and mutant PTPα proteins were expressed at similar levels (Figure 8A, lanes 2–4), much more wt PTPα than PTPα(C433S/C723S) was coprecipitated with Src (cf. lanes 6 and 7). Consistent with the proposed model, the Tyr789→Phe mutation also significantly reduced PTPα–Src association. (The residual PTPα in the immunoprecipitates is probably endogenous, which contains disproportionately more phosphorylated Tyr789, as discussed above.) This confirmed the first two predictions of the model.
Fig. 8. GST–Src SH2 fusion protein inhibits the in vivo association of Src with PTPα. (A) Endogenous Src was immunoprecipitated from lysates (1.5 mg of protein) prepared from the indicated induced HA-tagged PTPα expressor cell lines using anti-Src polyclonal antibody. Equal aliquots of each immunoprecipitate were immunoblotted with either anti-PTPα antibody 7-091 (panel a) or anti-Src mAb 327 (panel b). Immunoblots of whole-cell lysates prior to immunoprecipitation (30 μg of total cell protein) are shown in lanes 1–4. (B) In vivo association of wt PTPα with Src was assayed as described above except that endogenous Src was immunoprecipitated with anti-Src mAb 2-17, and the washed immunoprecipitates were either incubated alone (lane 2) or in the presence of the indicated amounts of purified GST (lanes 3 and 4) or GST–Src SH2 (lanes 5 and 6) fusion proteins for an additional 30 min at 4°C and rewashed prior to immunoblotting. All lanes are with lysates from wt PTPα overexpressor cells. Immunoblots of whole-cell lysate prior to immunoprecipitation (30 μg of protein) are shown in lane 1.
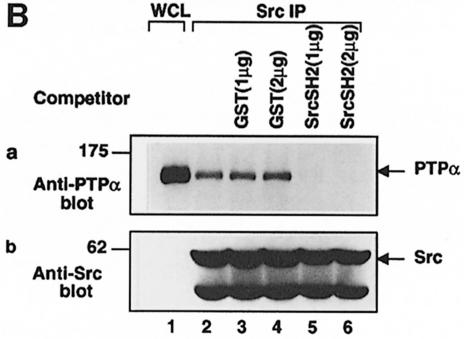
To test the hypothesis that the PTPα–Src association results from pTyr789–Src SH2 binding, we checked the prediction that the association can be competed by free Src SH2 domain (Figure 8B); as in Figure 8A, Src was immunoprecipitated with anti-Src mAb 2-17 from wt PTPα overexpressor cells, but here the immobilized complexes were incubated with or without purified GST or GST–Src SH2 fusion proteins before immunoblotting. As shown in Figure 8B, the addition of 1 μg of GST–Src SH2 (panel a, lane 5) completely abolished the association of Src and PTPα. No inhibition was observed with 2 μg of GST alone (Figure 8B, panel a, lane 4).
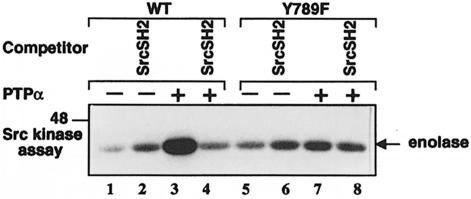
Inhibition of PTPα-mediated Src activation by competing Src SH2 domain
To test the final prediction, the in vitro Src activation assay shown in Figure 4 was repeated, except that competing GST–Src SH2 fusion protein was added to the (anti-HA) immunopurified wt PTPα and PTPα(Y789F) prior to incubation with Src immunoprecipitates from Src overexpressor cells (Figure 9). No effect was observed following pre-incubation with GST alone (data not shown). Src kinase activity was slightly increased by the presence of GST–Src SH2 in the absence of PTPα (Figure 9, cf. lanes 1 and 2, and 5 and 6). This probably reflects destabilization of the compact, negatively regulated form of Src by GST–Src SH2 competition with the intramolecular pTyr527–SH2 interaction; a similar effect has been observed using antibody that binds to the Src C-terminus (Cooper and King, 1986). Consistent with the results of Figure 4, an ∼5-fold increase in Src kinase activity was induced by pre-incubation with wt PTPα in the absence of competitor (Figure 9, cf. lanes 1 and 3), while only an ∼1.5-fold activation was induced by PTPα(Y789F) (cf. lanes 5 and 7). As predicted by the model, the wt PTPα-induced increase in Src activity was completely blocked by the presence of GST–Src SH2 fusion protein (Figure 9, cf. lanes 3 and 4), while competing fusion protein had little or no effect on the small increase in kinase activity induced by PTPα(Y789F) (cf. lanes 7 and 8).
Fig. 9. GST–Src SH2 fusion protein inhibits the activation of Src by wt PTPα in vitro. Eluates containing (anti-HA) immunopurified wt and mutant PTPα (or equivalent volumes of mock-purified eluates) were prepared from induced (+PTPα; lanes 3, 4, 7 and 8) and uninduced (–PTPα; lanes 1, 2, 5 and 6) wt PTPα (lanes 1–4) or PTPα(Y789F) (lanes 5–8) overexpressor cells, and were pre-incubated without (odd-numbered lanes) or with (even-numbered lanes) 5 μg of GST–Src SH2 fusion protein for 30 min at 4°C. The ability of 10% of each mixture to stimulate Src kinase activity was then tested as described in Figure 4A. The position of a molecular weight marker (in kDa) is indicated.
Differential inhibition of PTPα-mediated Src activation by Src and Grb2 SH2 domains
Inhibition of pTyr527 dephosphorylation by GST–Src SH2 is consistent with the displacement model, but does not exclude the possibility that inhibition was due to binding and protection of pTyr527 by the exogenous SH2 domain, rather than by blocking the pTyr789–SH2 association. The exclusion of this possibility required the use of two separate blocking reagents having different specificities for binding pTyr789 and pTyr527.
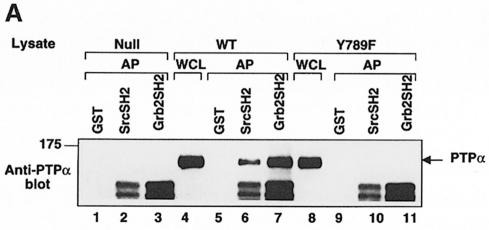
First, we explored the possibility that the Src and Grb2 SH2 domains might have appropriate specificity differences by comparing the abilities of GST fusion proteins containing either of these SH2 domains to affinity-precipitate immunopurified wt PTPα, PTPα(Y789F) and wt Src. For a more sensitive test of pTyr527 binding, we also assayed the affinity-precipitation of Src(W148R/Y416F): this has a mutation within the SH2 domain that weakens the competing intramolecular Src SH2–pTyr527 association, and a Tyr416→Phe mutation that ensures that pTyr527 is the only tyrosine that is significantly phosphorylated (Taylor et al., 1995). As could be predicted from the Grb2 SH2 binding consensus sequence (Songyang et al., 1993), we found that the Grb2 SH2 domain affinity-precipitated ∼3.3 times more wt PTPα than the Src SH2 domain (Figure 10A, lanes 5–7); consistent with the model of Figure 7, however, the Src SH2 domain was able to affinity-precipitate wt PTPα.
