Clinical Effect of Point Mutations in Myelodysplastic Syndromes (original) (raw)
. Author manuscript; available in PMC: 2011 Dec 30.
Published in final edited form as: N Engl J Med. 2011 Jun 30;364(26):2496–2506. doi: 10.1056/NEJMoa1013343
Abstract
BACKGROUND
Myelodysplastic syndromes are clinically heterogeneous disorders characterized by clonal hematopoiesis, impaired differentiation, peripheral-blood cytopenias, and a risk of progression to acute myeloid leukemia. Somatic mutations may influence the clinical phenotype but are not included in current prognostic scoring systems.
METHODS
We used a combination of genomic approaches, including next-generation sequencing and mass spectrometry–based genotyping, to identify mutations in samples of bone marrow aspirate from 439 patients with myelodysplastic syndromes. We then examined whether the mutation status for each gene was associated with clinical variables, including specific cytopenias, the proportion of blasts, and overall survival.
RESULTS
We identified somatic mutations in 18 genes, including two, ETV6 and GNAS, that have not been reported to be mutated in patients with myelodysplastic syndromes. A total of 51% of all patients had at least one point mutation, including 52% of the patients with normal cytogenetics. Mutations in RUNX1, TP53, and NRAS were most strongly associated with severe thrombocytopenia (P<0.001 for all comparisons) and an increased proportion of bone marrow blasts (P<0.006 for all comparisons). In a multivariable Cox regression model, the presence of mutations in five genes retained independent prognostic significance: TP53 (hazard ratio for death from any cause, 2.48; 95% confidence interval [CI], 1.60 to 3.84), EZH2 (hazard ratio, 2.13; 95% CI, 1.36 to 3.33), ETV6 (hazard ratio, 2.04; 95% CI, 1.08 to 3.86), RUNX1 (hazard ratio, 1.47; 95% CI, 1.01 to 2.15), and ASXL1 (hazard ratio, 1.38; 95% CI, 1.00 to 1.89).
CONCLUSIONS
Somatic point mutations are common in myelodysplastic syndromes and are associated with specific clinical features. Mutations in TP53, EZH2, ETV6, RUNX1, and ASXL1 are predictors of poor overall survival in patients with myelodysplastic syndromes, independently of established risk factors. (Funded by the National Institutes of Health and others.)
Myelodysplastic syndromes are clinically heterogeneous disorders for which treatments are tailored to the predicted prognosis for each patient. This makes the accurate prediction of the prognosis an essential component of the care of patients. Current prognostic scoring systems consider karyotypic abnormalities and certain clinical features to stratify patients with myelodysplastic syndromes into risk groups. Some karyotypic abnormalities, such as deletion of chromosome 5q, help establish the prognosis and may be associated with a specific clinical phenotype.1 However, more than half of patients with myelodysplastic syndromes have a normal karyotype, and patients with identical chromosomal abnormalities are often clinically heterogeneous.2,3 Single gene mutations are not currently used in prognostic scoring systems but are likely to be key drivers of clinical phenotypes and overall survival.4–6 An understanding of the clinical effects of mutations in various genes could improve the prediction of prognosis for patients with myelodysplastic syndromes and inform the selection of specific therapies.
The prognostic significance of some mutations in myelodysplastic syndromes has been described, but prior studies have generally examined small sample sets, have involved limited analyses of one or a small number of genes, or have focused exclusively on a particular subtype of myelodysplastic syndrome.7 Mutations in TP53, NRAS, RUNX1, TET2, and IDH1 and IDH2 have each been reported to influence overall survival in univariate analyses.8–15 Only mutations in the TP53 gene have been clearly associated with poor prognostic markers, such as a complex karyotype (having three or more chromosomal abnormalities), and have been reported to independently predict survival among patients at intermediate risk, as ascertained with the use of the International Prognostic Scoring System (IPSS). The IPSS assigns patients to one of four groups of increasing prognostic risk (low, intermediate 1, intermediate 2, and high) on the basis of the percentage of blasts in bone marrow, the karyotype, and the number of cytopenias present at the time of diagnosis (see Table 2 in the Supplementary Appendix, available with the full text of this article at NEJM.org).16,17 The interaction between the TP53 mutations and the genes more recently reported to be mutated in patients with myelodysplastic syndromes have not been studied in a large enough number of patients to establish their independent prognostic significance.18
To distinguish the independent contributions of mutations to the clinical phenotype and overall survival, we examined a large set of samples obtained from patients with myelodysplastic syndromes for the presence of somatic mutations in a broad spectrum of cancer-associated genes.
