Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution (original) (raw)
Abstract
Directed evolution is a valuable technique to improve enzyme activity in the absence of a priori structural knowledge, which can be typically enhanced via structure-guided strategies. In this study, a combination of both whole-gene error-prone polymerase chain reaction and site-saturation mutagenesis enabled the rapid identification of mutations that improved RmlA activity toward non-native substrates. These mutations have been shown to improve activities over 10-fold for several targeted substrates, including non-native pyrimidine- and purine-based NTPs as well as non-native d- and l-sugars (both α- and β-isomers). This study highlights the first broadly applicable high throughput sugar-1-phosphate nucleotidyltransferase screen and the first proof of concept for the directed evolution of this enzyme class toward the identification of uniquely permissive RmlA variants.
Keywords: Carbohydrate, Carbohydrate Biosynthesis, Carbohydrate Chemistry, Glycosylation, Nucleoside Nucleotide Analogs, Nucleoside Nucleotide Biosynthesis, Glycosyltransferase, Natural Product, Sugar Nucleotide
Introduction
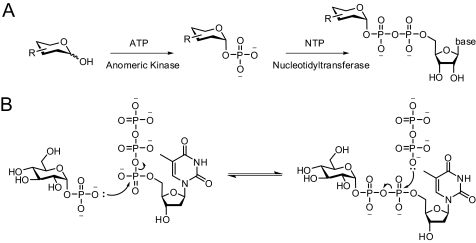
Glycosides are critical in cells, functioning in maintenance of cell integrity, molecular recognition, pathogen virulence, molecular defense, signaling, and information storage. The activities of these glycosides are often modulated by their sugar moieties, wherein even subtle alterations of the attached sugars can dramatically influence overall bioactivity (1–6). Among the emerging methods for altering the sugar substituents of complex glycosides, chemoenzymatic glycorandomization exploits the inherent or engineered substrate promiscuity of anomeric kinases and nucleotidyltransferases to provide a simple two-step route for the synthesis of diverse sugar nucleotide (NDP-sugar)3 libraries (Fig. 1A) (7). In chemoenzymatic glycorandomization, these unique sugar donors serve as substrates for permissive glycosyltransferases, thereby providing a rapid chemoenzymatic means to glycodiversify complex scaffolds (7–11). Such multienzyme, single vessel reactions complement conventional glycosylation methods and are also amenable to in vivo applications (12).
FIGURE 1.
A, sugar nucleotide synthesis via the glycorandomization pathway. B, the RmlA-catalyzed transformation.
The availability of diverse sugar nucleotides is key to this process, and given the challenges associated with their chemical synthesis (13–15), the enzymatic synthesis of sugar nucleotides is an attractive alternative (16, 17). Within this context, certain sugar-1-phosphate nucleotidyltransferases display some flexibility toward non-native sugar 1-phosphates and, in some cases, even non-native NTPs (18–25), the promiscuity of which can be further expanded via protein engineering (26–28). One of the most permissive and best studied nucleotidyltransferases to date is the glucose-1-phosphate thymidylyltransferase from Salmonella enterica typhimurium LT2 (RmlA; Fig. 1B) (24, 27, 28). Specifically, wild type RmlA was demonstrated to turn over a wide variety of sugar 1-phosphates, including deoxy, amino, and azido sugars, and even displayed weak activity with all eight ribo- and deoxyribonucleotides. Although structure-guided mutagenesis has led to some improvements in RmlA substrate range, this method requires access to accurate ligand-bound models and provides only a static representation (i.e. does not account for protein dynamics as a factor contributing to catalysis).
As a complement to structure-guided engineering, the directed evolution of enzymes has enabled studies to increase activity, enhance enantioselectivity, improve stability, broaden promiscuity, increase tolerance to organic solvents, and even engineer metabolic pathways (29–31). The process has also been specifically applied to two enzymes relevant to glycosylation, including those utilized for glycorandomization (32–35). To date, the main limitation for sugar-1-phosphate nucleotidyltransferase directed evolution has been a lack of a facile high-throughput screen. The one previous report of directed evolution for this enzyme class was restricted to native substrates and a substrate-inflexible biological readout (35). Herein we describe the application of a recently developed general high-throughput screen for sugar 1-phosphate toward RmlA directed evolution (36). In this study, epPCR and site-saturation techniques were combined to identify a number of improved variants for both the NTP and sugar 1-phosphate substrates. By combining the mutations identified, variants with up to 40-fold improvement in activity toward six non-native nucleotides, galactose-1-phosphate, and both the α- and β-anomers of l-fucose 1-phosphate were identified. Subsequent x-ray crystallography of key single amino acid mutants with enhanced NTP tolerance also revealed the mode of binding for non-natural NTPs. This study highlights the first application of nucleotidyltransferase directed evolution for expanding the activity toward non-native substrates.
EXPERIMENTAL PROCEDURES
General Materials and Methods
Chemicals were purchased from Sigma-Aldrich or Fisher and used without further purification. Oligonucleotides were obtained from either Integrated DNA Technologies (Coralville, IA) or the University of Wisconsin-Madison Biotechnology Center (Madison, WI) as “desalted” grade and used without any purification.
Liquid handling during high throughput screening was performed on a Beckman Coulter (Fullerton, CA) Biomek FX liquid handling system, and microtiter plate absorbance was obtained with a BMG Labtech (Durham, NC) FLUOstar Optima plate reader. Analytical and preparative HPLC was performed using a Varian ProStar HPLC system (Varian Inc., Palo Alto, CA).
Mutagenesis
Error-prone PCR was performed with the GeneMorph II kit from Stratagene (La Jolla, CA), using the manufacturer's protocols. For a mutation rate of 1–3 nucleotides/kb, 500–2000 ng of plasmid (75–325 ng amplified DNA) was used. Mutated genes were subsequently cloned into vectors via standard restriction enzyme digestion (NdeI/EcoRI) and ligation (T4 DNA ligase) or with the MEGAWHOP protocol, as described (37, 38). Site saturation and site-directed mutagenesis (supplemental Tables SI and SII) were performed using the megaprimer-based protocol of Tseng et al. (39).
Microtiter Plate Assay
The DNA from error-prone or site saturation libraries were desalted and directly transformed into electrocompetent BL21(DE3) cells (Lucigen, Middleton, WI), using manufacturer's protocols. The obtained colonies, along with empty vector and wild type controls, were inoculated into LB + kanamycin medium (50 mg/liter) in a 96-well deep well (2 ml) culture plate (MidSci, Valley Park, MO) for an overnight culture at 37 °C with mixing (350 rpm). Ten microliters from each well were transferred to 1 ml of LB + kanamycin medium in a fresh 96-well deep well plate, incubated at 37 °C for 3 h, at which time 100 μl/well of 4 mm isopropyl-β-d-thiogalactopyranoside was added to each well. After overnight (20–22 h) incubation at 28 °C and 350 rpm, the cells were pelleted (2000 × g, 10 min), decanted, and frozen at −20 °C.