Fig. 10. Differential inhibition of PTPα-mediated Src activation by Src and Grb2 SH2 domains. (A) Affinity-precipitation of wt PTPα and PTPα(Y789F) by the Src and Grb2 SH2 domains. Fifteen micrograms of either GST (lanes 1, 5 and 9), GST–Src SH2 (lanes 2, 6 and 10) or GST–Grb2 SH2 (lanes 3, 7 and 11) purified fusion protein were incubated without (lanes 1–3) or with (lanes 4–11) lysates (containing 1.5 mg of total cell protein) prepared from wt PTPα (lanes 4–7) or PTPα(Y789F) (lanes 8–11) induced overexpressor cells. Complexes were adsorbed on glutathione beads, and the proteins eluted from the washed beads were analyzed by immunoblotting for PTPα. Lanes 4 and 8 are anti-PTPα immunoblots of whole-cell lysates (30 μg of protein). The position of a molecular weight marker (in kDa) is indicated. (B) Affinity precipitation of wt Src and Src(W148R/Y416F) by the Src and Grb2 SH2 domains. As in (A) except that the GST fusion proteins were incubated with lysates (300 μg of protein) from wt Src overexpressor cells (lanes 2–4) or Src(W148R/Y416F) overexpressor cells (lanes 6–8), and the proteins eluted from the washed glutathione beads were immunoblotted with anti-Src antibody. Lanes 1 and 5 are anti-Src immunoblots of whole-cell lysates (5 μg of protein). (C) Anti-HA immunopurified wt PTPα from induced (+PTPα; lanes 2–10) PTPα overexpressor cells or an equivalent volume of mock-purified eluate from uninduced PTPα overexpressor cells (–PTPα; lane 1) was pre-incubated without (lanes 1 and 2) or with the indicated concentrations of either GST–Grb2 SH2 (lanes 3–7) or GST–Src SH2 (lanes 8–12) fusion proteins for 30 min at 4°C. (Note that the GST–Src SH2 concentrations are 10 times the corresponding GST–Grb2 SH2 concentrations.) Ten percent of each mixture was used with immunoprecipitated wt Src for in vitro dephosphorylation and Src kinase assays as in Figure 9.
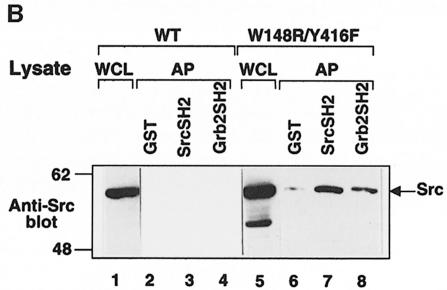
Neither SH2 domain affinity-precipitated PTPα(Y789F) (Figure 10A, lanes 10 and 11). Interestingly (and usefully), the Src SH2 domain affinity-precipitated pTyr527 more effectively than the Grb2 SH2 domain (Figure 10B); presumably because of competition from intramolecular pTyr527–Src SH2 binding, we were unable to detect affinity-precipitation of wt Src by either GST–SH2 fusion protein under the experimental conditions used (lanes 2–4), but GST–Src SH2 affinity-precipitated ∼2.2 times more Src(W148R/Y416F) than GST–Grb2 SH2 (lanes 6–8).
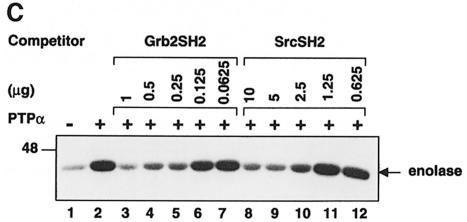
The relative abilities of the two GST–SH2 fusion proteins to inhibit the PTPα-induced activation of Src were measured in experiments similar to that of Figure 9. Titration showed that the Grb2 SH2 domain was >10 times more effective than the Src SH2 domain at blocking activation (Figure 10C). Since this matches the differential affinities for pTyr789 (and not for pTyr527), it implies that the SH2-mediated inhibition of PTPα-induced activation of Src primarily involves binding of pTyr789 rather than protection of pTyr527. Thus, the fourth prediction of the model was verified.
Discussion
We have shown that Tyr789→Phe mutation or dephos– phorylation of wt PTPα specifically blocks its ability to dephosphorylate Tyr527 in Src both in vitro and in vivo. As would then be predicted, the Tyr789→Phe mutation was found to abrogate PTPα's ability to transform NIH 3T3 cells. Overexpressed wt PTPα was tyrosine phosphorylated at only ∼40% of the level of endogenous PTPα, so that the net overexpression of tyrosine-phosphorylated wt PTPα was ∼7-fold. This is consistent (given the complexity within the cell) with the observed 3-fold increase in Src kinase activity. The lower level of tyrosine phosphorylation of PTPα may reflect titration of the endogenous Grb2, which is believed to protect pTyr789 from phosphatases (den Hertog et al., 1994; Su et al., 1996). Alternatively, it is possible that Src and other kinases that phosphorylate Tyr789 are limiting.
Mutation or dephosphorylation did not block PTPα's ability to dephosphorylate more accessible targets such as phosphotyrosines in Raytide peptide or MBP. More importantly, it also did not affect dephosphorylation of pTyr416 in Src, a physiological yet accessible target. This indicates that pTyr789 governs substrate specificity, not overall catalytic activity. We hypothesize that pTyr789 is needed to displace Tyr527 from the Src SH2 domain as outlined by the model in Figure 7. In this model, pTyr789, which is four amino acids from the PTPα C-terminus (Krueger et al., 1990; Matthews et al., 1990), displaces pTyr527, which is six amino acids from the Src C-terminus (Takeya and Hanafusa, 1983), in a required intermediate step in the dephosphorylation process.
As with the sequence downstream from pTyr527, the sequence downstream from pTyr789 (Y789ANF) is not a consensus sequence for high-affinity binding to the Src SH2 domain (Songyang and Cantley, 1995). Thus, we expect the binding of both of these phosphotyrosines to the Src SH2 domain to be relatively weak. Indeed, isothermal calorimetry measurements show that the phosphorylated pTyr789 peptide binds GST–Src SH2 with _K_d ≈ 6 μM, while the phosphorylated pTyr527 peptide binds with _K_d ≈ 14 μM (S.Showalter and L.Nicholson, personal communication). In comparison, a peptide corresponding to the high-affinity polyoma middle T pTyr324 binds with _K_d ≈ 0.2 μM (Bradshaw et al., 1998). Nonetheless, the strength of the pTyr789–Src SH2 binding is adequate to enable the Src SH2 domain to affinity-precipitate wt PTPα from cell lysates (Figure 10A). Consistent with consensus binding data (Songyang and Cantley, 1995), this affinity is ∼3.3-fold weaker than the pTyr789–Grb2 SH2 binding, which probably explains why it was not detected in prior, less sensitive, binding experiments (e.g. den Hertog et al., 1994). Predictions of the model, including pTyr789- and catalytic activity-dependent in vivo association of PTPα and Src as well as the ability of GST–SH2 fusion proteins to inhibit PTPα–Src association and Src activation, were verified. The fact that the affinities of pTyr527 and pTyr789 for the Src and Grb2 SH2 domains were complementary—pTyr527 bound more tightly to the Src SH2 domain than to the Grb2 SH2 domain, while pTyr789 bound more tightly to the Grb2 SH2 domain—made possible a differential-inhibition assay; this showed that free SH2 domains inhibited PTPα dephosphorylation of Src by binding to pTyr789, not to pTyr527.
Also in agreement with the displacement model, we found that wt PTPα was associated with Src, and that the phosphatase-defective PTPα(C433S/C723S) mutant bound significantly reduced amounts of Src. According to the model, PTPα will bind mostly to dephosphorylated Src in the intermediate state shown in Figure 7E, because binding to intermediates involving tyrosine-phosphorylated Src (i.e. as in Figure 7C and D) will be significantly reduced by competition from pTyr527. Since most Src in NIH 3T3 cells is phosphorylated at Tyr527 (Shenoy et al., 1992), there is little of the dephosphorylated form available for binding to phosphatase-defective PTPα. Since we predict that binding of dephosphorylated Src to wt PTPα will be transient, we expect only a relatively small fraction of Src and PTPα to be associated at any time, as observed.
