Comparative Proteomic Profiling of Atopic Dermatitis Patients Based on History of Eczema Herpeticum Infection and Staphylococcus aureus Colonization (original) (raw)
. Author manuscript; available in PMC: 2012 Jan 1.
Published in final edited form as: J Allergy Clin Immunol. 2011 Jan;127(1):186–19411. doi: 10.1016/j.jaci.2010.10.033
Abstract
Background
Atopic dermatitis is the most common inflammatory skin disorder in the general population worldwide and the majority of patients are colonized with Staphylococcus aureus. Eczema herpeticum is a disseminated herpes simplex virus infection that occurs in a small subset of patients.
Objectives
The goal was to conduct proteomic profiling of atopic dermatitis patients based on Staphylococcus aureus colonization status and history of eczema herpeticum. We hoped to identify new biomarkers for improved diagnosis and prediction of eczema herpeticum and Staphylococcus aureus susceptibility, and to generate new hypotheses regarding disease pathogenesis.
Methods
Skin taping was performed on nonlesional skin of non-atopic controls and on lesional and nonlesional skin of atopic dermatitis patients. Subjects were classified according to history of eczema herpeticum and Staphylococcus aureus colonization. Proteins were analyzed using mass spectrometry; diagnostic groups were compared for statistically significant differences in protein expression.
Results
Proteins related to the skin barrier (filaggrin-2, corneodesmosin, desmoglein-1, desmocollin-1, and transglutaminase-3) and generation of natural moisturizing factor (arginase-1, caspase-14, gamma-glutamyl cyclotransferase) were expressed at significantly lower levels in lesional versus nonlesional sites of atopic dermatitis patients with and without history of eczema herpeticum; epidermal fatty acid binding protein was expressed at significantly higher levels in patients with methicillin resistant Staphylococcus aureus.
Conclusion
This non-invasive, semi-quantitative profiling method has revealed novel proteins likely involved in the pathogenesis of atopic dermatitis. The lower expression of skin barrier proteins and enzymes involved in the generation of the natural moisturizing factor could further exacerbate barrier defects and perpetuate water loss from the skin. The greater expression of epidermal fatty acid binding protein, especially in patients colonized with methicillin-resistant Staphylococcus aureus, may perpetuate the inflammatory response via eicosanoid signaling.
Keywords: Atopic dermatitis, mass spectrometry, proteomics, natural moisturizing factor, eczema herpeticum, tape stripping, skin barrier, filaggrin-2, epidermal fatty acid binding protein, methicillin-resistant Staphylococcus aureus
INTRODUCTION
Atopic dermatitis (AD) is a chronic inflammatory skin disorder that affects nearly 17% of children and can persist into adulthood,1 significantly compromising quality of life.2 AD is a multifactorial skin disease characterized by defects in the skin barrier and immune system.3 Numerous factors modulate disease severity on an individual basis, including genetic susceptibility,4 immune response,5 and diverse environmental factors.3 AD patients are prone to skin infections, including eczema herpeticum (EH), a disseminated herpes simplex virus (HSV)-1 or -2 infection that occurs in a subset of AD patients.6 EH can be complicated by keratoconjunctivitis, viremia, meningitis and encephalitis.7 EH patients tend to have early onset AD, more severe disease, increased risk of asthma, increased allergen sensitization, increased TH2 polarity, and more frequent skin infections.8 Additionally, it has been shown that up to 90% of AD patients are colonized with Staphylococcus aureus (S. aureus)9 and 16% with methicillin-resistant S. aureus.10 Patients with a history of EH have a higher risk of MRSA colonization.8
In addition to an increased susceptibility to skin infection, AD patients have numerous abnormalities in their epidermis, which acts as a critical mechanical barrier against microbes and serves to maintain proper skin hydration.11 The epidermis is comprised of four distinct layers: basal (the deepest layer), spinous, granular, and cornified (the uppermost layer). Epidermal differentiation begins with the migration of proliferating keratinocytes from the basal layer and ends with their terminal differentiation into corneocytes (dead keratinocytes). The stratum corneum (SC) or cornified layer, is a flattened sheet of corneocytes tightly connected by corneodesmosomes and embedded in an intercellular matrix of non-polar lipids.12 This layer of dead cells is the key physical and permeability barrier against the environment and is continuously shed and renewed by differentiating keratinocytes. Recent work suggests that abnormal epidermal differentiation, including defective corneocyte compaction, cornification, and lipid release play a key role in the pathogenesis of AD.13
The goal of this exploratory, hypothesis-generating proteomics study funded by the NIH/NIAID Atopic Dermatitis Vaccinia Network (ADVN) was to identify unique patterns of biomarkers associated with AD pathogenesis and EH/S. aureus susceptibility. Samples were collected from non-atopic individuals and AD patients using tape stripping and proteomic profiling was performed. Samples were analyzed in triplicate via mass spectrometry, and a custom-designed, in-house Java Application was developed to process the data.14 Differences in protein expression between diagnostic groups were estimated and statistical significance evaluated based on a linear mixed model.
METHODS
Study Population and Design
Participants with AD and non-atopic healthy controls, aged 1 to 80 years, were enrolled at National Jewish Health. AD was diagnosed according to standardized criteria developed by the ADVN.8 A total of 65 participants were enrolled; 29 EH−, 21 EH+, and 15 non-atopic controls. Swabs were collected from nonlesional skin of all participants and lesional skin of AD participants to determine S. aureus colonization status (methicillin sensitive S. aureus [MSSA]/methicillin resistant S. aureus [MRSA]/no S. aureus colonization). Skin tapings were collected from nonlesional and lesional skin (if applicable). Participants were required to discontinue the use of topical medications for seven days and oral antibiotics for ten days prior to sample collection.