Cells were subsequently resuspended in 500 μl/well 50 mm sodium phosphate (pH 7.5) containing 2 mg/ml lysozyme, and the cell suspension was frozen at −80 °C. Frozen solutions were thawed in a warm water bath, and the insoluble portions were removed via centrifugation (6000 × g, 10 min). Ni2+-NTA-agarose slurry (Qiagen, Valencia, CA), washed and equilibrated to 25% (v/v) in a 50 mm sodium phosphate (pH 7.5) buffer containing 1.2 m NaCl, was added (100 μl/well) to a fresh 96-well deep well plate, followed by the cleared supernatant. Resin was incubated with protein for 10–20 min at room temperature, with occasional gentle mixing. The bound resin was centrifuged at 1000 × g for 5 min, followed by careful aspiration of the supernatant. Resin was washed twice with 1 ml/well each of 10 mm MOPS (pH 7.5). On the final wash, 400 μl/well was left behind from the aspiration.
For the assay, the purified resin slurry was thoroughly mixed, and 10 μl/well was added to a 2× master mix prealiquoted in 96-well PCR microplates (2× master mix: 4 mm sugar 1-phosphate, 5 mm (each) NTP, 200 mm MOPS (pH 7.5), 15 mm (sugar and single NTP reactions) or 25 mm (combined NTP reactions) MgCl2). The sealed PCR plates were incubated for the appropriate time (1 h for NTP, 2.5 h for galactose, 16 h for α-fucose, or 24 h for β-fucose), at which time 20 μl/well 2× alkaline phosphatase mix (100 mm sodium carbonate (pH 10), 2 mm ZnSO4, 100 units/ml alkaline phosphatase (Roche Applied Science)) was added. After 10–30 min at 37 °C, 120 μl/well of a freshly prepared 1:1 mixture of 2% _para_-hydroxybenzhydrazide (pHBH) in 0.5 m HCl and 50 mm EDTA in 2 m NaOH was added. Reactions were heated for 5 min in a dry well heat block at 95–100 °C under a silicone sealing cap mat (Applied Biosystems, Foster City, CA), cooled on ice, transferred to a 96-well microtiter assay plate (Nunc, Rochester, NY), and the absorbance was read at 410 nm.
Production and Purification of Identified RmlA Variants
RmlA mutants were produced as _N_-His6 fusion proteins using pET28a-based expression plasmids (Novagen, Madison, WI) in Escherichia coli BL21(DE3), and purified in a manner similar to previous methods (33). Protein concentrations were determined spectrophotometrically (calc. ϵ280 = 33,350 cm−1 m−1 for wild type and most mutants; 31,860 cm−1 m−1 for Y146F; 27,850 cm−1 m−1 for W224F; and 26,360 cm−1 m−1 for Y146F/W224F (40)). Protein concentrations obtained via Bradford assay were indistinguishable from those obtained spectrophotometrically.
Enzyme Kinetics
Assays with purified enzymes were performed using the previously described HPLC assay (28). Steady-state kinetic parameters were obtained by fixing either glucose 1-phosphate, UTP, or dTTP at a saturating concentration of 10 mm (34, 35) and titrating the compound of interest. At least six different concentrations in the range of 0.25 × Km to 4 × Km (1–40 mm for NTP, 1.5–50 mm for galactose 1-phosphate, 3–50 mm for fucose 1-phosphates) were assayed in replicate on at least three separate days for each titration.
X-ray Crystallography of RmlA Variants
Purified RmlA samples were concentrated to 12 mg/ml (Q83S) and 15 mg/ml (Q83D) with final concentration of dATP or dGTP for co-crystallization of 25 mm. Initial screens were performed with a local screen UW192 utilizing a Mosquito® dispenser (TTP labTech, Royston, UK) by the sitting drop method. Crystal growth was monitored by Bruker Nonius Crystal Farms at 20 and 4 °C.
Diffraction quality crystals of RmlA Q83S with dATP were obtained by mixing 2 μl of the sample solution and 2 μl of a reservoir solution (20% methyl ether PEG 5000, 120 mm MgCl2, 100 mm Tris, pH 8.5, and 1 mm suramine) at 20 °C using the hanging drop method. Diffraction quality crystals of RmlA Q83D with dGTP were generated by mixing 2 μl of the sample solution and 2 μl of a similar reservoir solution (10% methyl ether PEG 5000, 120 mm MgCl2, 100 mm Tris, pH 8.5, and 1 mm suramine) at 20 °C using the hanging drop method. Crystals were cryoprotected with 20% ethylene glycol and were flash-frozen by liquid nitrogen.
X-ray diffraction data were collected by the Life Science Collaborative Access Team at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) using an x-ray wavelength of 0.9794 Å. Data sets were indexed and scaled using HKL 2000 (41). Molrep was utilized for molecular replacement (42). For the RmlA Q83D/dGTP structure (Protein Data Bank entry 3PKQ), molecular replacement was used with the previous RmlA mutant structure (Protein Data Bank entry 1MP4) as a starting model. For the RmlA Q83S/dATP structure (Protein Data Bank entry 3PKP), molecular replacement was used with the RmlA Q83D/dGTP structure as a starting model. The structures were completed with alternating rounds of manual model building with COOT (43) and refinement with phenix.refine (44). The final round of Q83S structure refinement included eight translation-libration-screw rigid groups (45, 46). Noncrystallographic symmetry restraints were applied to both structures. Structure quality was assessed by Procheck (47) and Molprobity (48).
RESULTS AND DISCUSSION
Library Creation
Purely random mutagenesis techniques, such as epPCR, have several limitations. Random mutagenesis spreads mutations evenly throughout the protein, whereas productive mutations generally more often occur near the active site (49, 50). In addition, epPCR causes almost exclusively single nucleotide changes in any one codon, thereby restricting the possible amino acid substitutions for each position (51). As a result, researchers have increasingly turned to “semirational” techniques, such as structure or homology-guided saturation mutagenesis, to enhance the outcome of enzyme evolution studies (51, 52).
A similar strategy was adopted for this study. Of the 61 amino acid residues within 8 Å of the bound product, 32 were eliminated from consideration because they either orient away from the active site or are occluded by more proximal amino acids. Three additional residues, which appear to be directly involved in binding the invariant phosphates, were also eliminated. Based upon this assessment, 22 of the remaining residues were submitted to saturation mutagenesis using an NNK degenerate codon.
As a complement, epPCR libraries were also generated with a mutation rate of 1–3 nucleotides/gene. Higher mutation rates were found to be detrimental to protein folding/production, consistent with prior epPCR-based studies (53, 54).