METHODS
SAMPLES FROM STUDY PATIENTS
Bone-marrow–aspirate mononuclear cells and buccal-swab samples from patients with myelodysplastic syndromes were obtained from Rush University Medical Center, the University of Massachusetts Medical Center, and the M.D. Anderson Cancer Center. Samples were acquired from adult patients, who provided written informed consent, between 1994 and 2008 according to protocols approved by the institutional review board at each institution. Genomic DNA was isolated and used to produce whole genome amplified DNA (by means of a kit [Qiagen]). Amplified DNA was used for mutation discovery.
Demographic and clinical characteristics of the study patients are detailed in Table 1 in the Supplementary Appendix. The IPSS risk (low, intermediate 1, intermediate 2, or high) (Table 2 in the Supplementary Appendix) had been ascertained at the time of diagnosis. For those cases in which a sample was collected after diagnosis, the IPSS risk was recalculated on the basis of clinical variables at the time of sample collection. Only 17% of patients were assigned to an IPSS risk group that differed from the group assigned at the time of diagnosis. Survival analysis confirmed the prognostic validity of the IPSS risk recalculation in our sample set (Table 3 in the Supplementary Appendix). The median follow-up was 4.44 years (95% confidence interval [CI], 4.12 to 6.19), during which time 332 patients died and the data for 107 were censored at the last date they were known to be alive.
MASS-SPECTROMETRIC GENOTYPING
Genotyping of 953 mutations representing 111 genes was performed on amplified DNA with the use of iPlex extension-chemistry methods (Sequenom) and mass spectrometry, as previously described for the complete set of OncoMap assays.19,20 This technique was chosen for its high-throughput detection of recurrent oncogene mutations limited to well-characterized locations. All candidate mutations identified on mass-spectrometric genotyping were validated with the use of redesigned assays in nonamplified or independently amplified DNA specimens from the study patients with the use of homogeneous Mass-Extend (hME) chemistry, as described previously.21 This technique can reliably detect mutations present at a frequency of 10% or higher.
DNA SEQUENCING
Next-generation pyrosequencing of PCR-amplified exons of TET2, RUNX1, TP53, CDKN2A, PTEN, NPM1 exon 11, and CBL exons 8 and 9 was performed with the use of a next-generation sequencing platform (454 Life Sciences). Known single-nucleotide polymorphisms (SNPs), intronic polymorphisms more than six bases from a splice junction, and silent mutations were excluded from further analysis. ASXL1, EZH2, KDM6A, IDH1 exon 4, IDH2 exon 4, and ETV6 were analyzed by means of Sanger sequencing. Candidate mutations detected in whole-genome–amplified DNA were validated with the use of nonamplified DNA (see the Methods section, Table 4, and Fig. 1 in the Supplementary Appendix).
GERMLINE MUTATION ANALYSIS
Matched buccal DNA was available for 219 (49.9%) of the 439 samples analyzed in this study. Mutations listed in the National Center for Biotechnology Information’s SNP database (dbSNP, build 130), previously reported as germ line, or present in the buccal sample from any patient in our cohort were considered to be germ line and were excluded from further analysis (Table 5 in the Supplementary Appendix). A claim of intellectual property rights has been made for the list of mutated genes with independent prognostic significance identified in this analysis.
STATISTICAL ANALYSIS
We compared the characteristics of the study patients with mutations, using the appropriate statistical methods, as detailed in the Methods section in the Supplementary Appendix. Overall survival was measured from the time of sample collection to the time of death from any cause; data were censored at the time patients were last known to be alive. All P values were calculated with the use of two-sided tests. Overall survival was evaluated for all patients by means of unadjusted and adjusted Cox proportional-hazards regression modeling; models were adjusted for the IPSS risk group assigned at the time of sample collection. Details of the modeling strategy are provided in the Methods section in the Supplementary Appendix.
RESULTS
IDENTIFICATION OF NEW SOMATIC MUTATIONS
To define a set of somatically mutated genes in bone marrow aspirates 5 from patients with myelodysplastic syndromes, we first examined 191 samples for abnormalities in known oncogenes (Fig. 2 in the Supplementary Appendix). Using mass-spectrometric genotyping, we examined these samples for 953 recurrent mutations in 111 cancer-associated genes.20 We identified and validated mutations in 10 genes: NRAS, KRAS, BRAF, JAK2, GNAS, FLNB, MET, EGFR, CDH1, and PTPN11. Genotyping of germ-line DNA from buccal swabs revealed that the mutations in these 10 genes were somatic, except for those in MET (E168D in three samples), EGFR (T790M in one sample), and CDH1 (A617T in three samples). No buccal sample was available for the sole patient with a mutation in FLNB (R566Q), so this mutation could not be assumed to be somatic. These studies thus confirmed somatic mutations in five genes (NRAS, KRAS, BRAF, JAK2, and PTPN11) known to be mutated in patients with myelodysplastic syndromes and identified recurrent mutations in GNAS that have not been previously reported in hematologic cancers.