Proteomic analysis was conducted on skin tapings from a subset of participants gender and age matched (± 10 yrs and age=21 yrs cutoff) across groups based on the Spectra MRSA™ screening assay result (Remel): six EH+ patients colonized with MSSA, five EH+ patients colonized with MRSA, six EH+ patients with no S. aureus colonization, six EH− patients colonized with MSSA, six EH− patients colonized with MRSA, six EH− patients with no S. aureus colonization, and five non-atopic subjects with no S. aureus colonization and one non-atopic subject colonized with MSSA. The more accurate Kirby Bauer assay was then performed, and the results of this assay were used to assign S. aureus colonization status for analysis (See Table I).
Table I.
Demographics, S. aureus colonization status, and measures of disease severity by diagnostic group.
| EH− (n=18) | EH+ (n=17) | Non-atopic (n=6) | |
|---|---|---|---|
| Age (yrs) * | 30.6±18.4 | 22.0±16.1 | 30.7±14.1 |
| Gender n (%) | |||
| Male | 11 (61%) | 8 (47%) | 2 (33%) |
| Race n (%) | |||
| White | 15 (83%) | 12 (71%) | 6 (100%) |
| Black | 2 (11%) | 4 (24%) | 0 (0%) |
| Asian | 1 (6%) | 1 (6%) | 0 (0%) |
| Ethnicity n (%) | |||
| Hispanic | 0 (0%) | 2 (12%) | 0 (0%) |
| S. aureus colonization n (%)1 | |||
| MRSA | 5 (28%) | 3 (18%) | 0 (0%) |
| MSSA | 7 (39%) | 8 (47%) | 1 (17%) |
| No S. aureus | 6 (33%) | 6 (35%) | 5 (83%) |
| EASI Score*2 | 19.6±12.6 | 16.0±9.4 | -- |
| Rajka-Langeland Score*2 | 7.4±1.2 | 6.7±1.4 | -- |
| Total IgE KIU/L3** | 189.0 (52.8, 3680.0) | 1479.0 (409.0, 2949.0) | 17.1 (12.0, 22.2) |
The National Jewish Health institutional review board approved this study. Written informed consent was obtained from each participant or from the parent or legal guardian in the case of minors. Participants aged 7 to 17 years provided assent.
Skin Taping & Storage
Skin tapings were collected from lesional (mostly chronic/more than three days old) and nonlesional sites of AD subjects and from nonlesional skin of non-atopic subjects as described previously.14 Samples were “heat killed” in a water bath at 70°C for 30 min to eliminate risk of infectivity then frozen in a −80°C freezer. Lack of colony growth on blood agar plates was confirmed in preliminary test samples subjected to 70°C for 30 minutes.
Protein Extraction
Proteins were removed from the tape discs with an extraction buffer containing 0.01% PPS (3-[3-(1,1-bisalkyloxyethyl)pyridin-1-yl]propane-1-sulfonate).14 Extracts from tape discs corresponding to layers 1–5, 6–10, 11–15, and 16–20 were combined and processed as previously outlined.14
Protein Digestion
Proteins were digested as previously described14 then purified using Oasis HLB μElution Plate (30 μm), equipped with a vacuum manifold, according to manufacturer directions (Waters, Milford, MA).
Mass Spectrometry
Liquid chromatography and mass spectrometry were carried out as previously described.14 Samples were run in triplicate on an Agilent 1200 series HPLC (Agilent Technologies, Santa Clara, CA) and Agilent ETD ion trap (model 6340) mass spectrometer with an HPLC-chip.
Database Searching
Raw data were extracted and analyzed using the Spectrum Mill database searching program (Rev A.03.03.080 SR1; Agilent Technologies) as previously described.14 Data were searched against the SwissProt Homo Sapiens database (UniProt Release 14).14 Data were validated and protein identifications were considered significant if the following confidence thresholds were met: minimum of two peptides per protein, protein score >11, individual peptide scores of at least 7, and Scored Percent Intensity (SPI) of at least 70%.
Protein Selection
Protein database search results were compiled for the triplicate MS runs, pooled layers, subjects, and lesional/nonlesional sites via spectral counting using an in-house developed Java application (Sun Microsystems). To quantify relative protein amounts, spectral counts were calculated as the sum of the spectra matched to peptides corresponding to a protein in the database. Spectral counts were then normalized to the total number of spectra per MS run. Proteins were considered for statistical analysis only if they were present in two out of three technical replicates and if there were at least six non-zero values across treatment groups. Spectral counts for each selected protein were averaged across technical replicates and across pooled layers yielding one mean spectral count per tape stripping site (lesional/nonlesional sites for AD subjects and nonlesional sites for non-atopic subjects). All keratins were excluded from statistical analysis due to high homology, which rendered it impossible to distinguish isoforms with confidence.
Statistical Methods
Descriptive statistics are presented to characterize all subjects included in the analysis. Categorical data are presented as enumerations and percentages. Continuous data are presented as arithmetic mean ± standard deviation or as median (25th percentile, 75th percentile) if the distribution of the data is skewed.