Sensitivity Enhancements of the Plate-based Assay
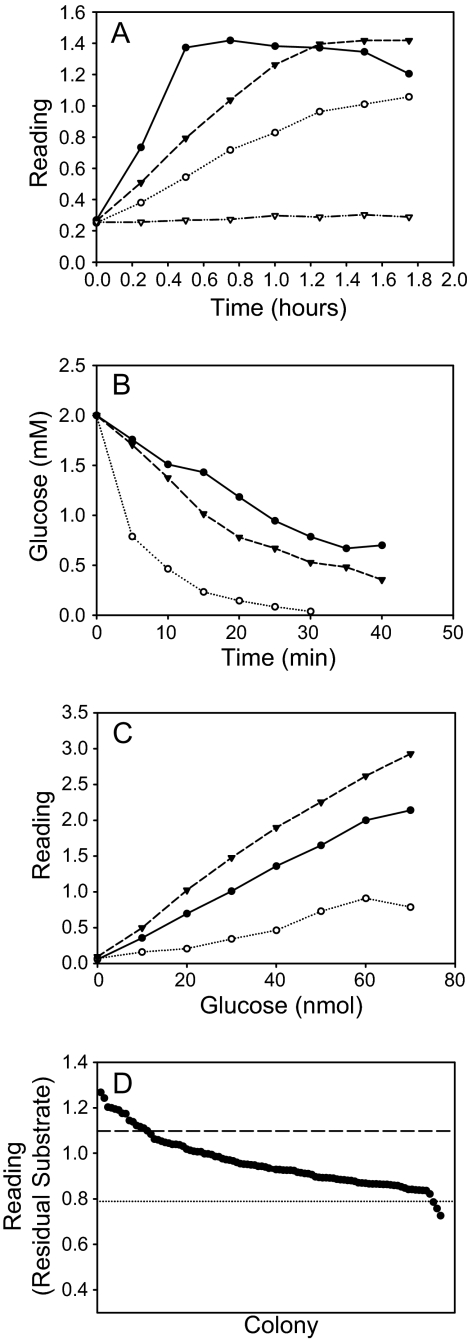
Although the previously published phosphatase/pHBH assay for residual sugar phosphates (36) works well for RmlA crude extracts and good substrates, contaminating phosphatase activity within the crude lysate is detrimental in the context of poor substrates. Specifically, various sugar 1-phosphates simply treated with crude lysates (without the addition of exogenous alkaline phosphatase) revealed nearly quantitative conversion of the sugar 1-phosphate to reducing sugar in 30–60 min (Fig. 2A). Thus, for non-native substrates requiring longer incubation times or more enzyme (i.e. more crude extract), the pHBH assay had to be adapted to a purified enzyme format.
FIGURE 2.
Plate-based enzyme assay. A, endogenous phosphatase activity in crude lysate. Sugar 1-phosphate (2 mm) (filled circles, Glc 1-phosphate; open circles, Gal-1-phosphate; filled triangles, Man 1-phosphate; open triangles, GlcNAc 1-phosphate) was reacted with BL21(DE3) crude lysate (at 2.5× concentration with respect to culture volume) in the absence of nucleotide. Without the addition of alkaline phosphatase, the reacted samples were tested directly for the presence of reducing sugar with the pHBH reagent in the plate form assay. B, comparison of solution phase and solid phase activities of WT RmlA (filled circles, 5 mm nominal RmlA on resin; open circles, 5 mm RmlA in solution; filled triangles, 1.7 mm RmlA in solution). C, the influence of Ni2+-NTA-agarose (filled circles, titration without resin; open circles, titration with resin) and EDTA (filled triangles, with resin in the presence of 25 mm EDTA) on pHBH reaction sensitivity. D, representative on-resin plate-based assay for RmlA variants screened against galactose 1-phosphate and UTP. The average activities of parent enzyme (WT RmlA) replicates (dotted line) are well separated from empty vector controls (dashed line). Wells with activity slightly greater than wild type are readily identifiable.
Initial attempts toward this goal utilized standard 96-well plate-based nickel affinity chromatography; however, although this method enabled the purification of the enzyme (and corresponding removal of contaminating phosphatase activity), salt in the elution buffer inhibited RmlA activity. Lowering salt concentration or changing the type of salt in the elution buffer failed to identify a satisfactory alternative (data not shown), and standard methods for buffer exchange (such as dialysis and gel filtration), although successful, are not conducive to high throughput applications.
Consistent with prior precedent for immobilized active nucleotidyltransferases (55), an examination of the activity of immobilized RmlA revealed specific activities (∼4-fold reduction), similar to purified enzyme in solution (Fig. 2B). However, initial pHBH assays revealed that the presence of the Ni2+-NTA-agarose greatly reduced assay sensitivity. However, consistent with the well documented detrimental effect of metal ions (such as Ni2+) on the pHBH reaction (36, 56), the addition of 50 mm EDTA to the sodium hydroxide reagent was sufficient to restore sensitivity (Fig. 2C).
Microtiter Plate Screening Conditions
To maximize sensitivity, reaction conditions that resulted in ∼25–50% turnover of the WT RmlA were selected for all screening. In this fashion, mutants that demonstrate ≥2-fold improvements (compared with WT RmlA) were readily discernable. Fig. 2D illustrates a typical reaction, highlighting the activities of both WT RmlA and inactive controls.
To identify mutations that specifically improve activity toward nucleotide substrates, ATP, CTP, and GTP with glucose-1-phosphate were tested, with the expectation that the ribo forms would serve as reasonable surrogates for the corresponding deoxyribo forms. To limit the number of preliminary screens, the target NTPs were screened as a combined equimolar mixture. Galactose 1-phosphate was selected as a primary sugar target for two reasons: (i) WT RmlA displays only moderate activity with galactose 1-phosphate (28, 34), and (ii) the sugar kinases commonly employed for glycorandomization are most proficient at generating galacto-configured sugar 1-phosphate analogs (34, 57). To further challenge the outcome in the context of galactose 1-phosphate, UTP was selected for the countersubstrate.
Although l-configured isomers are often found attached to microbial natural products (4–7), all reported RmlA sugar 1-phosphate substrates to date have been d-configured isomers. To explore the ability of engineered RmlA variants to accommodate l-configured sugars, two commercially available l-sugar 1-phosphates, α-l-fucose 1-phosphate and β-l-fucose 1-phosphate, differing only at their anomeric positions (supplemental Fig. S1), were also included as screening targets in the presence of dTTP (to maximize activity). Although β-l-fucose 1-phosphate has previously been shown to be a substrate for another glucose-1-phosphate thymidylyltransferase (20), both α and β forms were tested because the position of the anomeric phosphate is critical for RmlA catalysis, and it is unknown if the absolute ((R) versus (S)) or relative (axial versus equatorial) stereochemistry is most important in substrate positioning for RmlA.