We performed genomewide analysis of copy-number changes in a subgroup of 75 samples, using Affymetrix 6.0 SNP arrays. In addition to the known cytogenetic abnormalities, we identified a single case with a focal deletion encompassing ETV6, a gene that is recurrently involved in translocations in acute leukemia but is not known to be mutated in myelodysplastic syndromes (Fig. 3 in the Supplementary Appendix). Subsequent sequencing of this gene in other samples revealed several point mutations that were confirmed to be somatic on examination of matched buccal-swab DNA.
SURVEY OF MUTATIONS IN THE STUDY SAMPLES
To examine the clinical effects of mutations in patients with myelodysplastic syndromes, we evaluated all 439 samples for all the genes identified above, as well as a set of 13 genes previously reported to be mutated in hematologic cancers. The samples were from patients representative of the general population of patients with myelodysplastic syndromes (Table 1 in the Supplementary Appendix). A total of 70% of the patients were men (median age at the time of bone marrow aspirate collection, 70 years); 66% of the patients were in the IPSS low-risk or intermediate-1–risk groups, 58% had normal cytogenetic features, and 13% had a complex karyotype. These percentages are similar to those reported in published epidemiologic studies of patients with myelodysplastic syndromes.2,22–24
We identified mutations in 18 genes (Table 1). At least one mutation was present in 226 of the 439 samples (51.5%). (Missense mutations of KDM6A were found in three samples, but the mutations could not be confirmed as somatic and are not included in the totals listed above.)
Table 1.
Frequency of Mutation and Association with Median Survival.*
| Mutated Gene | No. of Samples (%) | Median Survival (95% CI) | P Value |
|---|---|---|---|
| γr | |||
| All samples | 439 (100) | 1.86 (1.60–2.14) | |
| TET2 | 90 (20.5) | 1.88 (1.26–2.55) | 0.48 |
| ASXL1 | 63 (14.4) | 1.33 (0.96–1.88) | 0.003 |
| RUNX1 | 38 (8.7) | 1.16 (0.77–1.53) | <0.001 |
| TP53 | 33 (7.5) | 0.65 (0.44–1.10) | <0.001 |
| EZH2 | 28 (6.4) | 0.79 (0.67–1.40) | <0.001 |
| NRAS | 16 (3.6) | 1.03 (0.44–1.98) | 0.006 |
| JAK2 | 13 (3.0) | 2.14 (1.02–3.12) | 0.96 |
| ETV6 | 12 (2.7) | 0.83 (0.62–2.29) | 0.04 |
| CBL | 10 (2.3) | 1.52 (0.14–1.71) | 0.02 |
| IDH2 | 9 (2.1) | 1.58 (0.50–2.14) | 0.03 |
| NPM1 | 8 (1.8) | 2.18 (0.59–2.74) | 0.43 |
| IDH1 | 6 (1.4) | 3.30 (0.35–9.52) | 0.52 |
| KRAS | 4 (0.9) | 0.89 (0.36–7.44) | 0.54 |
| GNAS | 3 (0.7) | ||
| PTPN11 | 3 (0.7) | ||
| BRAF | 2 (0.5) | ||
| PTEN | 1 (0.2) | ||
| CDKN2A | 1 (0.2) |
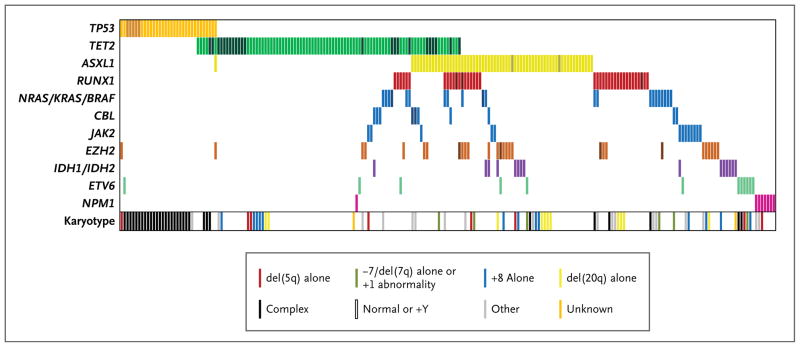
The frequency of coexisting somatic mutations can yield insights into the molecular circuitry of a cancer. Mutations of two or more genes were present in 79 samples (18.0%) (Fig. 1, and Table 6 in the Supplementary Appendix). As reported previously,25,26 mutations of genes involved in tyrosine-signaling pathways (JAK2, CBL, and NRAS_–_KRAS_–_BRAF) were largely mutually exclusive (Fig. 1). TET2 mutations, in contrast, overlapped with lesions in nearly every other mutated gene, suggesting that TET2 mutations have a pathogenic role that is at least partially independent of other abnormalities.