To compare protein levels between diagnostic groups, normalized mean spectral counts were modeled using a linear mixed model with random intercepts to account for the correlation of multiple samples (lesional/nonlesional) for a single subject. The predictors of interest were diagnostic group, S. aureus colonization status (as measured by the Kirby Bauer assay) and sample type, but age and gender were included in the model to account for the study design. Comparisons between diagnostic groups within sample type (EH− lesional vs EH+ lesional, EH− nonlesional vs EH+ nonlesional, EH− nonlesional vs non-atopic nonlesional, EH+ nonlesional vs non-atopic nonlesional) and between sample types within diagnostic group (EH− lesional vs EH− nonlesional, EH+ lesional vs EH+ nonlesional) were made based on model-based estimates of normalized mean spectral count differences and corresponding _P_-values. For AD subjects only, similar analyses were also performed on MRSA lesional vs MSSA lesional, MRSA nonlesional vs MSSA nonlesional, MRSA lesional vs no S. aureus lesional, MRSA nonlesional vs no S. aureus nonlesional, MSSA lesional vs no S. aureus lesional, and MSSA nonlesional vs no S. aureus nonlesional. To account for multiple comparisons, the Benjamini-Hochberg method was used to control false discovery rate (FDR). To control FDR at a level of 0.05 for the EH analysis, comparisons with a _P_-value less than 0.0040 were considered significant. Likewise for the S. aureus analysis, comparisons with a _P_-value less than 0.0005 were considered significant. However, for exploratory purposes, any comparison with a _P_-value less than 0.05 was considered to be of interest.
To examine the impact of other covariates on the EH comparisons of interest, the model was expanded to control for severity (EASI Score and Rajka-Langeland Score) and total IgE. A sensitivity analysis for the potential effect of race was conducted, in which the EH comparisons were repeated excluding all black subjects.
RESULTS
Demographics, S. aureus colonization status, and measures of disease severity
Table I presents descriptive statistics characterizing the study sample by diagnostic group. Samples were analyzed from 18 EH− patients (five MRSA, seven MSSA, six no S. aureus), 17 EH+ patients (three MRSA, eight MSSA, six no S. aureus), and six non-atopic controls (one MSSA, five no S. aureus). Table E I lists the body locations of skin tapings by diagnostic group (see Online Repository).
Complete List of Identified Proteins
One hundred fifty-three proteins were identified in two out of three technical replicates in layers 1–5, 6–10, 11–15, or 16–20 for at least one biological sample (see Online Repository Table E II). Proteins identified included blood proteins, keratins, skin barrier proteins, and immune-related proteins, among others.
Statistical Analysis of Protein Levels Between EH Diagnostic Groups and Sample Types
Seventy-one proteins were analyzed for differences in protein expression between diagnostic groups and sample types (see Online Repository Table E III). Model-based estimates of normalized mean spectral count differences and corresponding _P_-values are presented for each comparison. Comparisons highlighted in gray met the experiment-wise threshold for statistical significance (P ≤ 0.004) while comparisons with an asterisk did not meet the threshold but were of interest for exploratory purposes. These results are also presented in Table II for proteins related to the skin barrier and generation of natural moisturizing factor. Again, comparisons highlighted in gray met the experiment-wise threshold for statistical significance (P ≤ 0.004) while comparisons with a superscript did not meet the threshold but were of interest for exploratory purposes..
Table II.
Comparisons of protein expression between EH diagnostic groups and sample types for proteins related to the skin barrier and generation of natural moisturizing factor.
| Accession Number | Protein Name | EH− Lesional vs EH+ Lesional | EH− NL vs EH+ NL | EH− NL vs NA NL | EH+ NL vs NA NL | EH− Lesional vs EH− NL | EH+ Lesional vs EH+ NL |
|---|---|---|---|---|---|---|---|
| P05089 | Arginase-1 | 0.35 | 4.05 | 3.55 | −0.50 | −16.49 | −12.791 |
| Q13867 | Bleomycin hydrolase | −2.41 | −2.52 | −4.24 | −1.72 | −3.931 | −4.051 |
| P31944 | Caspase-14 | −3.09 | −0.51 | −0.65 | −0.14 | −23.54 | −20.97 |
| Q15517 | Corneodesmosin | −0.00 | 1.18 | 5.39 | 4.21 | −7.68 | −6.51 |
| Q08554 | Desmocollin-1 | −0.46 | −3.29 | 14.65 | 17.941 | −18.02 | −20.85 |
| Q02413 | Desmoglein-1 | −2.07 | 2.68 | 15.53 | 12.85 | −24.74 | −20.002 |