Screening Results
To minimize the chances of a false negative and to correct for potential variability in expression levels, a multitiered screening approach was used. Specifically, the four most active “hits” from each plate, identified using the primary pHBH screen, were reinoculated from the reserved culture plates into fresh growth plates and were re-expressed and reassayed (supplemental Fig. S2). During the secondary assay, the active mutants were tested against each NTP individually. Completion of this screening program revealed a fair overlap between hits, especially for the NTP targets. A final cohort of 76 hits, representing all of the unique starting colonies, were tested against each substrate and sequenced. This analysis revealed many of the hits from the site saturation library to be degenerate, leaving a total of 37 distinct variants (supplemental Table SIII), with 49 distinct mutations at 38 different sites within the protein (supplemental Fig. S3).
Preliminary Hit Characterization
To confirm identified hits, a selection of the top hits were produced in E. coli, purified, and tested in an HPLC-based specific activity assay at the substrate concentrations employed during screening (Tables 1 and 2). Although not all of the putative hits held up under this final analysis (∼40% had activities less than WT), at least one mutation per substrate was identified with a demonstrable improvement in activity.
TABLE 1.
Specific activity of selected NTP mutants
Shown is specific activity in min−1 (values greater than wild type underlined with the two best mutants for each category in boldface type) using the following assay conditions: 2 mm sugar 1-phosphate, 2.5 mm NTP, 100 mm MOPS, pH 7.5, 7.5 mm MgCl2, 1 unit/ml inorganic pyrophosphatase, 30-min incubation, 37 °C. The amount of enzyme used varied based on substrate (2.5 μm for ATP and CTP, 1.25 μm for GTP, 0.125 μm for dCTP, 0.0625 μm for dATP and dGTP, and 0.01 μm for dTTP).
| dTTP | ATP | CTP | GTP | dATP | dCTP | dGTP | |
|---|---|---|---|---|---|---|---|
| WT | 1970 | 3.1 | 4.3 | 4.5 | 110 | 69 | 127 |
| Q27A | 1040 | 7.4 | 13.0 | 7.9 | 7 | 6 | 8 |
| Q27P | 790 | 6.7 | 8.5 | 10.1 | 0 | 0 | 4 |
| Q27S | 410 | 7.1 | 13.2 | 15.1 | 16 | 18 | 14 |
| Q27T | 1070 | 8.3 | 8.5 | 8.4 | 0 | 2 | 5 |
| Q27V | 1120 | 10.2 | 10.9 | 12.9 | 0 | 0 | 5 |
| M36T | 0 | 0.1 | 0.1 | 0.1 | 0 | 0 | 3 |
| Q83Aa | 850 | 8.7 | 2.7 | 1.5 | 382 | 39 | 19 |
| Q83Da | 300 | 1.2 | 0.7 | 14.1 | 44 | 14 | 134 |
| Q83Ea | 2620 | 0.5 | 2.2 | 5.9 | 10 | 38 | 127 |
| Q83Na | 180 | 9.6 | 2.6 | 0.5 | 263 | 77 | 9 |
| Q83S | 690 | 11.3 | 2.8 | 5.7 | 375 | 41 | 61 |
| P86T | 50 | 8.5 | 6.9 | 2.1 | 1 | 8 | 7 |
| Q91A | 1990 | 5.8 | 6.8 | 18.2 | 275 | 144 | 449 |
| Q91G | 1500 | 2.0 | 2.7 | 16.1 | 57 | 25 | 181 |
| Q91H | 1240 | 1.5 | 0.9 | 3.5 | 33 | 9 | 48 |
| Q91P | 310 | 0.9 | 0.4 | 2.0 | 21 | 4 | 27 |
| Q91S | 2240 | 5.3 | 5.2 | 49.8 | 137 | 63 | 690 |
| Q91T | 2080 | 9.0 | 7.0 | 51.5 | 375 | 134 | 753 |
| A106V | 1210 | 3.3 | 5.9 | 6.5 | 117 | 102 | 137 |
| A137T | 640 | 0.9 | 1.5 | 2.3 | 44 | 33 | 58 |
| Y146F | 1020 | 2.5 | 4.0 | 4.8 | 70 | 39 | 69 |
| W224F | 1530 | 0.2 | 0.4 | 0.3 | 16 | 4 | 15 |
| W224Ha | 1710 | 0.0 | 0.1 | 0.0 | 15 | 0 | 8 |
| M22V/L89I | 720 | 1.1 | 6.9 | 1.5 | 7 | 17 | 26 |
| M22V | 1660 | 0.7 | 1.1 | 1.2 | 14 | 9 | 23 |
| L89I | 400 | 0.7 | 4.8 | 1.0 | 6 | 15 | 21 |
| L89V | 820 | 0.8 | 5.7 | 1.7 | 8 | 18 | 24 |
| L89Ta | 1270 | 0.8 | 3.3 | 1.1 | 5 | 11 | 18 |
| S237N/I270V | 660 | 3.6 | 5.9 | 7.3 | 111 | 74 | 143 |
| S237N | 350 | 1.9 | 3.0 | 3.3 | 58 | 24 | 78 |
| I270V | 1180 | 0.7 | 1.2 | 1.3 | 25 | 15 | 30 |
| W74L/K121R | 1080 | 3.4 | 5.0 | 5.2 | 76 | 50 | 74 |
| W74L | 820 | 2.9 | 4.4 | 4.8 | 69 | 51 | 79 |
| K121R | 1250 | 0.6 | 1.1 | 1.1 | 16 | 10 | 21 |
| P84S/F151L | 1300 | 3.0 | 1.6 | 2.3 | 96 | 15 | 55 |
| P84S | 1570 | 2.3 | 1.4 | 2.1 | 84 | 20 | 55 |
| F151L | 1000 | 1.3 | 2.2 | 4.8 | 86 | 53 | 103 |
TABLE 2.
Specific activity of selected sugar mutants
Shown is specific activity in min−1 (values greater than wild type underlined with the two best mutants for each category in boldface type) using the following assay conditions: 2 mm sugar 1-phosphate, 2.5 mm NTP, 100 mm MOPS, pH 7.5, 7.5 mm MgCl2, 1 unit/ml inorganic pyrophosphatase, 30-min incubation, 37 °C. The amount of enzyme used varied based on substrate (12.5 μm for α-fucose/dTTP and β-fucose/dTTP, 5 μm for galactose/UTP, 0.5 μm for galactose/dTTP, and 0.01 μm for glucose/dTTP).