Figure 1. Mutations and Cytogenetic Abnormalities in 223 Samples with at Least One Mutation.
Mutations in the 11 most frequently mutated gene groups are shown by colored bars. Each column represents 1 of the 223 samples with a mutation in one or more of the genes listed. Darker bars indicate samples with two or more distinct mutations in that gene group. The karyotype of each of the 223 samples is also shown.
ASSOCIATIONS OF MUTATIONS WITH OVERALL SURVIVAL
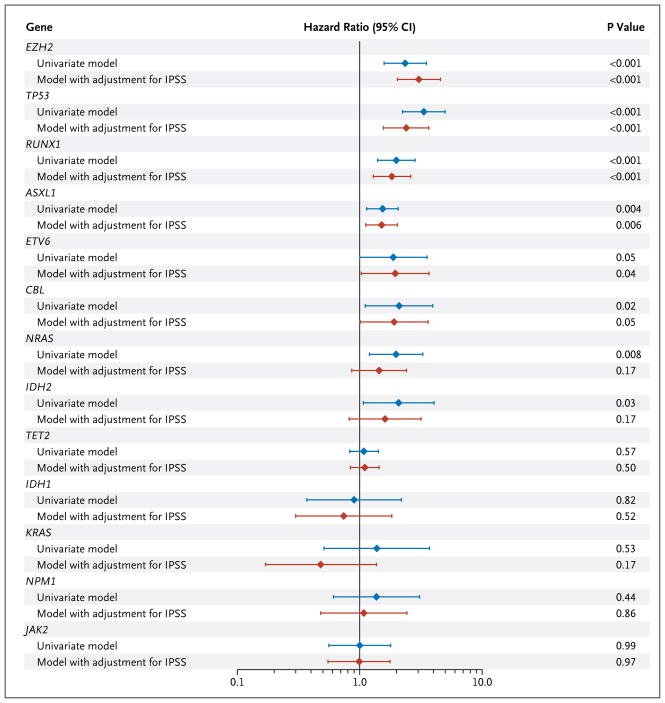
Abnormalities in seven genes were significantly associated with poor overall survival in univariate analyses (Fig. 2, and Fig. 4 in the Supplementary Appendix). Mutations in six genes — ASXL1, RUNX1, TP53, EZH2, CBL, and ETV6 — were significant predictors of poor overall survival, after adjustment for IPSS risk group, and were found in 74 of 255 samples (29.0%) with normal cytogenetic features.
Figure 2. Hazard Ratios for Death from Any Cause, According to Presence (vs. Absence) of Mutation in Each of Seven Genes.
Results are shown, on a log10 scale, for univariate analyses as well as for analyses with adjustment for the International Prognostic Scoring System (IPSS) risk category (based on the percentage of blasts in bone marrow, the karyotype, and the number of cytopenias) (for details, see Table 2 in the Supplementary Appendix). CI denotes confidence interval.
ASSOCIATIONS OF MUTATIONS WITH CYTOGENETIC FEATURES AND CYTOPENIAS
The prognostic significance of point mutations in patients with myelodysplastic syndromes may be driven by the association of these mutations with risk factors, including karyotype, blast proportion, and cytopenias, which are captured by existing clinical risk scoring systems such as the IPSS. For each mutation, we therefore compared the clinical characteristics of patients who had the mutation with those of patients who did not have the mutation.
The mutated genes most strongly associated with a specific karyotype group were TET2 and TP53. Mutations of TET2 were overrepresented in samples with normal cytogenetic features (P = 0.005) (Table 7 in the Supplementary Appendix), whereas TP53 mutations were strongly associated with a complex karyotype (P<0.001). Eight of the 33 TP53 mutant samples (24.2%) had abnormalities of chromosome 17 (P<0.001), suggesting that mutation and chromosomal loss are frequently found together, abrogating the activity of wild-type TP53. In contrast, mutations of EZH2, which lies on the distal portion of chromosome 7q, were not associated with 7q deletions.