Our results agree with Harder et al. (1998), who have previously shown that wt PTPα and Src can be co-immunoprecipitated from A431 carcinoma cells transfected to overexpress PTPα. Some of their data (e.g. Figure 10, Harder et al., 1998) also suggest reduced association of a catalytic-defective PTPα mutant with Src, but this was not consistently observed in their experiments. [Src has significantly elevated activity in A431 cells (Osherov and Levitski, 1994). This presumably results from reduced Tyr527 phosphorylation levels, which would result in a decrease in the difference between the binding of wt and catalytic-defective PTPα to Src in this cell type.]
The difference between the ability of wt PTPα and PTPα(Y789F) to transform NIH 3T3 cells and to activate Src in vivo was demonstrated using both untagged and HA-tagged proteins. No effect of the epitope tag was detected, and we think it is unlikely that it affected other biochemical experiments that, for technical reasons, were performed using only HA-tagged proteins. In any case, the presence of the tag would, if anything, be expected to weaken rather than strengthen pTyr789–Src SH2 interactions.
The PTPα tested in these experiments was immunoprecipitated from NIH 3T3 cells and thus was tyrosine phosphorylated. However, in apparent contradiction with the displacement model, it has previously been reported that PTPα expressed in bacteria, which would not contain phosphotyrosine, can dephosphorylate pTyr527 in intact Src (Zheng et al., 1992; den Hertog et al., 1993). Notably, these dephosphorylation reactions occurred at pH 6.0 with no salt, and it is possible that these artificial conditions may destabilize the pTyr527–SH2 interaction. Indeed, Fang et al. (1994b) found that bacterially expressed PTPα could not effectively dephosphorylate pTyr527 when tested at pH 7.4 and 150 mM NaCl. Their experiment, conducted under more physiological conditions, probably better reflects the biologically relevant situation.
den Hertog et al. (1994) report that Src-catalyzed tyrosine phosphorylation of bacterially expressed wt PTPα, but not of PTPα(Y789F), reduced the activity of wt PTPα by ∼50%, and therefore suggested that phosphorylation of Tyr789 reduces PTPα activity. This contrasts with our finding that PTPα(Y789F) has the same in vitro activity as wt PTPα on unprotected substrates. Since the relevant experimental conditions are not described in den Hertog et al. (1994), we can not resolve this discrepancy. One possibility is that the pTyr789-mediated binding of wt PTPα and Src promotes Src-catalyzed phosphorylation of additional sites that suppress PTPα activity. [Indeed, Src can phosphorylate multiple sites in PTPα (den Hertog et al., 1994).]
den Hertog et al. (1993, 1996) have also reported that overexpressed PTPα(Y789F), like wt PTPα, is able to induce neuronal differentiation of P19 embryonal carcinoma cells. The contrast between this observation and the inability of PTPα(Y789F) to transform NIH 3T3 fibroblasts suggests that PTPα may act on a substrate other than Src in P19 cells, and that this action may be sufficient to induce differentiation. [The ability of PTPα(Y789F) to activate Src in P19 cells was not reported.] In contrast with the results in P19 cells, Su et al. (1996) found that overexpression of wt PTPα, but not catalytically defective PTPα, inhibited the induction of neurite outgrowth from PC12 cells by acidic fibroblast growth factor, and that overexpression of PTPα(Y789F) enhanced outgrowth. (In this case as well, the effects on Src activity were not reported.) Clearly, the role of PTPα in neuronal differentiation is complex and more investigation is needed.
We expect that the pTyr789–SH2 binding will act as a tether that will increase the local effective concentra– tion of the D1 catalytic domain in the vicinity of pTyr527, [D1]eff, to ∼1 mM. Theoretical kinetic analysis (unpublished results) suggests that this entropic reduction will decrease the _K_m by the factor _K_B/[D1]eff, where _K_B is the pTyr789–Src SH2 binding constant (∼6 μM; S.Showalter and L.Nicholson, personal communication). Thus, we expect approximately a two orders of magnitude reduction in _K_m, which is consistent with the experimental observations.
Although the three-dimensional structure of complete PTPα is not known, the structure of the D1 catalytic domain has been determined (Bilwes et al., 1996). Its size, and those of the other PTPα regions, is represented to approximate scale in Figure 7. The fact that the N-termini of both Src and PTPα are localized to the plasma membrane, in combination with the locations and sizes of the interacting pTyr789–SH2 and pTyr527–D1 partners, implies that PTPα and Src must interact in a head-to-tail configuration in which at least one of the molecules is adjacent to the plasma membrane (although this need not be PTPα, as indicated in Figure 7). Bilwes et al. (1996) have found that the active site of D1 is blocked by dimerization in crystals and have suggested that D1 dimerization between complete PTPα proteins may be a physiologically important mechanism for constraining their activity. In support of this hypothesis, Jiang et al. (1999) have shown that a PTPα mutant that dimerizes because of the introduction of a cysteine in its extracellular domain is not effective at dephosphorylating Src. If this model is correct, the PTPα monomer shown in Figure 7C may in some cases be a PTPα dimer. In this case, the initial interaction of one pTyr789 with an Src SH2 and the subsequent interactions between the bodies of PTPα and Src and/or the plasma membrane may require and/or facilitate disruption of the D1 dimer. The need for facilitation could help restrict PTPα activity to appropriate targets. Attachment of PTPα to the plasma membrane (which confines the entropic degrees of freedom to a plane) may be needed to stabilize the D1–D1 dimerization. If so, this specificity mechanism would not be observed in reactions in vitro.
The head-to-tail arrangement shown in Figure 7 may also sterically prevent PTPα from dephosphorylating pTyr416 at the same time that pTyr527 dephosphorylation is being promoted. Since Tyr416 phosphorylation increases Src activity (Kmiecik and Shalloway, 1987; Piwnica-Worms et al., 1987; Kmiecik et al., 1988), this would enable PTPα to accomplish the difficult task of dephosphorylating the negative regulatory site in Src while not dephosphorylating the positive regulatory site.
The proposed model is similar to models that have been proposed for the association of SHP-1 with Src (Falet et al., 1996) and for the association of CD45 with Lck (Autero et al., 1994). SHP-1, a cytosolic PTP containing two SH2 domains that is expressed at highest levels in hematopoietic cells (for review, see Neel and Tonks, 1997), can dephosphorylate Tyr527 in vitro (Somani et al., 1997). Multiple experiments indicate that it may physiologically activate Src in some cell types (Li et al., 1994, 1995; Somani et al., 1997). SHP-1 co-immunoprecipitates with Src in a phosphotyrosine-dependent manner (Falet et al., 1996; Somani et al., 1997), and it has been suggested that their binding is mediated by the N-proximal SH2 domain of SHP-1, since it can bind tyrosine-phosphorylated c-Src in vitro (Somani et al., 1997). However, like PTPα, SHP-1 is tyrosine phosphorylated in vivo at a Grb2 consensus binding site (Yeung et al., 1992) and can bind Grb2 in vivo (Lorenz et al., 1994) and the Src SH2 domain in vitro. It has therefore also been suggested that the SHP-1 binding may involve displacement of pTyr527 from the Src SH2 domain (Falet et al., 1996). CD45 dephosphorylates the Src family member Lck (Ostergaard et al., 1989; Hurley et al., 1993), but not Fyn (Bhandari et al., 1998; although, see Mustelin et al., 1992 for contrasting data) or Src (Hurley et al., 1993). It can be tyrosine phosphorylated in vivo (Stover et al., 1991), and tyrosine phosphorylation within the second catalytic domain (similar to D2 in Figure 7) stimulates CD45 catalytic activity and its ability to bind Lck in vitro and in vivo (Autero et al., 1994). Based on this, Autero et al. (1994) have proposed a displacement model similar to that of Figure 7. The model differs in detail, in that an internal phosphorylation site, head-to-head (rather than head-to-tail) binding and the D2 (rather than the D1) domain are proposed to be involved. Other PTPs may act by similar displacement mechanisms: for example, den Hertog et al. (1993) have pointed out that, similar to PTPα, PTPɛ and PTPμ each contain a potential tyrosine phosphorylation site five residues from their C-termini and that PTPβ contains one site 17 residues from its C-terminus. PTPα also contains a number of tyrosines near its C-terminus (Krueger et al., 1990). It seems quite likely that PTPα dephosphorylates Fyn (Bhandari et al., 1998), and possibly other Src family members, using the same mechanism. In agreement with this, we have found that both Tyr789→Phe and phosphatase-inactivating mutations greatly reduce PTPα binding to Fyn (data not shown), just as they do with Src. This differs from Bhandari et al. (1998) who found that phosphatase-defective PTPα bound Fyn to the same extent as wt PTPα when both Fyn and PTPα were transiently overexpressed in COS cells. One possible explanation for the discrepancy is that the transiently overexpressed wt PTPα was not significantly tyrosine phosphorylated in COS cells (the extent of tyrosine phosphorylation was not reported), so that only pTyr789-independent residual binding was measured. Alternatively, the transiently overexpressed Fyn may not have been tyrosine phosphorylated (e.g. by the endogenous level of Csk) at its Tyr527 homolog; in this case the model of Figure 7 would predict no difference between binding of Fyn to wt and catalytically defective PTPα. Further studies to compare the levels of tyrosine phosphorylation in the associated and unassociated proteins are needed.