| α-Fuc/dTTP | β-Fuc/dTTP | Gal/UTP | Gal/dTTP | Glu/dTTP | |
|---|---|---|---|---|---|
| WT | 0.13 | 0.25 | 1.1 | 24 | 1970 |
| Q27A | 1.7 | 2 | 1040 | ||
| Q27S | 1.2 | 7 | 410 | ||
| Q27T | 1.4 | 2 | 1070 | ||
| Q27V | 6.1 | 2 | 1120 | ||
| M36T | 0.00 | 0.04 | 0.2 | 2 | 0 |
| P86T | 0.06 | 0.11 | 0.5 | 9 | 50 |
| A106V | 0.11 | 0.23 | 1.6 | 21 | 1210 |
| A137T | 0.08 | 0.13 | 0.8 | 20 | 640 |
| Y146F | 0.19 | 0.92 | 1.4 | 41 | 1020 |
| W224F | 0.39 | 0.58 | 0.1 | 16 | 1530 |
| W224Ha | 0.00 | 0.00 | 0.0 | 0 | 1710 |
| M22V/L89I | 0.09 | 0.27 | 2.6 | 37 | 720 |
| M22V | 0.03 | 0.15 | 0.3 | 17 | 1660 |
| L89I | 0.06 | 0.25 | 1.7 | 23 | 400 |
| L89V | 0.51 | 0.07 | 4.2 | 17 | 820 |
| L89Ta | 0.10 | 0.05 | 4.7 | 12 | 1270 |
| S237N/I270V | 0.15 | 0.40 | 2.4 | 32 | 660 |
| S237N | 0.06 | 0.25 | 1.1 | 18 | 350 |
| I270V | 0.03 | 0.14 | 0.6 | 14 | 1180 |
| W74L/K121R | 0.07 | 0.13 | 1.5 | 13 | 1080 |
| P84S/F151L | 0.14 | 0.25 | 0.2 | 20 | 1300 |
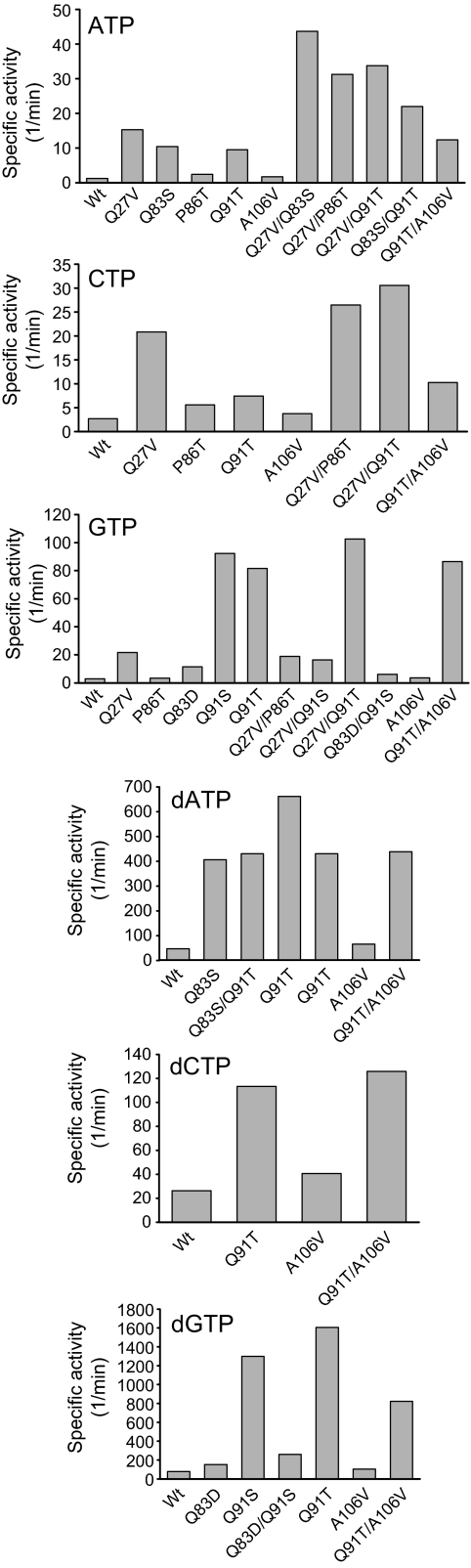
With regard to NTP improvements, Table 1 reveals the most productive substitutions to occur at a specific series of glutamines (Gln27, Gln83, and Gln91) wherein small polar (Ser and Thr) or small hydrophobic (Ala and Val) substitutions provide the greatest desired improvement. Although subtle amino acid substitutions at one or more of these positions lead to some purine (ATP/GTP) versus pyrimidine (CTP)- and/or ribose (NTP) versus deoxyribose (dNTP)-based preferences, this analysis suggests that the best NTP “generalist” derives from the Gln91 series with small side chain substitutions (Q91T > Q91S > Q91A). It should be noted that a mutation equivalent to Q27S in a homologous enzyme was previously reported to improve activity toward UTP (26). In addition, Gln83 mutations (including Q83S) had previously been identified as important for enhancing RmlA activity with purine nucleotides (28).
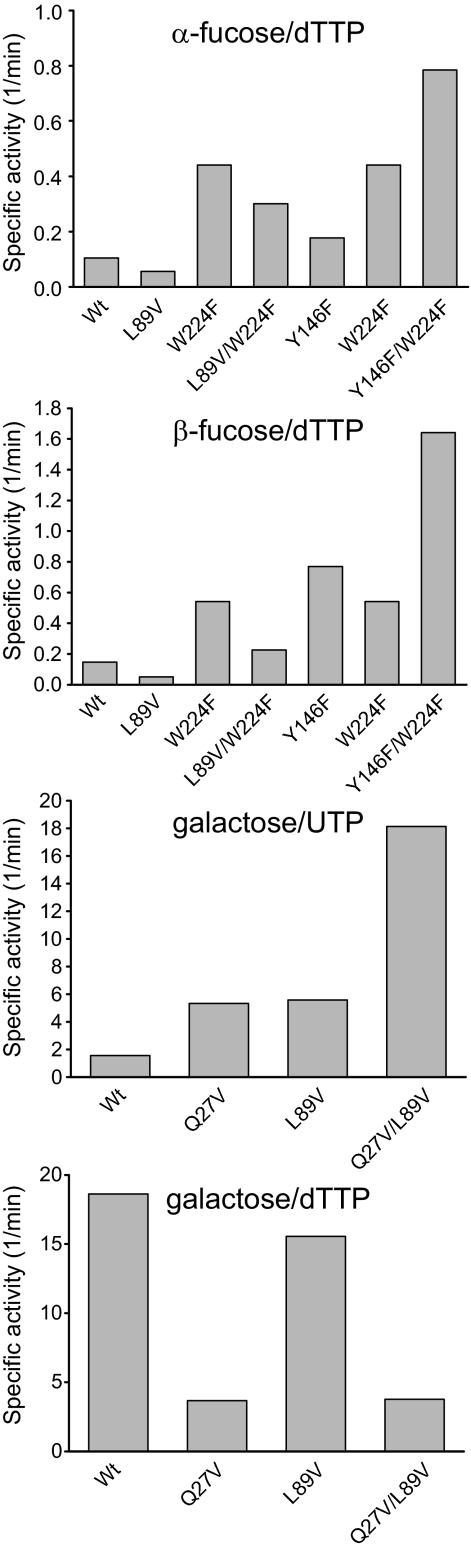
Table 2 suggests sugar 1-phosphate improvements to be more case-specific, with the most productive l-sugar 1-phosphate substitutions occurring at Leu89, Tyr146, and Trp224 with small (Val) or planar (Phe) hydrophobic substitutions preferred. In contrast, the double mutant M22V/L89I provides the best “general” C-4 epimer selectivity switch from glucose to galactose wherein the single mutations are roughly additive. In addition, galactose-directed mutants may be nucleotide-sensitive, with some improvements being specific to UDP/Gal 1-phosphate (Q27V) and some to dTDP/Gal 1-phosphate (Y146F).