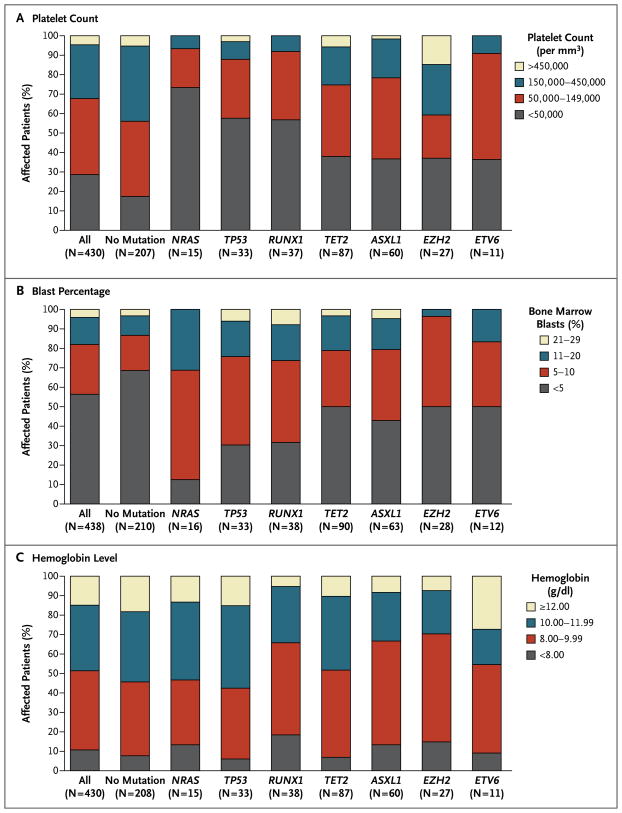
Myelodysplastic syndromes are characterized by ineffective hematopoiesis and impaired differentiation, leading to peripheral-blood cytopenias, but the contribution of specific genotypes to particular cytopenias is unknown. We observed that mutations of RUNX1, TP53, and NRAS were each strongly associated with severe thrombocytopenia (P<0.001 for each gene) (Fig. 3A). Patients with mutations of these genes, as compared with patients who did not have such mutations, were also more likely to have an elevated blast percentage (Fig. 3B), but the two groups did not differ significantly with respect to neutropenia or anemia (Fig. 3C). These findings show that the association of some mutations with poor survival may be indirectly captured by the IPSS because of their associations with cytopenias, blast percentage, and karyotype.
Figure 3. Proportions of Patients with Mutations, According to Platelet Count, Blast Percentage, and Hemoglobin Level.
Data are shown for the platelet count (Panel A), percentage of blasts in bone marrow aspirate (Panel B), and hemoglobin level (Panel C) at the time of bone marrow sample collection. The numbers in parentheses along the x axis indicate the number of patients with a mutation in the gene (patients could have >1 mutated gene). Mutations in NRAS, TP53, and RUNX1 were significantly associated with severe thrombocytopenia (defined as <50,000 platelets per cubic millimeter) (P<0.001 for each comparison) (Panel A) and elevated blast percentage (defined as ≥5%) (P<0.001, P = 0.005, and P = 0.003 for mutations in the three genes, respectively) (Panel B).
ASSOCIATIONS FROM THE MULTIVARIABLE SURVIVAL MODEL
Mutations in multiple genes were associated with overall survival in univariate analyses (Table 1). However, these mutations often occur together in the same patient, and several are associated with established prognostic markers. To determine the relative contribution of mutation status to overall survival, we generated a multivariable Cox model, using a stepwise variable-selection procedure incorporating age, sex, IPSS classification, and mutation status for the 13 most frequently mutated genes identified in this study.
As expected, age and IPSS risk group were strongly associated with overall survival (Table 2). But mutations in TP53, EZH2, ETV6, RUNX1, and ASXL1 also emerged as independent predictors of survival. Mutation of NRAS, previously reported as a marker of poor prognosis, did not influence survival in this model, most likely owing to strong associations between oncogenic NRAS mutations and components of the IPSS. EZH2 mutations, which have not previously been associated with known prognostic markers, retained a strong association with survival in our model. This analysis indicates that evaluation of the mutation status of TP53, EZH2, ETV6, RUNX1, and ASXL1 would add the most information to clinical prognostic scores as currently assessed in patients with myelodysplastic syndromes.