| Q5D862 | Filaggrin-2 | 0.20 | 7.511 | −15.67 | −23.19 | −10.63 | −3.31 |
| O75223 | Gamma-glutamylcyclotransferase | 3.55 | 6.39 | 14.433 | 8.04 | −12.86 | −10.02 |
| Q08188 | Protein-glutamine gamma-glutamyltransferase E | −2.64 | −6.48 | −6.10 | 0.39 | −11.07 | −14.91 |
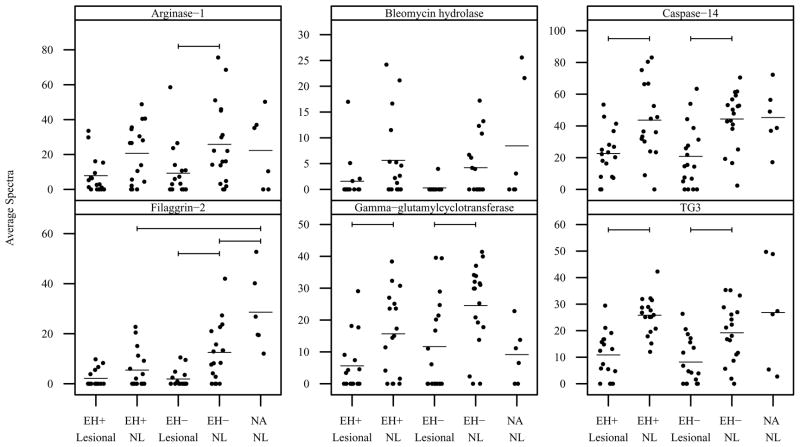
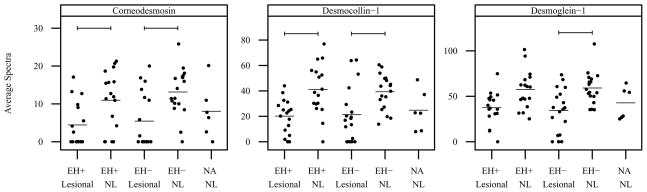
Vertical scatter plots of normalized mean spectral count data are presented by diagnostic category and sample type in Figures 1 and 2 for proteins related to the skin barrier and generation of natural moisturizing factor. Statistically significant comparisons are denoted by bars. Arginase-1 was expressed at significantly lower levels in lesional vs nonlesional sites in EH− subjects (Figure 1). Bleomycin hydrolase was expressed at lower levels in lesional vs nonlesional sites in both EH− and EH+ subjects; however the difference was not statistically significant (Figure 1). Caspase-14 and protein-glutamine gamma-glutamyltransferase E (also known as transglutaminase 3 or TG3) were expressed at significantly lower levels in lesional vs nonlesional sites in both EH− and EH+ subjects (Figure 1). Filaggrin-2 was expressed at significantly lower levels in lesional vs nonlesional sites in EH− subjects, and in EH+/EH− nonlesional vs nonatopics (Figure 1). Gamma-glutamylcyclotranserase (GGCT) was expressed at significantly lower levels in lesional vs nonlesional sites in both EH+ and EH− subjects (Figure 1). Corneodesmosin and desmocollin-1 were also expressed at significantly lower levels in lesional vs nonlesional sites in both EH− and EH+ subjects (Figure 2). Desmoglein-1 was expressed at significantly lower levels in lesional vs nonlesional sites in EH− subjects and was expressed at lower levels in EH+ subjects; however the difference in EH+ subjects was not statistically significant (Figure 2).
Figure 1. Mean spectral count by diagnostic group and sample type for proteins related to generation of natural moisturing factor.
Vertical scatter plots of the mean spectral count for all subjects by diagnostic group and sample type. Horizontal bars denote group means. Horizontal brackets denote significant differences between groups (_P_-value threshold = 0.0040). NL=nonlesional.
Figure 2. Mean spectral count by diagnostic group and sample type for proteins related to the skin barrier.
Vertical scatter plots of the mean spectral count for all subjects by diagnostic group and sample type. Horizontal bars denote group means. Horizontal brackets denote significant differences between groups (_P_-value threshold = 0.0040). NL=nonlesional.
The results of these comparisons did not change appreciably when controlling for severity (as measured by EASI Score and Rajka-Langeland Score) or total IgE or when excluding black subjects from analysis (data not shown).
Statistical Analysis of Protein Levels between S. aureus Diagnostic Groups
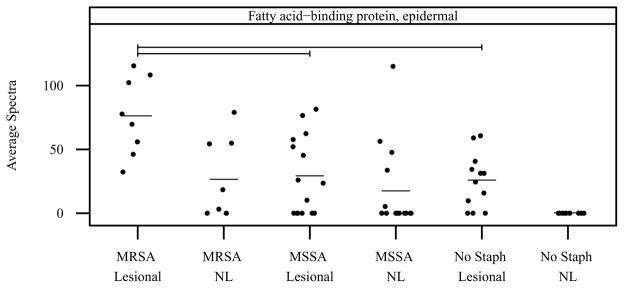
Model-based estimates of normalized mean spectral count differences and corresponding _P_-values are presented in Table E IV for each S. aureus comparison. E-fabp was expressed at significantly higher levels in MRSA lesional vs MSSA lesional (50.16 mean spectra, _P_-value = 2.4E-04) and MRSA lesional vs no S. aureus lesional sites (55.65 mean spectra, P value = 1.3E-04). Vertical scatter plots of normalized mean spectral count data are presented by diagnostic category and sample type in Figure 3 for e-fabp. Statistically significant comparisons are denoted by bars.
Figure 3. Mean spectral count by S. aureus infection group and sample type for e-fabp.
Vertical scatter plots of the mean spectral count for all subjects by S. aureus infection group and sample type. Horizontal bars denote group means. Horizontal brackets denote significant differences between groups (_P_-value threshold = 0.0005). NL=nonlesional.
DISCUSSION
These studies reveal decreased levels of proteins related to the skin barrier (filaggrin-2, corneodesmosin, desmoglein-1, desmocollin-1, and transglutaminase-3) and generation of natural moisturizing factor (arginase-1, caspase-14, gamma-glutamyl cyclotransferase) in lesional versus nonlesional sites of EH+ and EH− AD patients. Epidermal fatty acid binding protein was expressed at significantly higher levels in patients with methicillin resistant Staphylococcus aureus as compared to patients with methicillin sensitive Staphylococcus aureus. No significant differences were found between AD patients with and without a history of eczema herpeticum, but many proteins neared the significance threshold, notably filaggrin-2.