Targeted Mutant Combinations
Key active mutations for targeted substrates were combined via site-directed mutagenesis to test for further gains in activity. Specifically, this study focused upon two series of combinations: (i) improving the ATP/CTP activities of the “NTP generalist” (substituted at Gln91) via combining with advantageous Gln27 mutations and (ii) improving targeted sugar 1-phosphate activities through combining the most productive mutations at Gln27, Leu89, Tyr146, and Trp224. In most cases, the combination of two mutations enhanced activity as expected (Figs. 3 and 4). One noticeable exception was Q91S, which failed to combine profitably with either Q27V (in the case of GTP) or Q83D (for GTP and dGTP). It should be noted that the specific activity observed for β-l-fucose-1-phosphate and dTTP with Y146F/Y224F (1.6 min−1; Fig. 4), compares favorably with other Glc 1-phosphate thymidylyltransferases reported to turn over this l-sugar 1-phosphate (0.45–1.4 min−1) (24), and this level of activity is sufficient for bulk production of dTDP-l-fucose-1-phosphates (supplemental material).
FIGURE 3.
Specific activity of combined mutants toward NTPs. The top mutations from each assay were combined, and the specific activity of the combination was compared with that of the individual mutations.
FIGURE 4.
Specific activity of combined mutants toward sugar 1-phosphates. The top mutations from each assay were combined, and the specific activity of the combination was compared with those of the individual mutations.
To examine if anomeric flexibility is a general feature of RmlA, the specific activities of WT RmlA and Y146/W224F with β-d-Glc 1-phosphate and dTTP were compared. Y146F/W224F displayed a small amount of turnover (0.043 min−1; 16-fold lower than the anomerically similar α-l-Fuc 1-phosphate and 46,000-fold lower than wild type RmlA with dTTP and α-d-Glc 1-phosphate). No activity was detected for the wild type enzyme and β-d-glucose 1-phosphate to the limit of sensitivity (0.007 min−1).
Table 3 highlights the single substrate kinetic parameter for key mutants. Notably, no appreciable change in Km was observed for any of the mutants characterized, despite the fact that the screening conditions were in the Km regime. Instead, the improvements in activity were confined primarily to _k_cat and _k_cat/Km with overall improvements in _k_cat/Km (for the screened reactions) ranging from 6-fold (for Q27V/Q91T and CTP) to 40-fold (for Q27V/Q91T and GTP). Determined parameters for non-screened dNTPs revealed more modest improvements in _k_cat/Km, ranging from 1.4-fold (for dCTP and Q91T) to 9-fold (for dATP and Q83S/Q91T). Notably, the observed _k_cat values for dATP and dGTP are in the range for WT RmlA and native dTTP substrate (1850 ± 40 min−1) (28). Likewise, _k_cat improvements for the targeted sugar 1-phosphates ranged from 11- to 28-fold, with the greatest improvement observed for β-l-Fuc 1-phosphate/dTTP (Y146F/W224F).
TABLE 3.
Pseudo-first order kinetic constants
Kinetics were performed with 0.05–25 μm enzyme, [NTP] + 5 mm MgCl2, 10 units/ml inorganic pyrophosphatase, and fixed cosubstrate concentrations (10 mm), in 100 mm HEPES, pH 7.5, at 37 °C, and conversion was detected via HPLC.
| _k_catapp | K _m_app | _k_cat/K _m_app | |
|---|---|---|---|
| min_−_1 | mm | min_−_1 mm_−_1 | |
| ATP | |||
| Wild type | 10.5 ± 0.8 | 4.7 ± 1.2 | 2.3 ± 0.5 |
| Q27V/Q83S | 1100 ± 200 | 20 ± 9 | 55 ± 13 |
| CTP | |||
| Wild type | 29 ± 2 | 11 ± 2 | 2.6 ± 0.3 |
| Q27V/Q91T | 260 ± 20 | 16 ± 3 | 15.8 ± 1.7 |
| GTP | |||
| Wild type | 25.1 ± 1.4 | 5.3 ± 1.0 | 4.8 ± 0.7 |
| Q27V/Q91T | 650 ± 40 | 3.4 ± 0.7 | 190 ± 30 |
| dATP | |||
| Wild type | 700 ± 50 | 5.9 ± 1.4 | 120 ± 20 |
| Q83S/Q91T | 1840 ± 70 | 1.7 ± 0.3 | 1080 ± 150 |
| dCTP | |||
| Wild type | 300 ± 30 | 7 ± 2 | 43 ± 9 |
| Q91T | 600 ± 60 | 10 ± 2 | 62 ± 10 |
| dGTP | |||
| Wild type | 650 ± 50 | 3.9 ± 1.0 | 160 ± 30 |
| Q91T | 2850 ± 150 | 3.6 ± 0.7 | 790 ± 130 |
| Gal (UTP) | |||
| Wild type | 9.5 ± 0.4 | 6.3 ± 1.0 | 1.5 ± 0.2 |
| Q27V/L89V | 104 ± 15 | 4.6 ± 2.6 | 23 ± 10 |
| α-Fuc (dTTP) | |||
| Wild type | 0.154 ± 0.007 | 6.2 ± 1.0 | 0.025 ± 0.003 |
| Y146F/W224F | 2.71 ± 0.18 | 10 ± 2 | 0.26 ± 0.04 |
| β-Fuc (dTTP) | |||
| Wild type | 0.323 ± 0.011 | 8.7 ± 1.0 | 0.037 ± 0.003 |
| Y146F/W224F | 8.9 ± 0.8 | 11 ± 3 | 0.80 ± 0.14 |
Structural Basis for Sugar 1-Phosphate Improvements
From a structural perspective, most of the key mutations have straightforward explanations. Two of the identified mutations for Fuc 1-phosphate (Y146F and W224F) are located near C6 of the hexose in the active site (based upon bound NDP-Glc). Although the specific structural impacts of Y146F or W224F have not been determined, previous structural characterization of the W224H mutant revealed an astonishing active site side chain rearrangement and main chain distortion that creates an expansive gap in the substrate-binding pocket surrounding C6. The precedent for such mutants to accommodate significant C6 steric bulk, in conjunction with a bias for β-l-Fuc 1-phosphate over α-l-Fuc 1-phosphate is consistent with β-l-Fuc 1-phosphate adopting the less favored α-d-like 4C1 chair conformation (with the 6″-methyl in the axial position) within the active site. A similar induced fit mechanism has been put forth to explain the ability of engineered anomeric sugar kinases to accept both d- and l-sugars (34). That identical mutations also provide an advantage for α-l-Fuc 1-phosphate activity and may suggest contributions from a median chair conformation potentially accessible from both α- and β-l-Fuc 1-phosphate (e.g. boat, skew, or envelope) or even the possibility that the sugar may completely flip over in the active site.