Table 2.
Hazard Ratios for Death in a Multivariable Model.*
| Risk Factor | Hazard Ratio (95% CI) | P Value |
|---|---|---|
| Age ≥55 yr vs. <55 yr | 1.81 (1.20–2.73) | 0.004 |
| IPSS risk group | ||
| Intermediate-1 vs. low | 2.29 (1.69–3.11) | <0.001 |
| Intermediate-2 vs. low | 3.45 (2.42–4.91) | <0.001 |
| High vs. low | 5.85 (3.63–9.40) | <0.001 |
| Mutational status | ||
| TP53 mutation present vs. absent | 2.48 (1.60–3.84) | <0.001 |
| EZH2 mutation present vs. absent | 2.13 (1.36–3.33) | <0.001 |
| ETV6 mutation present vs. absent | 2.04 (1.08–3.86) | 0.03 |
| RUNX1 mutation present vs. absent | 1.47 (1.01–2.15) | 0.047 |
| ASXL1 mutation present vs. absent | 1.38 (1.00–1.89) | 0.049 |
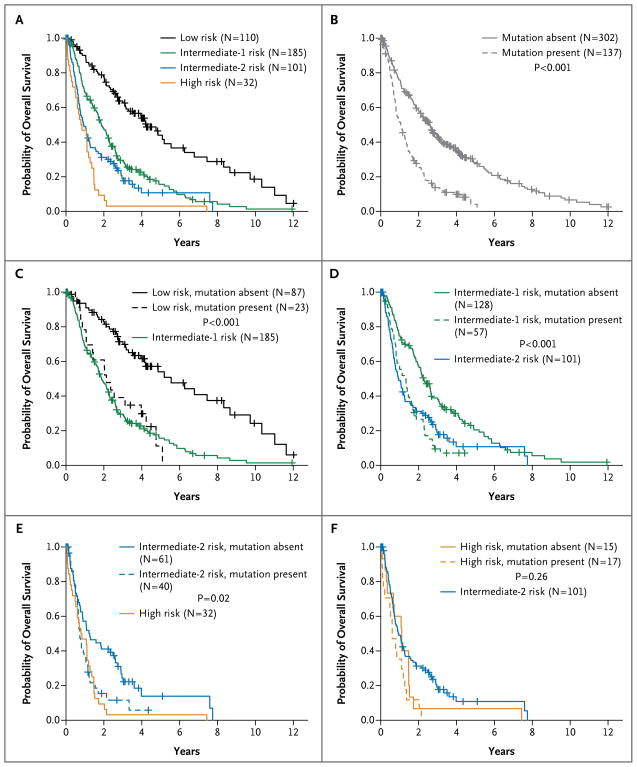
One approach to the integration of mutational-analysis results into the IPSS would be to include a variable for mutations in one or more of these five prognostic genes (Fig. 4). In all patients except those at highest risk, the presence of these mutations is associated with an overall survival similar to that of patients in the next-highest IPSS risk group.
Figure 4. Overall Survival, According to International Prognostic Scoring System (IPSS) Risk Category and Mutational Status.
Panel A shows the overall survival of patients within each IPSS risk group. Panel B shows the overall survival of patients with mutations in one or more of the five prognostic genes (TP53, EZH2, ETV6, RUNX1, or ASXL1) as compared with patients without such mutations. Panels C through F show the overall survival of patients according to the presence and absence of prognostic mutations and according to IPSS risk group. In Panels C, D, and E, the overall-survival curve for patients in the next-highest IPSS risk group is included for the purpose of comparison. In Panel F, the comparison curve is for patients in the next-lowest IPSS risk group. P values were calculated for the log-rank comparison of overall survival between patients with mutations and those without mutations for the given IPSS risk group. The IPSS risk classification, which is based on the percentage of blasts in bone marrow, the karyotype, and the number of cytopenias (Table 2 in the Supplementary Appendix), was recalculated for 428 of the 439 samples at the time of bone marrow sample collection (the IPSS classification could not be recalculated for 11 samples).
DISCUSSION
In a broad survey of mutations in 439 primary DNA samples from patients with myelodysplastic syndromes, we identified point mutations in 18 genes, including 2 (ETV6 and GNAS) that have not previously been reported to harbor mutations in such patients. We found that several of these genetic lesions correlate strongly with features of the clinical phenotype, including specific cytopenias, blast percentage, cytogenetic abnormalities, and overall survival. In a multivariable analysis that included clinical features and other mutations, mutations in five genes — TP53, EZH2, ETV6, RUNX1, and ASXL1 — were independently associated with decreased overall survival. Mutations in one or more of these genes were present in 137 of the 439 patients (31.2%). These findings indicate that mutations in specific genes help explain the clinical heterogeneity of myelodysplastic syndromes and that the identification of these abnormalities would improve the prediction of prognosis in patients with myelodysplastic syndromes.