The displacement mechanism may not be required in all cases for pTyr527 dephosphorylation. Fang et al. (1994b) found that a chimeric, myristylation-peptide/ChPTPα protein (which contained most of the ChPTPλ cytoplasmic region) was much better at dephosphorylating pTyr527 and activating Src than an analogous PTPα chimera, and thus suggested that ChPTPλ may physiologically activate Src family members. However, the ChPTPλ cell-type-specific expression profile [low in brain and chicken embryo fibroblasts, abundant in spleen, intestine and pre-B cells (Fang et al., 1994b)] does not match that of Src. Furthermore, the relatively low pTyr527 dephosphorylating activity of the PTPα chimera tested may not accurately reflect the wild-type activity, since the chimera did not include the activating phosphorylation sites, Ser180 and 204, and it was not reported whether Tyr789 was phosphorylated. Nonetheless, the ability of ChPTPλ to efficiently dephosphorylate Src is interesting since this PTP does not contain a C-proximal tyrosine (Fang et al., 1994a; DDBJ/EMBL/GenBank accession No. CAA79972) that could be involved in a displacement mechanism like pTyr789. It is possible that a more centrally located tyrosine is involved, as proposed for CD45. Alternatively, ChPTPλ may weaken the protective pTyr527–Src SH2 interaction by other means: for example, displacement of the intramolecular SH3–helix association in Hck by intermolecular interaction with a polyproline helix in the HIV-1 Nef protein indirectly disrupts the pTyr527–homolog/SH2 binding in this Src family member (Moarefi et al., 1997), and the Src SH2–pTyr527 interaction can be weakened by N-region serine/threonine phosphorylation (Shenoy et al., 1992; Bagrodia et al., 1994; Stover et al., 1994). Further investigation of the potential of ChPTPλ for physiological interaction with Src is needed.
Control of pTyr789 phosphorylation introduces another mechanism by which PTPα activity and Src-family signaling pathways can be regulated. Moreover, phosphorylation of Tyr789 by Src (den Hertog et al., 1994) provides a potential positive-feedback loop that could temporarily ‘latch’ incoming signals to either Src or PTPα. In addition to the role suggested for PTPα dimerization (Bilwes et al., 1996; Jiang et al., 1999), PTPα–Grb2 binding might also be involved; den Hertog et al. (1994) suggest that most of the tyrosine-phosphorylated PTPα in NIH 3T3 cells is bound to Grb2 through pTyr789. Since this will block pTyr789–Src SH2 interaction, free Grb2 may act to regulate this pathway negatively. In this scenario, the binding of Grb2 by activated peptide growth factor receptors may release this regulation and indirectly activate Src-family pathways. This physiological role for the Grb2–pTyr789 binding is an alternative to the possibility that it participates in the regulation of Ras signaling pathways (den Hertog et al., 1994; den Hertog and Hunter, 1996). We have suggested previously that transient activation of a membrane-bound PTP is involved in the activation of Src family members during mitosis (Bagrodia et al., 1993; Chackalaparampil et al., 1994). It will be interesting to see whether changes in either Grb2 association or tyrosine phosphorylation of PTPα are involved in the mitotic activation.
Materials and methods
Antibodies
Plasmid LRP-pGEX-KG (a generous gift from E.Wong) expresses fusion protein GST–PTPα(B20), which contains GST and residues 165–793 from mouse PTPα including the complete cytoplasmic domain residues (Matthews et al., 1990; Sap et al., 1990). GST–PTPα(B20) was purified from transformed Escherichia coli strain DH5α cells using glutathione–Sepharose by standard procedures. Rabbits were injected s.c. with 100 μg of GST–PTPα(B20) in complete Freund's adjuvant and were boosted two weeks later with 100 μg of protein in incomplete Freund's adjuvant. Productive bleeds generally appeared after 6 weeks. Anti-PTPα antibodies were purified using a GST–PTPα(B20) affinity column. Antibodies recognized the GST–PTPα(B20) fusion protein as well as a GST fusion protein containing the D1 and D2 domains of human PTPα. Anti-PTPα 7-091 was used in subsequent experiments.
Monoclonal antibodies against the HA epitope (12CA5; Field et al., 1988) and Src (mAb 327; Lipsich et al., 1983) were purified from Balb/c mouse ascites fluid using protein A–Sepharose beads (Amersham Pharmacia). For some experiments mAb 12CA5 was crosslinked to protein A–Sepharose beads with dimethylpimelimidate (Sigma). Anti-HA rat mAb 3F10 was from Roche Molecular Biochemicals, anti-Src mAb 2-17 from Quality Biotech, and anti-pTyr mAb 4G10 from Upstate Biotechnology.
PTPα inducible expression plasmids
DNA fragments encoding an HA epitope tag at the 3′ end of wt or mutant human PTPα were generated by PCR with Vent polymerase (New England Biolabs). PTPα-HA was amplified from plasmid pXJ41PTPα (Zheng et al., 1992) using the 5′ primer 5′-CGCCAAGCTTGG_CCA_– CCATGGATTCCTGGTTCATTCTTGT-3′ and the 3′ primer 5′-TG– TCAAGCTTCA_TGCGTAGTCTGCACGTCGTATGGGTA_CTTGAAGT– TGGCATAATCTGA-3′. The 5′ primer contains a _Hin_dIII site (underlined), a Kozak consensus sequence CCACC (Kozak, 1987) and the start codon (bold). The 3′ primer contains the coding sequence (italics) for the HA epitope YPYDVPDYA (Wilson et al., 1984), the stop codon (bold) and a _Hin_dIII site (underlined). PTPα(C433S/C723S)-HA was generated from plasmid PTPα-D1SD2S (Lim et al., 1997). PTPα(Y789F)-HA was generated using pXJ41PTPα as a template with the 3′ primer 5′-TGTCAAGCTTCA_TGCGTAGTCTGGCACGTCGTATGGGTA_CTT– GAAGTTGGCAAAATCTGA-3′. The substitution ATA→**AAA** (bold italics) results in a Tyr789→Phe substitution. _Hin_dIII-cleaved PCR products were ligated into pTet-Splice (Life Technologies) to make plasmids pTPTPα, pTPTPα(CCSS) and pTPTPα(Y789F). The plasmids contain a cytomegalovirus promoter under control of regulatory sequences from the tet operon such that, when cotransfected with the transactivator pTet-tTAk (Life Technologies), they express protein only in the absence of doxycycline (Shockett et al., 1995).