Although the specific structural contributions of L89V or Q27V have not been determined, Leu89 and Gln27 both interact with the nucleoside substructure of the NTP/NDP-sugar. In addition, previous structural characterization of the L89T mutant revealed this mutation to relieve sugar 1-phosphate C2, C3, and C4 steric constraints via a potential adjustment or “slipping” of the sugar base in the enlarged active site pocket (58). Mutations at Gln27 may translate to expanding the active site in a similar manner and thereby provide additional accommodations for novel sugar 1-phosphates and/or NTPs.
Structural Basis of Nucleotide Improvements
Gln83 is specifically involved in binding the nucleoside base substructure of the NTP/NDP-sugar and thereby contributes to base specificity. In contrast, Gln91 is found within the secondary shell, where it hydrogen-bonds with Gln83 but does not directly contact the substrate. To further probe the effect of the Gln83 mutations on nucleotide specificity, x-ray structures of RmlA Q83S with dATP and Q83D with dGTP were solved to a resolution of 2.6 and 2.4 Å, respectively (supplemental Table SIV). The overall D2 symmetric tetrameric conformations were found to be similar to previous wild type and other mutant RmlA structures (58). Q83S was found to have two tetramers per asymmetric unit compared with one tetramer per asymmetric unit for Q83D. Although the Gln83-containing loops (residues 83–87) of both mutant structures were not as clearly defined due to high B-factors and perhaps incomplete occupancy of dGTP in the Q83D (80–86%), key contacts and the mutational influence upon non-native nucleotides can be confirmed in both structures (Fig. 5). In the Q83S mutant with dATP bound, a serine hydroxyl group forms a weak hydrogen bond to the N1 and N6 of adenine (distance: 3.1 Å with N1, 3.2 Å with N6). In the Q83D mutant with dGTP bound, the side chain oxygens of aspartate show weaker hydrogen bond interactions with N1 of guanine (distance: Oδ1, 2.9 Å with N1).
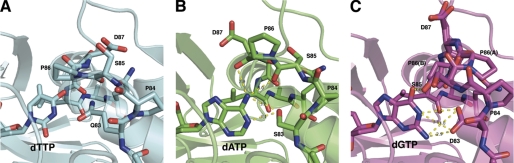
FIGURE 5.
Nucleotide recognition region in the WT RmlA wild type bound to dTTP (Protein Data Bank entry 1IIM) (A), RmlA Q83S bound to dATP (B), and RmlA Q83D bound to dGTP (C). Residues in the loop region (Gln83–Gln91) are shown in sticks. The distances between nucleotide and mutated residues are indicated by yellow dotted lines.
There are several features that distinguish the binding from the previous wild-type structures (Protein Data Bank entry 1IIM). First, in the mutant proteins, the purine rings of dATP and dGTP are tilted about 45° toward the mutated residue due to the new interactions. Second, the loop containing residues 83–89 shows a variety of conformations and possesses higher average _B_-factor than other regions. In the Q83S structure, Pro84, Ser85, and Pro86 shift outward toward the position of the nucleotide, creating more space for a larger substrate. Ser85 shows a completely different rotamer conformation compared with the wild type. Also, the backbone oxygen of Ser85 and Pro86 interact with adenine N6 via hydrogen bonding. Meanwhile, in the Q83D structure, we assigned alternative conformations for Ser85, Pro86, and Asp87 due to the apparent disorder. One conformer has the loop shifted outward toward the position of the nucleotide, creating more space for larger substrate, and Pro86 has a flipped conformation. The other conformer is similar to the wild type and Q83S structures but has a _cis_-proline (Pro86). In both conformers, the hydroxyl group of the Ser85 side chain interacts with O6 of guanine. The Q83S and Q83D structures also have different rotamer conformations of Glu87. Furthermore, the loop with residue 83 lacks interactions with Gln91, consistent with the lack of an additive effect within certain Gln83/Gln91 double mutants that have been assayed (e.g. Q83D/Q91S). Based on these observations, introduction of the Gln83 mutation not only creates hydrogen bonds to the new substrates but also makes a wider active site and contributes to a more flexible binding loop.
CONCLUSION
This study highlights the first application of nucleotidyltransferase directed evolution for expanding the activity toward non-native substrates. This work further illuminates the plasticity of this unique catalyst and enables, for the first time, the preparative syntheses of a set of non-native fucose nucleotides. The high throughput tools presented and key nucleotidyltransferase residues identified are likely to contribute to the future engineering of nucleotidyltransferases and related biocatalysts.
Acknowledgments
We thank the University of Wisconsin-Madison School of Pharmacy Analytical Instrumentation Center and the University of Wisconsin-Madison Chemistry Department Mass Spectrometry Facility for analytical support.
*
This work was supported, in whole or in part, by National Institutes of Health (NIH) Molecular Bioscience Training Grant GM07215 (to R. M., trainee) and NIH Grant AI552218 (to J. S. T. in support of R. M.), NIH Grant U54 GM074901 (to John L. Markley, Principal Investigator, for G. N. P., C. A. B., and A. C.), and NIH Molecular Biophysics Training Grant GM08293 (to A. C., trainee). The Life Sciences Collaborative Access Team was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under Contact W-31-102-ENG-38. J. S. T. is a co-founder of Centrose (Madison, WI).
The atomic coordinates and structure factors (codes 3PKQ and 3PKP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
3
The abbreviations used are:
NDP-sugar
nucleotide diphosphosugar
epPCR
error-prone polymerase chain reaction
pHBH
_para_-hydroxybenzhydrazide.