By analyzing copy-number alterations with the use of SNP arrays and oncogene mutations, by means of high-throughput genotyping, we identified new mutations in ETV6 and GNAS. Both translocations and mutations of ETV6 have been identified in acute myeloid leukemia, and translocations have been described in rare cases of myelodysplastic syndromes, but ETV6 mutations have not previously been reported in myelodysplastic syndromes.27,28 We also identified three samples with activating mutations of amino acid R201 in GNAS, the gene encoding the GSα subunit of the heterotrimeric GS-protein complex. Identical somatic activating mutations of GNAS have been identified in several types of solid tumors but not in hematologic cancers.29–32 In general, our data support the idea that activating mutations of oncogenes are relatively infrequent in myelodysplastic syndromes. Our survey of more than 900 mutations in 111 cancer-associated genes identified only 6 mutated oncogenes, which were present in less than 10% of samples.
Prognostically significant somatic mutations occurred in patients in all risk groups. Most patients with EZH2 or ASXL1 mutations had low or intermediate-1 risk according to the IPSS (86% and 73%, respectively). The presence (vs. the absence) of EZH2 mutations was strongly associated with decreased overall survival in the stepwise, multivariable model that considered age, sex, IPSS risk group, and the presence of other mutations (hazard ratio for death, 2.13 [95% CI, 1.36 to 3.33]). The presence (vs. the absence) of mutations of ASXL1 carried a more modest hazard ratio for death (1.38 [95% CI, 1.00 to 1.89]), but ASXL1, as the second most commonly mutated gene identified in this study, contributed additional risk to the greatest number of patients. Therefore, lower-risk patients with myelodysplastic syndromes who have EZH2 and ASXL1 mutations may require more aggressive treatment than would be predicted by the IPSS.
In contrast, TP53 mutations were observed mainly in patients with intermediate-2 or high risk according to the IPSS (79%) and were strongly associated with thrombocytopenia, an elevated blast proportion, and a complex karyotype. Even though these measures are integrated into the IPSS, TP53 mutations remained strongly associated with shorter overall survival after adjustment for IPSS risk group (P<0.001), indicating that these mutations adversely affect survival through other means (Fig. 5 in the Supplementary Appendix). Furthermore, patients with mutant TP53 and a complex karyotype had a paucity of mutations in other genes, suggesting that this group could be considered to have a distinct molecular subclass of myelodysplastic syndromes with a unique pathogenic mechanism.
TET2 mutations were the most prevalent genetic abnormality identified in our sample set. These mutations were not strongly associated with clinical features such as cytopenias or blast proportion, findings that are consistent with the observation that TET2 mutations occur in diverse myeloid cancers, including myeloproliferative neoplasms, that are not characterized by defects in hematopoietic differentiation. More than one quarter (26%) of the samples with at least one TET2 mutation had two distinct TET2 mutations, suggesting that biallelic loss of wild-type TET2 contributes to the pathogenesis of myelodysplastic syndromes in some cases. In contrast to previously reported findings in smaller sample sets, neither monoallelic nor biallelic mutations were associated with IPSS risk group or overall survival (Fig. 6 in the Supplementary Appendix).11,33 Furthermore, analysis of the mutant-allele burden in samples with mutations of TET2 and other genes showed that TET2 mutations are not always present at the greatest frequency, which would be expected if they were exclusively involved in early pathogenic events (Fig. 7 in the Supplementary Appendix). TET2 mutations were not exclusive of abnormalities in other epigenetic regulators such as the chromatin-modifying genes ASXL1 and EZH2.34–36 Mutations in ASXL1 and EZH2 had associations with clinical phenotypes, including overall survival, that differed from those of TET2 mutations, suggesting that these genes drive distinct and additive aspects of cellular transformation to myelodysplastic syndromes.
Each of the prognostically significant mutations most likely alters the biologic characteristics and phenotype of myelodysplastic syndromes in unique ways, as is the case for cytogenetic abnormalities, with complex interactions among combinations of genetic and epigenetic lesions. Nevertheless, a simplified prognostic scoring scheme has great clinical value. One approach to improving the IPSS would be to include one additional variable: the presence or absence of a mutation in any of the five genes with independent prognostic significance. The presence of such a mutation would reclassify patients into the next highest IPSS risk group.