To construct an expression plasmid for wt PTPα without the HA tag, the coding sequence in pTPTPα extending from the _Bam_HI site near codon 468 to the _Hin_dIII site downstream from the stop codon was amplified by PCR using the 5′ primer 5′-TTGTGAGCCGGATCC–GGGCACA-3′ and the 3′ primer 5′-TGTTGAAGCTTACTTGAAGTTGGCATAATC-3′. The 5′ primer contains the _Bam_HI site (underlined); the 3′ primer contains the stop codon (bold), no HA epitope sequence, and a _Hin_dIII site (underlined). The PCR product was cleaved with _Bam_HI and _Hin_dIII, mixed with the 1.44 kb gel-purified _Hin_dIII–_Bam_HI restriction fragment from pTPTPα, which contained the 5′ coding sequence (i.e. down to codon 468), and ligated into pTet-Splice to construct plasmid pNTPTPα. Plasmid pNTPTPα(Y789F), which overxpresses PTPα(Y789F) without an HA tag, was constructed similarly except that the 3′ PCR primer was 5′-TGTTGAAGCTTACTTGAAGTTGGC**AAAATC-3′. This contains the substitution ATA→AAA** (bold italics), which results in a Tyr789→Phe substitution.
Cell lines, cell culture and transfection
Wild-type chicken Src overexpressor cells [NIH(pMcsrc/foc)B], chicken Src(Y527F) overexpressor cells [NIH(pMcsrcF527/foc)B] and v-_src_-transformed cells [NIH(pMvsrc/foc/ep)A1] have been described previously (Johnson et al., 1985; Kmiecik and Shalloway, 1987).
Cell lines that inducibly express HA-tagged wt PTPα, PTPα(C433S/C723S), PTPα(Y789F) or nothing (control) were created by co-transfecting plasmids pTPTPα, pTPTPα(CCSS), pTPTPα(Y789F) or pTet-Splice into NIH 3T3 cells with pTet-tTAk and the G418-resistance plasmid pSV2neo (Southern and Berg, 1982). Similarly, plasmids pNTPTPα and pNTPTPα(Y789F) were used to generate cell lines inducibly expressing untagged wt PTPα and PTPα(Y789F). Transfection was with LipofectAMINE (Life Technologies) using 3 μg of PTPα expression plasmid, 3 μg of pTet-tTAk, 0.3 μg of pSV2neo and 30 μl of LipofectAMINE. Cells were selected for resistance to 0.5 mg/ml G418 in the presence of 5 ng/ml doxycycline (Sigma), and G418-resistant colonies were screened by immunoblotting with anti-PTPα and anti-HA antibodies for inducible expression of PTPα. Selected lines were recloned and homogeneity of expression was verified by immunofluorescence with anti-HA mAb 3F10. Cells were maintained in monolayer culture in complete Dulbecco's modified Eagle's medium (DMEM; Mediatech) containing 10% calf serum (Life Technologies) and 5 ng/ml doxycycline.
Analysis of PTPα expression
To induce PTPα expression, cells were washed with DMEM, trypsinized, plated into medium lacking doxycycline, and grown for 16–18 h (80–90% confluence) before harvesting. Uninduced cells were treated in the same way except that doxycycline was included in the medium. Cells were washed twice with ice-cold PBS and lysed in 1 ml of lysis buffer [50 mM HEPES pH 7.2, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 1% NP-40, 1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride (PMSF)] for 20 min at 4°C with rocking. Whole-cell lysates were clarified by centrifugation for 30 min at 28 000 g at 4°C and total cell protein concentrations were determined using the Bio-Rad DC protein assay.
Cell lysates (20 μg of protein) or immunoprecipitates were resolved by 9 or 10% SDS–PAGE. Proteins were transferred to Immobilon-P polyvinylidene membrane (Millipore) and blocked with either TBST (Tris-buffered saline, 0.1% Tween 20) containing 3% bovine serum albumin (BSA) for mAbs or PBST (PBS, 0.05% Tween 20) containing 3% non-fat milk for polyclonal antibodies. Membranes were incubated with primary antibodies for 1 h at room temperature at the following dilutions: anti-Src mAb 327 (1:3000); anti-pTyr mAb 4G10 (1:3000); anti-HA mAb 12CA5 (1:2000); anti-PTPα polyclonal antibody 7-091 (1:2500), then either peroxidase-conjugated anti-rabbit or anti-mouse-IgG (Life Technologies) at 1:10 000. Proteins were visualized by chemiluminescence (NEN Life Science Products).
Subcellular fractionation
Cells were washed twice with ice-cold PBS and scraped into 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF. Cells were pelleted, resuspended in 1 ml of TN (50 mM Tris–HCl pH 8.0, 10 mM NaCl, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF, 2 mM MgCl2 and 1 mM Na3VO4), homogenized in a small dounce, and centrifuged at 1000 g for 10 min at 4°C to remove unbroken cells and nuclei. The homogenate was centrifuged at 100 000 g for 15 min at 4°C to isolate the cytosolic fraction (supernatant), which was stored on ice until needed. The pellets were rinsed once with TN, solubilized in TN supplemented with 1% NP-40 and 150 mM NaCl on ice for 60 min, and clarified by centrifugation at 100 000 g for 15 min at 4°C. This second supernatant was the membrane fraction.
Focus formation and growth in soft agarose
Cells were assayed for focus formation by mixing 500 cells of each type to be tested with 5×105 normal NIH 3T3 cells, plating in 100-mm tissue culture dishes in media without doxycycline, and scoring for focus formation after 16 days (Johnson et al., 1985). Cells were assayed for colony formation in 0.3% soft agarose (without doxycycline) as described previously (Shalloway et al., 1984).
Src kinase assay
Cells (80–90% confluence) were lysed in RIPA buffer essentially as described (Shenoy et al., 1992). Clarified lysates containing either 100 μg (Src overexpressor cells) or 1 mg (cells that did not overexpress Src) of protein were incubated with anti-Src mAb 327 for 2 h at 4°C, and immune complexes were bound to GammaBind Sepharose beads (Amersham Pharmacia) for an additional 1 h with mixing. Pellets were washed with RIPA buffer raised to 0.5 M NaCl, and twice with regular RIPA buffer. Immunoprecipitates were assayed for their ability to phosphorylate acid-denatured enolase as described in Bagrodia et al. (1993). Results were visualized by autoradiography or quantitated by PhosphorImager analysis (Molecular Dynamics).
Immunopurification of inducible PTPα proteins and in vitro dephosphorylation of Src
Clarified lysates from PTPα overexpressor and control cell lines (1 mg of protein) were incubated with 25 μl of anti-HA-conjugated protein A– Sepharose beads for 2 h at 4°C. The beads were washed, and proteins were eluted in 0.1 ml of 0.1 M glycine pH 2.5, then neutralized to pH 7.2–7.4 with 5 μl of 1 M Tris–HCl pH 8.0.
Overexpressed wt chicken Src or Src(Y527F) was immunoprecipitated from lysates (containing 100 μg of protein) from NIH(pMcsrc/foc)B or NIH(pMcsrcF527/foc)B cells, respectively, as described above. Immunoprecipitates were washed with phosphatase buffer (50 mM imidazole pH 7.2, 5 mM dithiothreitol), and resuspended in 200 μl of phosphatase buffer. Twenty microlitres of each resuspended immunoprecipitate and 5 μl of immunopurified PTPα (or control) eluate were incubated in a final volume of 50 μl of phosphatase buffer for 30 min at 30°C with occasional shaking. Each pellet was washed three times with RIPA buffer to remove PTPα proteins, resuspended in 200 μl of kinase buffer, and 10 μl of the resuspension was used in the Src kinase assay described above.
In vitro phosphatase assay with Raytide and MBP
Immunoprecipitated v-Src was used to phosphorylate 100 μg of Raytide (Calbiochem) or 300 μg of MBP (Sigma) in 300 μl of kinase buffer for 16 h at 25°C. Reactions were stopped by adding 0.3 ml of 20% (w/v) trichloroacetic acid (TCA) and 100 μg of BSA. Samples were centrifuged, and the precipitates were washed three times with 0.5 ml of 20% TCA and resuspended in 100 μl of 0.2 M Tris–HCl pH 8.0.