REFERENCES
- 1.Lepenies B., Yin J., Seeberger P. H. (2010) Curr. Opin. Chem. Biol. 14, 404–411 [DOI] [PubMed] [Google Scholar]
- 2.Kiessling L. L., Splain R. A. (2010) Annu. Rev. Biochem. 79, 619–653 [DOI] [PubMed] [Google Scholar]
- 3.Boltje T. J., Buskas T., Boons G. J. (2009) Nat. Chem. 1, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thibodeaux C. J., Melançon C. E., 3rd, Liu H. W. (2008) Angew. Chem. Int. Ed. Engl. 47, 9814–9859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kren V., Rezanka T. (2008) FEMS Microbiol. Rev. 32, 858–889 [DOI] [PubMed] [Google Scholar]
- 6.Luzhetskyy A., Méndez C., Salas J. A., Bechthold A. (2008) Curr. Top. Med. Chem. 8, 680–709 [DOI] [PubMed] [Google Scholar]
- 7.Langenhan J. M., Griffith B. R., Thorson J. S. (2005) J. Nat. Prod. 68, 1696–1711 [DOI] [PubMed] [Google Scholar]
- 8.Gantt R. W., Goff R. D., Williams G. J., Thorson J. S. (2008) Angew. Chem. Int. Ed. Engl. 47, 8889–8892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C., Griffith B. R., Fu Q., Albermann C., Fu X., Lee I. K., Li L., Thorson J. S. (2006) Science 313, 1291–1294 [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Albermann C., Fu X., Thorson J. S. (2006) J. Am. Chem. Soc. 128, 16420–16421 [DOI] [PubMed] [Google Scholar]
- 11.Fu X., Albermann C., Zhang C., Thorson J. S. (2005) Org. Lett. 7, 1513–1515 [DOI] [PubMed] [Google Scholar]
- 12.Williams G. J., Yang J., Zhang C., Thorson J. S. (2011) ACS Chem. Biol. 6, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner G. K., Pesnot T., Field R. A. (2009) Nat. Prod. Rep. 26, 1172–1194 [DOI] [PubMed] [Google Scholar]
- 14.Weijers C. A., Franssen M. C., Visser G. M. (2008) Biotechnol. Adv. 26, 436–456 [DOI] [PubMed] [Google Scholar]
- 15.Rupprath C., Schumacher T., Elling L. (2005) Curr. Med. Chem. 12, 1637–1675 [DOI] [PubMed] [Google Scholar]
- 16.White-Phillip J., Thibodeaux C. J., Liu H. W. (2009) Methods Enzymol. 459, 521–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bülter T., Elling L. (1999) Glycoconj. J. 16, 147–159 [DOI] [PubMed] [Google Scholar]
- 18.Beaton S. A., Huestis M. P., Sadeghi-Khomami A., Thomas N. R., Jakeman D. L. (2009) Chem. Commun., 238–240 [DOI] [PubMed] [Google Scholar]
- 19.Timmons S. C., Hui J. P., Pearson J. L., Peltier P., Daniellou R., Nugier-Chauvin C., Soo E. C., Syvitski R. T., Ferrières V., Jakeman D. L. (2008) Org. Lett. 10, 161–163 [DOI] [PubMed] [Google Scholar]
- 20.Huestis M. P., Aish G. A., Hui J. P., Soo E. C., Jakeman D. L. (2008) Org. Biomol. Chem. 6, 477–484 [DOI] [PubMed] [Google Scholar]
- 21.Timmons S. C., Mosher R. H., Knowles S. A., Jakeman D. L. (2007) Org. Lett. 9, 857–860 [DOI] [PubMed] [Google Scholar]
- 22.Bae J., Kim K. H., Kim D., Choi Y., Kim J. S., Koh S., Hong S. I., Lee D. S. (2005) Chembiochem 6, 1963–1966 [DOI] [PubMed] [Google Scholar]
- 23.Ko K. S., Zea C. J., Pohl N. L. (2004) J. Am. Chem. Soc. 126, 13188–13189 [DOI] [PubMed] [Google Scholar]
- 24.Jiang J., Albermann C., Thorson J. S. (2003) Chembiochem 4, 443–446 [DOI] [PubMed] [Google Scholar]
- 25.Thorson J. S., Kelly T. M., Liu H. W. (1994) J. Bacteriol. 176, 1840–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakeman D. L., Young J. L., Huestis M. P., Peltier P., Daniellou R., Nugier-Chauvin C., Ferrières V. (2008) Biochemistry 47, 8719–8725 [DOI] [PubMed] [Google Scholar]
- 27.Williams G. J., Gantt R. W., Thorson J. S. (2008) Curr. Opin. Chem. Biol. 12, 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretti R., Thorson J. S. (2007) J. Biol. Chem. 282, 16942–16947 [DOI] [PubMed] [Google Scholar]
- 29.Khersonsky O., Tawfik D. S. (2010) Annu. Rev. Biochem. 79, 471–505 [DOI] [PubMed] [Google Scholar]
- 30.Dougherty M. J., Arnold F. H. (2009) Curr. Opin. Biotechnol. 20, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tracewell C. A., Arnold F. H. (2009) Curr. Opin. Chem. Biol. 13, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kittl R., Withers S. G. (2010) Carbohydr. Res. 345, 1272–1279 [DOI] [PubMed] [Google Scholar]
- 33.Williams G. J., Zhang C., Thorson J. S. (2007) Nat. Chem. Biol. 3, 657–662 [DOI] [PubMed] [Google Scholar]
- 34.Hoffmeister D., Yang J., Liu L., Thorson J. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13184–13189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salamone P. R., Kavakli I. H., Slattery C. J., Okita T. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moretti R., Thorson J. S. (2008) Anal. Biochem. 377, 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyazaki K., Takenouchi M. (2002) BioTechniques 33, 1033–1034, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki K. (2003) Methods Mol. Biol. 231, 23–28 [DOI] [PubMed] [Google Scholar]
- 39.Tseng W. C., Lin J. W., Wei T. Y., Fang T. Y. (2008) Anal. Biochem. 375, 376–378 [DOI] [PubMed] [Google Scholar]
- 40.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 42.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 43.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 46.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 47.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 48.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morley K. L., Kazlauskas R. J. (2005) Trends Biotechnol. 23, 231–237 [DOI] [PubMed] [Google Scholar]
- 50.Chica R. A., Doucet N., Pelletier J. N. (2005) Curr. Opin. Biotechnol. 16, 378–384 [DOI] [PubMed] [Google Scholar]
- 51.Miyazaki K., Arnold F. H. (1999) J. Mol. Evol. 49, 716–720 [DOI] [PubMed] [Google Scholar]
- 52.Lutz S., Patrick W. M. (2004) Curr. Opin. Biotechnol. 15, 291–297 [DOI] [PubMed] [Google Scholar]
- 53.Bloom J. D., Meyer M. M., Meinhold P., Otey C. R., MacMillan D., Arnold F. H. (2005) Curr. Opin. Struct. Biol. 15, 447–452 [DOI] [PubMed] [Google Scholar]
- 54.Voigt C. A., Mayo S. L., Arnold F. H., Wang Z. G. (2001) J. Cell. Biochem. Suppl. 37, 58–63 [DOI] [PubMed] [Google Scholar]
- 55.Nahalka J., Liu Z., Chen X., Wang P. G. (2003) Chemistry 9, 372–377 [DOI] [PubMed] [Google Scholar]
- 56.Lever M., Powell J. C., Killip M., Small C. W. (1973) J. Lab. Clin. Med. 82, 649–655 [PubMed] [Google Scholar]
- 57.Yang J., Fu X., Liao J., Liu L., Thorson J. S. (2005) Chem. Biol. 12, 657–664 [DOI] [PubMed] [Google Scholar]
- 58.Barton W. A., Biggins J. B., Jiang J., Thorson J. S., Nikolov D. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13397–13402 [DOI] [PMC free article] [PubMed] [Google Scholar]