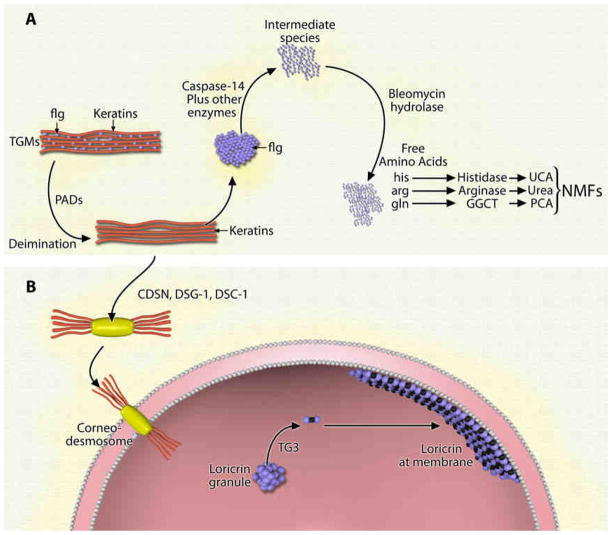
Recent studies indicate that defects in skin barrier proteins are highly associated with the development of AD. Loss of function mutations in filaggrin can be found in approximately 20% of AD patients.15 Filaggrin, a member of the fused S100 family of S100 Ca2+-binding proteins, is synthesized in the granular layer as a large 400 kDa precursor termed profilaggrin.16 Profilaggrin is stored within keratohyalin granules in the granular layer;17 as calcium levels rise during differentiation, it undergoes extensive processing including dephosphorylation and cleavage into filaggrin monomers.17 In the cornified layer, transglutaminases crosslink filaggrin to keratin 1 and 10 to form the insoluble keratin matrix crucial to the development of the skin barrier (see Figure 4).18 Next, the cross-linked filaggrin monomers undergo further post-translation modification (deimination/citrullination) via the calcium dependent enzyme peptidylarginine deiminase (PAD).18 This deimination results in disruption of the filaggrin/keratin crosslinking, setting the stage for filaggrin degradation into NMF. NMF refers to a mixture of primarily filaggrin derived hygroscopic amino acids including arginine, glutamine, and histidine and their derivatives citrulline/urea, PCA (2-pyrrolidone-5-carboxylic acid), and UCA (urocanic acid), respectively (see Figure 4).3
Figure 4. a) Generation of Natural Moisturizing Factor.
In the cornified layer, transglutaminases (TGMs) crosslink filaggrin (flg) to K1/K10 to form keratin bundles. The cross-linked filaggrin monomers undergo further post-translational modification (deimination) via the calcium dependent enzyme peptidylarginine deiminase (PAD). Deiminated filaggrin is released from the complex and further processed by caspase-14 and other enzymes into an intermediate species, which is further processed by bleomycin hydrolase into free amino acids (arg, his, gln). Arginine (arg) is processed by arginase-1 to urea, glutamine (gln) is processed by gamma-glutamyl cyclotranserase (GGCT) to pyroglutamic acid (PCA), and histidine (his) is processed by histidase into urocanic acid (UCA). NMF refers to this mixture of primarily filaggrin derived hygroscopic amino acids and their derivatives. b) Corneodesmosome Formation and Reinforcement of Corneocyte Membrane. Upon release of filaggrin monomers, keratins bind to desmoglein-1 (dsg-1), desmocollin-1 (dsc-1) and corneodesmosin (CDSN) to form corneodesmosomes, which tightly bind adjacent corneocytes. Simultaneously, transglutaminase-3 (TG3) binds loricrin to small proline rich proteins, and this complex further reinforces the inner membrane of the corneocyte.
Numerous enzymes are involved in the processing of profilaggrin to filaggrin to NMF as reviewed by Sandilands et al and Candi et al.17,18 In this exploratory study, three filaggrin/NMF processing enzymes were found to be expressed at significantly lower levels in EH+ and/or EH− lesional AD skin as compared to nonlesional AD skin (caspase-14, gamma glutamylcyclotransferase (GGCT), and arginase-1). Additionally, bleomycin hydrolase (BH), while not statistically significant, showed a trend towards lower expression in EH+/EH− lesional vs nonlesional skin. Caspase-14 is an enzyme required for the processing of deiminated filaggrin, and homozygous null mice lacking caspase-14 display mild barrier defects characterized by increased transepidermal water loss, decreased SC hydration and abnormal filaggrin degradation.19 The neutral cysteine protease bleomycin hydrolase (BH) is important in the final breakdown of partially processed and deiminated filaggrin peptides into amino acids which are components of the NMF.20 Gamma-glutamyl cyclotransferase (GGCT) catalyzes the formation of pyroglutamic acid or PCA which is the most abundant NMF found in the stratum corneum.21 Arginase-1, an enzyme in the urea cycle, hydrolyzes L-arginine into L-ornithine and urea.22 Arginine is a significant amino acid component of filaggrin18 and is released upon filaggrin degradation. Decreased expression of arginase-1 may decrease urea generation, a hygroscopic component of the NMF.23 As a whole, these data indicate altered filaggrin processing in lesional skin which may further exacerbate the disease process via abnormal corneocyte development and a decrease in the amount of NMF which is crucial to skin hydration. Restoration of NMF components using creams and moisturizers containing urea or PCA has been shown to alleviate the symptoms of AD and/or reduce the risk of relapse as reviewed by Loden.24 The ability of these NMF-based creams to restore the skin barrier further highlights the critical role of NMFs in skin barrier integrity.