PTPα immunoprecipitates were washed three times with lysis buffer and once with phosphatase buffer, then resuspended in 150 μl of phosphatase buffer. Ten microlitres of the resuspended immunoprecipitate was incubated in a total volume of 25 μl of phosphatase buffer with 2 μg of tyrosine-phosphorylated Raytide peptide or 6 μg of tyrosine-phosphorylated MBP for 30 min at 30°C. Released radioactive phosphate was measured by scintillation counting according to Krueger et al. (1990).
Src–PTPα in vivo association assay
PTPα overexpressor cells were lysed in the lysis/binding buffer (50 mM Tris–HCl pH 7.2, 50 mM NaCl, 10% glycerol, 2 mM EDTA, 50 mM NaF, 1% NP-40, 1 mM Na3VO4, 10 μg/ml aprotinin and 10 μg/ml leupeptin) used by Harder et al. (1998). Clarified lysates were combined with either 3 μl of anti-Src polyclonal antiserum 4260 or 0.2 μl of anti-Src mAb 2-17, as specified, for 2 h at 4°C, and immune complexes were bound to protein A–Sepharose (4260) or GammaBind Sepharose (mAb 2-17) beads for an additional 1 h with inversion. Immunoprecipitates were washed three times with 0.8 ml of binding buffer, divided into equal aliquots, and immunoblotted as indicated.
SH2 affinity-precipitation of PTPα proteins
GST and the GST–Src SH2 and GST–Grb2 SH2 fusion proteins were purified from bacteria transfected with pGEX2T (Guan and Dixon, 1991) or pGEX2T-derived plasmids encoding GST–Src SH2 (Bibbins et al., 1993) or GST–Grb2 SH2 (Rozakis-Adcock et al., 1992) fusion proteins as described (Taylor et al., 1995).
Expressor cells were lysed in binding buffer raised to 150 mM NaCl. Lysates were clarified by centrifugation and aliquots (1.5 mg of total cell protein in 1 ml) were incubated with 15 μg of the indicated GST fusion proteins for 1 h at 4°C. Complexes were adsorbed to 10 μl (packed volume) of glutathione–Sepharose 4B beads for 1 h at 4°C with inversion, washed three times, and immunoblotted as described above.
Acknowledgments
Acknowledgements
We thank S.Showalter and L.Nicholson for unpublished isothermal calorimetry measurements, Ed Wong for LRP-pGEX-KG, K.Bibbins for the GST–Src SH2 and T.Pawson for the GST–Grb2 SH2 expression plasmids, Stephen Taylor for many helpful conversations and insights, and both him and Michael Dehn for reviewing the manuscript. This study was supported by grant CA32317 from the National Cancer Institute.
Note added in proof
Lammers et al. have reported recently that (in NIH 3T3 cells) they can not overexpress wt PTPα, but can overexpress a mutant corresponding to Y789F [Lammers,R., Lerch,M.M. and Ullrich,A. (2000) The carboxyl–terminal tyrosine residue of protein-tyrosine phosphatase α mediates association with focal adhesion plaques. J. Biol. Chem., 275, 3391–3396]. Moreover, they have reported previously that wt PTPα is able to rescue BHK-IR cells from insulin-induced growth changes, but that the mutant PTPα can not. Furthermore, they observe differences between the in vivo tyrosine phosphorylation patterns induced by overexpression of the wt and mutant proteins even though, as here, they detect no differences in their ability to dephosphorylate non-Src substrates [Lammers,R., Moller,P.H. and Ullrich,A. (1998) Mutant forms of the protein tyrosine phosphatase α show differential activities towards intracellular substrates. Biochem. Biophys. Res. Commun., 242, 32–38]. Our results suggest that these differences may result from (i) the reduced ability of the mutant to dephosphorylate/activate Src family members and (ii) a consequent reduction of toxicity due to phosphatase overexpression in their cell system.
References
- Alema S., Cassalbore, P., Agostini, E. and Tato, F. (1985) Differentiation of PC12 phaeochromocytoma cells induced by v-src oncogene. Nature, 316, 557–559. [DOI] [PubMed] [Google Scholar]
- Autero M., et al. (1994) Tyrosine phosphorylation of CD45 phospho– tyrosine phosphatase by p50_csk_ kinase creates a binding site for p56_lck_ tyrosine kinase and activates the phosphatase. Mol. Cell. Biol., 14, 1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagrodia S., Chackalaparampil, I., Kmiecik, T.E. and Shalloway, D. (1991) Altered tyrosine 527 phosphorylation and mitotic activation of p60c-src. Nature, 349, 172–175. [DOI] [PubMed] [Google Scholar]
- Bagrodia S., Taylor, S.J. and Shalloway, D. (1993) Myristylation is required for Tyr-527 dephosphorylation and activation of pp60c-src in mitosis. Mol. Cell. Biol., 13, 1464–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagrodia S., Laudano, A.P. and Shalloway, D. (1994) Accessibility of the c-src SH2-domain for binding is increased during mitosis. J. Biol. Chem., 269, 10247–10251. [PubMed] [Google Scholar]
- Bhandari V., Lim, K.L. and Pallen, C.J. (1998) Physical and functional interactions between receptor-like protein-tyrosine phosphatase α and p59_fyn_. J. Biol. Chem., 273, 8691–8698. [DOI] [PubMed] [Google Scholar]
- Bibbins K.B., Boeuf, H. and Varmus, H.E. (1993) Binding of the Src SH2 domain to phosphopeptides is determined by residues in both the SH2 domain and the phosphopeptides. Mol. Cell. Biol., 13, 7278–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes A.M., den Hertog, J., Hunter, T. and Noel, J.P. (1996) Structural basis for inhibition of receptor protein-tyrosine phosphatase-α by dimerization. Nature, 382, 555–559. [DOI] [PubMed] [Google Scholar]
- Bjelfman C., Meyerson, G., Cartwright, C.A., Mellstrom, K., Hammerling, U. and Pahlman, S. (1990) Early activation of endogenous pp60_src_ kinase activity during neuronal differentiation of cultured human neuroblastoma cells. Mol. Cell. Biol., 10, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter C.A. and Wagner, E. (1988) Expression of c-src and c-abl in embryonal carcinoma cells and adult mouse tissues. Exp. Cell Res., 179, 214–221. [DOI] [PubMed] [Google Scholar]
- Bradshaw J.M., Grucza, R.A., Ladbury, J.E. and Waksman, G. (1998) Probing the ‘two-pronged plug two-holed socket’ model for the mechanism of binding of the Src SH2 domain to phosphotyrosyl peptides: a thermodynamic study. Biochemistry, 37, 9083–9090. [DOI] [PubMed] [Google Scholar]
- Brown M.T. and Cooper, J.A. (1996) Regulation, substrates and functions of src. Biochim. Biophys. Acta, 1287, 121–149. [DOI] [PubMed] [Google Scholar]
- Cartwright C.A., Eckhart, W., Simon, S. and Kaplan, P.L. (1987) Cell transformation by pp60c–src mutated in the carboxy-terminal regulatory domain. Cell, 49, 83–91. [DOI] [PubMed] [Google Scholar]
- Chackalaparampil I., Bagrodia, S. and Shalloway, D. (1994) Tyrosine dephosphorylation of pp60c-src is stimulated by a serine/threonine phosphatase inhibitor. Oncogene, 9, 1947–1955. [PubMed] [Google Scholar]
- Clark E.A. and Brugge, J.S. (1993) Redistribution of the activated pp60c–src to integrin-dependent cytoskeletal complexes in thrombin-stimulated platelets. Mol. Cell. Biol., 13, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.A. and King, C.S. (1986) Dephosphorylation or antibody binding to the carboxy terminus stimulates pp60c–src. Mol. Cell. Biol., 6, 4467–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog J. and Hunter, T. (1996) Tight association of GRB2 with receptor protein-tyrosine phosphatase α is mediated by the SH2 and C-terminal SH3 domains. EMBO J., 15, 3016–3027. [PMC free article] [PubMed] [Google Scholar]
- den Hertog J., Pals, C.E.G.M., Peppelenbosch, M.P., Tertoolen, L.G.J., de Laat, S.W. and Kruijer, W. (1993) Receptor protein tyrosine phosphatase α activates pp60c-src and is involved in neuronal differentiation. EMBO J., 12, 3789–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog J., Tracy, S. and Hunter, T. (1994) Phosphorylation of receptor protein-tyrosine phosphatase α on Tyr789, a binding site for the SH3–SH2-SH3 adaptor protein GRB-2 in vivo. EMBO J., 13, 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog J., Sap, J., Pals, C.E.G.M., Schlessinger, J. and Kruijer, W. (1995) Stimulation of receptor protein-tyrosine phosphatase α activity and phosphorylation by phorbol ester. Cell Growth Differ., 6, 303–307. [PubMed] [Google Scholar]
- Falet H., Ramos-Morales, F., Bachelot, C., Fischer, S. and Rendu, F. (1996) Association of the protein tyrosine phosphatase PTP1C with the protein tyrosine kinase c-Src in human platelets. FEBS Lett., 383, 165–169. [DOI] [PubMed] [Google Scholar]
- Fang K.D., Barker, K., Sudo, M. and Hanafusa, H. (1994a) A trans– membrane protein-tyrosine phosphatase contains spectrin-like repeats in its extracellular domain. J. Biol. Chem., 169, 14056–14063. [PubMed] [Google Scholar]
- Fang K.D., Sabe, H., Saito, H. and Hanafusa, H. (1994b) Comparative study of three protein-tyrosine phosphatases. J. Biol. Chem., 269, 20194–20200. [PubMed] [Google Scholar]
- Field J., Nikawa, J., Broek, D., MacDonald, B., Rodgers, L., Wilson, I.A., Lerner, R.A. and Wigler, M. (1988) Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol., 8, 2159–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M. and Bujard, H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.L. and Dixon, J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione _S_-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Harder K.W., Moller, N.P.H., Peacock, J.W. and Jirik, F.R. (1998) Protein-tyrosine phosphatase α regulates Src family kinases and alters cell–substratum adhesion. J. Biol. Chem., 273, 31890–31900. [DOI] [PubMed] [Google Scholar]
- Hurley T.R., Hyman, R. and Sefton, B.M. (1993) Differential effects of expression of the CD45 tyrosine protein phosphatase on the tyrosine phosphorylation of the lck, fyn and c-src tyrosine protein kinases. Mol. Cell. Biol., 13, 1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., den Hertog, J., Su, J., Noel, J., Sap, J. and Hunter, T. (1999) Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-α. Nature, 401, 606–610. [DOI] [PubMed] [Google Scholar]
- Johnson P.J., Coussens, P.M., Danko, A.V. and Shalloway, D. (1985) Overexpressed pp60csrc can induce focus formation without complete transformation of NIH 3T3 cells. Mol. Cell. Biol., 5, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S., Covic, L., Wyss, A. and Ballmer-Hofer, K. (1991) Association of p60c–src with polyoma virus middle-T antigen abrogating mitosis-specific activation. Nature, 350, 431–433. [DOI] [PubMed] [Google Scholar]
- Kaplan K.B., Swedlow, J.R., Morgan, D.O. and Varmus, H.E. (1995) c-Src enhances the spreading of _src_–/– fibroblasts on fibronectin by a kinase-independent mechanism. Genes Dev., 9, 1505–1517. [DOI] [PubMed] [Google Scholar]
- Kaplan R., Morse, B., Huebner, K., Croce, C., Howk, R., Ravera, M., Ricca, G., Jaye, M. and Schlessinger, J. (1990) Cloning of three human tyrosine phosphatases reveals a multigene family of receptor-linked protein-tyrosine-phosphatases expressed in brain. Proc. Natl Acad. Sci. USA, 87, 7000–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer R.A., Sachsenmaier, C., Cooper, J.A. and Soriano, P. (1999) Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J., 18, 2459–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiecik T.E. and Shalloway, D. (1987) Activation and suppression of pp60c–src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell, 49, 65–73. [DOI] [PubMed] [Google Scholar]
- Kmiecik T.E., Johnson, P.J. and Shalloway, D. (1988) Regulation by the autophosphorylation site in overexpressed pp60c–src. Mol. Cell. Biol., 8, 4541–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger N.X., Streuli, M. and Saito, H. (1990) Structural diversity and evolution of human receptor-like protein tyrosine phosphatases. EMBO J., 9, 3241–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.Y., Gaits, F., Ragab, A., Ragab-Thomas, J.M.F. and Chap, H. (1994) Translocation of an SH2-containing protein tyrosine phosphatase (SH-PTP1) to the cytoskeleton of thrombin-activated platelets. FEBS Lett., 343, 89–93. [DOI] [PubMed] [Google Scholar]
- Li R.Y., Gaits, F., Ragab, A., Ragab-Thomas, J.M.F. and Chap, H. (1995) Tyrosine phosphorylation of an SH2-containing protein tyrosine phosphatase is coupled to platelet thrombin receptor via a pertussis toxin-sensitive heterotrimeric G-protein. EMBO J., 14, 2519–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.L., Lai, D.S.Y., Kalousek, M.B., Wang, Y. and Pallen, C.J. (1997) Kinetic analysis of two closely related receptor-like protein-tyrosine-phosphatases, PTPα and PTPɛ. Eur. J. Biochem., 245, 693–700. [DOI] [PubMed] [Google Scholar]
- Lipsich L.A., Lewis, A.J. and Brugge, J.S. (1983) Isolation of monoclonal antibodies that recognize the transforming proteins of avian sarcoma viruses. J. Virol., 48, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz U., Ravichandran, K.D., Pei, D., Walsh, C.T., Burakoff, S.J. and Neel, B.G. (1994) Lck-dependent tyrosyl phosphorylation of the phosphotyrosine phosphatase SH-PTP1 in murine T cells. Mol. Cell. Biol., 14, 1824–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch S.A., Brugge, J.S. and Levine, J.M. (1986) Induction of altered c-src product during neural differentiation of embryonal carcinoma cells. Science, 234, 873–876. [DOI] [PubMed] [Google Scholar]
- Matthews R.J., Cahir, E.D. and Thomas, M.L. (1990) Identification of an additional member of the protein-tyrosine-phosphatase family: evidence for alternative splicing in the tyrosine phosphatase domain. Proc. Natl Acad. Sci. USA, 87, 4444–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi I., LaFevre-Bernt,M., Sicheri,F., Huse,M., Lee,C.H., Kuriyan,J. and Miller,W.T. (1997) Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature, 13, 385, 650–653. [DOI] [PubMed] [Google Scholar]
- Mustelin T., Pessa-Morikawa, T., Autero, M., Gassman, M., Andersson, L.C., Gahmberg, C.G. and Burn, P. (1992) Regulation of the p59_fyn_ protein tyrosine kinase by the CD45 phosphotyrosine phosphatase. Eur. J. Immunol., 22, 1173–1178. [DOI] [PubMed] [Google Scholar]
- Neel B.G. and Tonks, N.K. (1997) Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol., 9, 193–204. [DOI] [PubMed] [Google Scholar]
- Osherov N. and Levitzki, A. (1994) Epidetermal-growth-factor-dependent activation of the Src-family kinases. Eur. J. Biochem., 225, 1047–1053. [DOI] [PubMed] [Google Scholar]