In addition to lower expression of enzymes involved in NMF generation, our current work revealed lower expression of three proteins directly linked to the skin barrier and corneodesmosome structure. Corneodesmosomes, comprised of desmoglein-1, desmocollin-1, and corneodesmosin, bind keratins to the cellular membrane and serve to tightly attach adjacent corneocytes (Figure 4).18 Through a tightly controlled process, corneodesmosomes are proteolytically degraded in the uppermost layers of the stratum corneum to allow desquamation. The remaining keratins are covalently attached to the cell envelope and provide mechanical resistance.25 Simultaneously, the cytosolic enzyme transglutaminase-3 (protein-glutamine γ-glutamyltransferase E or TG3) mediates the crosslinking of loricrin to small proline rich proteins; this complex further reinforces the cell membrane (Figure 4). The significantly lower expression of desmocollin-1, desmoglein-1, corneodesmosin, and TG3 in lesional skin could be indicative of inappropriate desquamation26 or abnormal differentiation3 both of which have been found in AD.13
Filaggrin-2 (flg-2) is one of five genes in the S100 fused-type protein gene cluster.27,28 It is closely homologous to filaggrin, but the precise role of flg-2 in skin biology is unknown. The lower expression of flg-2 in both EH+/EH− nonlesional skin vs non-atopic nonlesional skin and in EH− lesional vs nonlesional skin indicate a potential role in the maintenance of the skin barrier. Furthermore, there was a trend towards lower levels of flg-2 in EH+ NL as compared to EH− NL (_P_=0.02) skin. Unpublished work reported in a patent by Jens-Michael Schroder indicates potent antimicrobial activity of the C-terminal of flg-2 against the soil bacterium, Pseudomonas.29 Our observed lower expression of flg-2 in EH+ nonlesional skin is consistent with previous work showing an association of impaired skin barrier and decreased antimicrobial activity in subjects prone to EH.30,31 Further work needs to be done on this potentially important skin barrier protein and antimicrobial peptide as a biomarker distinguishing EH+ vs EH− subjects.
In our present study, e-fabp was elevated in MRSA lesional vs MSSA lesional and MRSA lesional vs. no S. aureus lesional. Elevated levels of fatty acid binding protein in AD patients have been reported in other studies,14,32 but none evaluated concurrent S. aureus colonization status. Fatty acid binding proteins are abundant intracellular proteins that bind and transport otherwise insoluble long-chain fatty acids.33 E-fabp is found in the basal and granular cell layers in normal human skin34 and appears to be essential for normal keratinocyte differentiation.35 It has been proposed that fatty acid binding proteins may serve as master regulators of inflammatory and metabolic signaling pathways.36 In support of this, e-fabp has been shown to bind and stabilize leukotriene A4 (LTA4) and may modulate the production or metabolism of bioactive eicosanoids which have been found in the urine of AD patients.37,38 In addition to a potential role in eicosanoid signaling, e-fabp may also serve as an antioxidant protein and has been shown to bind 4-hydroxynonenal, a highly reactive aldehyde by-product of lipid peroxidation.39 Increased levels of urinary oxidative stress markers have been found in AD patients40 as well as direct evidence of oxidative stress in the stratum corneum in AD.41 It is possible that oxidative stress may induce e-fabp, and this may be exacerbated in AD patients with MRSA infection. Additionally, liver fabp has been shown to be a critical host factor for malaria42 and increases the intracellular growth of chlamydia.43 Further studies are needed to determine whether e-fabp may promote S. aureus growth or serve as a host factor for infection.
In conclusion, we have found lower expression of skin barrier proteins in lesional skin of AD patients. These proteins are involved in the generation of the NMF, corneodesmosomes, and antimicrobial host defense. These changes may reflect defective differentiation of corneocytes in AD and promote susceptibility to skin infection. The findings presented here support recent work highlighting broad defects in epidermal cornification in AD.13 In addition, increased e-fabp levels in MRSA infected AD patients may indicate aberrant eicosanoid signaling, oxidative stress, or e-fabp may serve as a host factor for S. aureus colonization.
Key Messages.
- The lower levels of four enzymes involved in the generation of the natural moisturizing factor in lesional atopic dermatitis skin are a novel finding. This defect could perpetuate the dry skin cycle and predispose patients to infection.
- The lower levels of key skin barrier proteins involved in the generation of corneodesmosomes may result in decreased corneocyte adhesion. The lower levels of filaggrin-2 in atopic dermatitis lesions may indicate a skin barrier defect or a decrease in antimicrobial peptides.
- The higher levels of epidermal fatty acid binding protein in patients colonized with methicillin-resistant Staphylococcus aureus as compared to patients colonized with methicillin sensitive Staphylococcus aureus or no S. aureus may represent a protective mechanism to elevated oxidative stress or may perpetuate the inflammatory response.
Acknowledgments
The authors thank Maureen Sandoval for her invaluable assistance in the preparation of this manuscript and in final reference formatting. We also wish to thank the nursing staff in the CTRC for their help in patient recruitment and sample collection as well as Aaron Holliday and Serae Thomas at Rho Federal Systems Division, Inc for their work coordinating the study. Technical assistance was provided by the National Jewish Health Mass Spectrometry Core Facility.