- Ostergaard H.L., Shackelford, D.A., Hurley, T.R., Johnson, P., Hyman, R., Sefton, B.M. and Trowbridge, I.S. (1989) Expression of CD45 alters phosphorylation of the _lck_-encoded tyrosine protein kinase in murine lymphoma T-cell lines. Proc. Natl Acad. Sci. USA, 86, 8959–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. (1995) Protein modules and signalling networks. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- Piwnica-Worms H., Saunders, K.F., Roberts, T.M., Smith, A.E. and Cheng, S.H. (1987) Tyrosine phosphorylation regulates the biochemical and biological properties of pp60c–src. Cell, 49, 75–82. [DOI] [PubMed] [Google Scholar]
- Ponniah S., Wang, D.Z.M., Lim, K.L and Pallen, C.J. (1999) Targeted disruption of the tyrosine phosphatase PTPα leads to constitutive downregulation of the kinases Src and Fyn. Curr. Biol., 9, 535–538. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M., McGlade, J., Mbamalu, G., Pelicci, C., Daly, R., Li, W. and Pawson, T. (1992) Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature, 360, 689–692. [DOI] [PubMed] [Google Scholar]
- Sap J., D'Eustachio, P., Givol, D. and Schlessinger, J. (1990) Cloning and expression of a widely expressed receptor tyrosine phosphatase. Proc. Natl Acad. Sci. USA, 87, 6112–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaapveld R., Wieringa, B. and Hendriks, W. (1997) Receptor-like protein tyrosine phosphatases: alike and yet so different. Mol. Biol. Rep., 24, 247–262. [DOI] [PubMed] [Google Scholar]
- Shalloway D. and Taylor, S.J. (1997) Src: more than the sum of its parts. Trends Cell Biol., 7, 215–217. [DOI] [PubMed] [Google Scholar]
- Shalloway D., Coussens, P.M. and Yaciuk, P. (1984) Overexpression of the c-src protein does not induce transformation of NIH 3T3 cells. Proc. Natl Acad. Sci. USA, 81, 7071–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S., Choi, J.-K., Bagrodia, S., Copeland, T.D., Maller, J.L. and Shalloway, D. (1989) Purified maturation promoting factor phosphorylates pp60c–src at the sites phosphorylated during fibroblast mitosis. Cell, 57, 763–774. [DOI] [PubMed] [Google Scholar]
- Shenoy S., Chackalaparampil, I., Bagrodia, S., Lin, P.-H. and Shalloway, D. (1992) Role of p34_cdc2_-mediated phosphorylations in two-step activation of pp60c–src during mitosis. Proc. Natl Acad. Sci. USA, 89, 7237–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockett P., Difillipantonio, M., Hellman, N. and Schatz, D.G. (1995) A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc. Natl Acad. Sci. USA, 92, 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F., Moerefi, I. and Kuriyan, J. (1997) Crystal structure of the Src family tyrosine kinase Hck. Nature, 385, 602–609. [DOI] [PubMed] [Google Scholar]
- Somani A.-K., Bignon, J.S., Mills, G.B., Siminovitch, K.A. and Branch, D.R. (1997) Src kinase activity is regulated by the SHP-1 protein-tyrosine phosphatase. J. Biol. Chem., 272, 21113–21119. [DOI] [PubMed] [Google Scholar]
- Songyang Z. and Cantley, L.C. (1995) Recognition and specificity in protein tyrosine kinase-mediated signalling. Trends Biochem. Sci., 20, 470–475. [DOI] [PubMed] [Google Scholar]
- Songyang Z., et al. (1993) SH2 domains recognize specific phospho– peptide sequences. Cell, 72, 767–778. [DOI] [PubMed] [Google Scholar]
- Sorge L.K., Levy, B.T. and Maness, P.F. (1984) pp60c–src is develop– mentally regulated in the neural retina. Cell, 36, 249–257. [DOI] [PubMed] [Google Scholar]
- Southern P.J. and Berg, P. (1982) Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet., 1, 327–341. [PubMed] [Google Scholar]
- Stover D.R., Charbonneau, H., Tonks, N.K. and Walsh, K.A. (1991) Protein-tyrosine-phosphatase CD45 is phosphorylated transiently on tyrosine upon activation of Jurkat T cells. Proc. Natl Acad. Sci. USA, 88, 7704–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover D.R., Liebetanz, J. and Lydon, N.B. (1994) Cdc2-mediated modulation of pp60c–src activity. J. Biol. Chem., 269, 26885–26889. [PubMed] [Google Scholar]
- Streuli M., Krueger, N.X., Thai, T., Tang, M. and Saito, H. (1990) Distinct functional roles of the two intracellular phosphatase like domains of the receptor-linked protein tyrosine phosphatases LCA and LAR. EMBO J., 9, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Batzer, A. and Sap, J. (1994) Receptor tyrosine phosphatase R-PTP-α is tyrosine-phosphorylated and associated with the adaptor protein Grb2. J. Biol. Chem., 269, 18731–18734. [PubMed] [Google Scholar]
- Su J., Yang, L.-T. and Sap, J. (1996) Association between receptor protein-tyrosine phosphatase RPTPα and the Grb2 adaptor. J. Biol. Chem., 271, 28086–28096. [DOI] [PubMed] [Google Scholar]
- Su J., Muranjan, M. and Sap, J. (1999) Receptor protein tyrosine phosphatase α activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr. Biol., 9, 505–511. [DOI] [PubMed] [Google Scholar]
- Takeya T. and Hanafusa, H. (1983) Structure and sequence of the cellular gene homologous to the RSV src gene and the mechanism for generating the transforming virus. Cell, 32, 881–890. [DOI] [PubMed] [Google Scholar]
- Taylor S.M., Anafi, M., Pawson, T. and Shalloway, D. (1995) Functional interaction between c-Src and its mitotic target, Sam 68. J. Biol. Chem., 270, 10120–10124. [DOI] [PubMed] [Google Scholar]
- Thomas A.M. and Brugge, J.S. (1997) Cellular functions regulated by Src family kinases. Annu. Rev. Cell Biol., 13, 513–609. [DOI] [PubMed] [Google Scholar]
- Tracy S., van der Geer, P. and Hunter, T. (1995) The receptor-like protein-tyrosine phosphatase, RPTPα, is phosphorylated by protein kinase C on two serines close to the inner face of the plasma membrane. J. Biol. Chem., 270, 10587–10594. [DOI] [PubMed] [Google Scholar]
- Wang Y. and Pallen, C.J. (1991) The receptor-like protein tyrosine phosphatase HPTPα has two active catalytic domains with distinct substrate specificities. EMBO J., 10, 3231–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.A., Niman, H.L., Houghten, R.A., Cherenson, A.R., Connolly, M.L. and Lerner, R.A. (1984) The structure of an antigenic determinant in a protein. Cell, 37, 767–778. [DOI] [PubMed] [Google Scholar]
- Wu L., Buist, A., den Hertog, J. and Zhang, Z.-Y. (1997) Comparative kinetic analysis and substrate specificity of the tandem catalytic domains of the receptor-like protein-tyrosine phosphatase α. J. Biol. Chem., 272, 6994–7002. [DOI] [PubMed] [Google Scholar]
- Xu W., Harrison, S.C. and Eck, M.J. (1997) Three-dimensional structure of the tyrosine kinase c-Src. Nature, 385, 595–602. [DOI] [PubMed] [Google Scholar]
- Yeung Y.-G., Berg, K.L., Pixley, F.J., Angeletti, R.H. and Stanley, E.R. (1992) Protein tyrosine phosphatase-1C is rapidly phosphorylated in tyrosine in macrophages in response to colony stimulating factor-1. J. Biol. Chem., 267, 23447–23450. [PubMed] [Google Scholar]
- Zheng X.M., Wang, Y. and Pallen, C.J. (1992) Cell transformation and activation of pp60c–src by overexpression of a protein tyrosine phosphatase. Nature, 359, 336–339. [DOI] [PubMed] [Google Scholar]