Funding: NIH/NIAID Contracts N01-AI-40029, N01-AI-40033, R01 AR41256, and NCRR S10RR023703.
ABBREVIATIONS
AD
atopic dermatitis
ADVN
NIH/NIAID Atopic Dermatitis and Vaccinia Network
EH−
AD without a history of eczema herpeticum
EH+
AD with a history of eczema herpeticum
BH
bleomycin hydrolase
CDSN
corneodesmosin
dsc-1
desmocollin-1
dsg-1
desmoglein-1
e-fabp
epidermal fatty acid binding protein
FDR
false discovery rate
flg-2
filaggrin-2
GGCT
gamma-glutamyl cyclotransferase
HSV
herpes simplex virus
MRSA
methicillin resistant Staphylococcus aureus
MSSA
methicillin sensitive Staphylococcus aureus
NA
non-atopic
NMF
natural moisturizing factor
PAD
peptidylarginine deiminase
PCA
2-pyrrolidone-5-carboxylic acid
PPS
3-[3-(1,1-bisalkyloxyethyl)pyridin-1-yl]propane-1-sulfonate
SC
stratum corneum
S. aureus
Staphylococcus aureus
transglutaminase-3/TG3
protein-glutamine gamma-glutamyltransferase E
UCA
urocanic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–94. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 2.Zuberbier T, Orlow SJ, Paller AS, Taieb A, Allen R, Hernanz-Hermosa JM, et al. Patient perspectives on the management of atopic dermatitis. J Allergy Clin Immunol. 2006;118:226–32. doi: 10.1016/j.jaci.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 3.Proksch E, Folster-Holst R, Brautigam M, Sepehrmanesh M, Pfeiffer S, Jensen JM. Role of the epidermal barrier in atopic dermatitis. J Dtsch Dermatol Ges. 2009;7:899–910. doi: 10.1111/j.1610-0387.2009.07157.x. [DOI] [PubMed] [Google Scholar]
- 4.Baurecht H, Irvine AD, Novak N, Illig T, Buhler B, Ring J, et al. Toward a major risk factor for atopic eczema: meta-analysis of filaggrin polymorphism data. J Allergy Clin Immunol. 2007;120:1406–12. doi: 10.1016/j.jaci.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 5.De Benedetto A, Agnihothri R, McGirt LY, Bankova LG, Beck LA. Atopic dermatitis: a disease caused by innate immune defects? J Invest Dermatol. 2009;129:14–30. doi: 10.1038/jid.2008.259. [DOI] [PubMed] [Google Scholar]
- 6.Wetzel S, Wollenberg A. Eczema herpeticatum. Hautarzt. 2004;55:646–52. doi: 10.1007/s00105-004-0744-1. [DOI] [PubMed] [Google Scholar]
- 7.Wollenberg A, Zoch C, Wetzel S, Plewig G, Przybilla B. Predisposing factors and clinical features of eczema herpeticum: a retrospective analysis of 100 cases. J Am Acad Dermatol. 2003;49:198–205. doi: 10.1067/s0190-9622(03)00896-x. [DOI] [PubMed] [Google Scholar]
- 8.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–9. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breuer K, SHA, Kapp A, Werfel T. Staphylococcus aureus: colonizing features and influence of an antibacterial treatment in adults with atopic dermatitis. Br J Dermatol. 2002;147:55–61. doi: 10.1046/j.1365-2133.2002.04872.x. [DOI] [PubMed] [Google Scholar]
- 10.Suh L, Coffin S, Leckerman KH, Gelfand JM, Honig PJ, Yan AC. Methicillin-resistant Staphylococcus aureus colonization in children with atopic dermatitis. Pediatr Dermatol. 2008;25:528–34. doi: 10.1111/j.1525-1470.2008.00768.x. [DOI] [PubMed] [Google Scholar]
- 11.Fluhr JW, Feingold KR, Elias PM. Transepidermal water loss reflects permeability barrier status: validation in human and rodent in vivo and ex vivo models. Exp Dermatol. 2006;15:483–92. doi: 10.1111/j.1600-0625.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 12.Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp Dermatol. 2008;17:1063–72. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 13.Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol. 2009;124:1235–44. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 14.Broccardo CJ, Mahaffey SB, Strand M, Reisdorph NA, Leung DY. Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. J Allergy Clin Immunol. 2009;124:1113–5. doi: 10.1016/j.jaci.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez E, Baurecht H, Herberich E, Wagenpfeil S, Brown SJ, Cordell HJ, et al. Meta-analysis of filaggrin polymorphisms in eczema and asthma: robust risk factors in atopic disease. J Allergy Clin Immunol. 2009;123:1361–70. doi: 10.1016/j.jaci.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Presland R. Function of filaggrin and caspase-14 in formation and maintenance of the epithelial barrier. Dermatolgica Sinica. 2009;27:1–14. [Google Scholar]
- 17.Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. J Cell Sci. 2009;122:1285–94. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 19.Denecker G, Hoste E, Gilbert B, Hochepied T, Ovaere P, Lippens S, et al. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat Cell Biol. 2007;9:666–74. doi: 10.1038/ncb1597. [DOI] [PubMed] [Google Scholar]
- 20.Kamata Y, Taniguchi A, Yamamoto M, Nomura J, Ishihara K, Takahara H, et al. Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J Biol Chem. 2009;284:12829–36. doi: 10.1074/jbc.M807908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oakley AJ, Yamada T, Liu D, Coggan M, Clark AG, Board PG. The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle. J Biol Chem. 2008;283:22031–42. doi: 10.1074/jbc.M803623200. [DOI] [PubMed] [Google Scholar]
- 22.Kezic S, Kammeyer A, Calkoen F, Fluhr JW, Bos JD. Natural moisturizing factor components in the stratum corneum as biomarkers of filaggrin genotype: evaluation of minimally invasive methods. Br J Dermatol. 2009;161:1098–104. doi: 10.1111/j.1365-2133.2009.09342.x. [DOI] [PubMed] [Google Scholar]
- 23.Wu G, Morris SM., Jr Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336 ( Pt 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loden M. Role of topical emollients and moisturizers in the treatment of dry skin barrier disorders. Am J Clin Dermatol. 2003;4:771–88. doi: 10.2165/00128071-200304110-00005. [DOI] [PubMed] [Google Scholar]
- 25.Wiren K, Nohlgard C, Nyberg F, Holm L, Svensson M, Johannesson A, et al. Treatment with a barrier-strengthening moisturizing cream delays relapse of atopic dermatitis: a prospective and randomized controlled clinical trial. J Eur Acad Dermatol Venereol. 2009;23:1267–72. doi: 10.1111/j.1468-3083.2009.03303.x. [DOI] [PubMed] [Google Scholar]
- 26.Elias PM, Schmuth M. Abnormal skin barrier in the etiopathogenesis of atopic dermatitis. Curr Opin Allergy Clin Immunol. 2009;9:437–46. doi: 10.1097/ACI.0b013e32832e7d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, Jacob D, et al. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007;8:R107. doi: 10.1186/gb-2007-8-6-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Z, Hansmann B, Meyer-Hoffert U, Glaser R, Schroder JM. Molecular identification and expression analysis of filaggrin-2, a member of the S100 fused-type protein family. PLoS One. 2009;4:e5227. doi: 10.1371/journal.pone.0005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroder J-M. Ifapsoriasin fragments as antimicrobial peptides. Patent pending [Google Scholar]
- 30.Boguniewicz M, Leung DY. Recent insights into atopic dermatitis and implications for management of infectious complications. J Allergy Clin Immunol. 125:4–13. doi: 10.1016/j.jaci.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–13. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamane Y, Moriyama K, Yasuda C, Miyata S, Aihara M, Ikezawa Z, et al. New horny layer marker proteins for evaluating skin condition in atopic dermatitis. Int Arch Allergy Immunol. 2009;150:89–101. doi: 10.1159/000210385. [DOI] [PubMed] [Google Scholar]
- 33.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe R, Fujii H, Yamamoto A, Hashimoto T, Kameda K, Ito M, et al. Immunohistochemical distribution of cutaneous fatty acid-binding protein in human skin. J Dermatol Sci. 1997;16:17–22. doi: 10.1016/s0923-1811(97)00615-4. [DOI] [PubMed] [Google Scholar]
- 35.Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–27. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makowski L, Hotamisligil GS. Fatty acid binding proteins--the evolutionary crossroads of inflammatory and metabolic responses. J Nutr. 2004;134:2464S–8S. doi: 10.1093/jn/134.9.2464S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickinson Zimmer JS, Voelker DR, Bernlohr DA, Murphy RC. Stabilization of leukotriene A4 by epithelial fatty acid-binding protein in the rat basophilic leukemia cell. J Biol Chem. 2004;279:7420–6. doi: 10.1074/jbc.M311404200. [DOI] [PubMed] [Google Scholar]
- 38.Oymar K, Aksnes L. Increased levels of urinary leukotriene E4 in children with severe atopic eczema/dermatitis syndrome. Allergy. 2005;60:86–9. doi: 10.1111/j.1398-9995.2005.00658.x. [DOI] [PubMed] [Google Scholar]
- 39.Bennaars-Eiden A, Higgins L, Hertzel AV, Kapphahn RJ, Ferrington DA, Bernlohr DA. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277:50693–702. doi: 10.1074/jbc.M209493200. [DOI] [PubMed] [Google Scholar]
- 40.Omata N, Tsukahara H, Ito S, Ohshima Y, Yasutomi M, Yamada A, et al. Increased oxidative stress in childhood atopic dermatitis. Life Sci. 2001;69:223–8. doi: 10.1016/s0024-3205(01)01124-9. [DOI] [PubMed] [Google Scholar]
- 41.Niwa Y, Sumi H, Kawahira K, Terashima T, Nakamura T, Akamatsu H. Protein oxidative damage in the stratum corneum: Evidence for a link between environmental oxidants and the changing prevalence and nature of atopic dermatitis in Japan. Br J Dermatol. 2003;149:248–54. doi: 10.1046/j.1365-2133.2003.05417.x. [DOI] [PubMed] [Google Scholar]
- 42.Mikolajczak SA, Jacobs-Lorena V, MacKellar DC, Camargo N, Kappe SH. L-FABP is a critical host factor for successful malaria liver stage development. Int J Parasitol. 2007;37:483–9. doi: 10.1016/j.ijpara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Burczynski F, Anderson J, Zhong G. Effect of host fatty acid-binding protein and fatty acid uptake on growth of Chlamydia trachomatis L2. Microbiology. 2007;153:1935–9. doi: 10.1099/mic.0.2006/003491-0. [DOI] [PubMed] [Google Scholar]