As our study shows, somatic mutations in several genes are associated with distinct effects on cytopenias, blast proportion, the likelihood of co-occurrence with other molecular lesions, and overall survival. It will soon be possible for clinicians to detect a broad range of point mutations in peripheral blood with the use of sensitive genotyping methods, which will not only improve prognostication in myelodysplastic syndromes but also facilitate the diagnosis of these disorders, the evaluation of disease progression, and the monitoring of response to treatment. The integration of mutation assessment in diagnostic classification and prognostic scoring systems has the potential to parse diverse myelodysplastic syndromes into a set of discrete diseases with predictable clinical phenotypes, prognosis, and responses to therapy.
Supplementary Material
Supplement1
Acknowledgments
Supported by grants from the National Institutes of Health (NIH) (5R01 HL082945 and P01 CA108631), the Starr Cancer Consortium, and the Burroughs–Wellcome Fund (Career Awards for Medical Scientists) (all to Dr. Ebert) and a grant from the NIH (T32 HL007623) and a Huber–Foster Fellowship (both to Dr. Bejar).
We thank Laura MacConaill, Sarah Kehoe, Charles Hatton, Adriana Heguy, Randall MacAuley, Bernd Boidol, Bennett Caughey, and Marie McConkey for assistance with genetic analyses and Todd Golub, David Steensma, and Gary Gilliland for helpful advice.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–9. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core data-set of 2124 patients. Blood. 2007;110:4385–95. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 3.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–15. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–88. [Erratum, Blood 1998;91:1100.] [PubMed] [Google Scholar]
- 5.Garcia-Manero G, Shan J, Faderl S, et al. A prognostic score for patients with lower risk myelodysplastic syndrome. Leukemia. 2008;22:538–43. doi: 10.1038/sj.leu.2405070. [DOI] [PubMed] [Google Scholar]
- 6.Malcovati L, Germing U, Kuendgen A, et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J Clin Oncol. 2007;25:3503–10. doi: 10.1200/JCO.2006.08.5696. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen-Bjergaard J, Andersen MK, Andersen MT, Christiansen DH. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2008;22:240–8. doi: 10.1038/sj.leu.2405078. [DOI] [PubMed] [Google Scholar]
- 8.Horiike S, Kita-Sasai Y, Nakao M, Taniwaki M. Configuration of the TP53 gene as an independent prognostic parameter of myelodysplastic syndrome. Leuk Lymphoma. 2003;44:915–22. doi: 10.1080/1042819031000067620. [DOI] [PubMed] [Google Scholar]
- 9.Padua RA, Guinn BA, Al-Sabah AI, et al. RAS, FMS and p53 mutations and poor clinical outcome in myelodysplasias: a 10-year follow-up. Leukemia. 1998;12:887–92. doi: 10.1038/sj.leu.2401044. [DOI] [PubMed] [Google Scholar]
- 10.Paquette RL, Landaw EM, Pierre RV, et al. N-ras mutations are associated with poor prognosis and increased risk of leukemia in myelodysplastic syndrome. Blood. 1993;82:590–9. [PubMed] [Google Scholar]
- 11.Kosmider O, Gelsi-Boyer V, Cheok M, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–91. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 12.Chen CY, Lin LI, Tang JL, et al. RUNX1 gene mutation in primary myelodysplastic syndrome—the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br J Haematol. 2007;139:405–14. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 13.Steensma DP, Gibbons RJ, Mesa RA, Tefferi A, Higgs DR. Somatic point mutations in RUNX1/CBFA2/AML1 are common in high-risk myelodysplastic syndrome, but not in myelofibrosis with myeloid metaplasia. Eur J Haematol. 2005;74:47–53. doi: 10.1111/j.1600-0609.2004.00363.x. [DOI] [PubMed] [Google Scholar]
- 14.Pardanani A, Patnaik MM, Lasho TL, et al. Recurrent IDH mutations in high-risk myelodysplastic syndrome or acute myeloid leukemia with isolated del(5q) Leukemia. 2010;24:1370–2. doi: 10.1038/leu.2010.98. [DOI] [PubMed] [Google Scholar]
- 15.Kosmider O, Gelsi-Boyer V, Slama L, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodys-plastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–6. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko H, Misawa S, Horiike S, Nakai H, Kashima K. TP53 mutations emerge at early phase of myelodysplastic syndrome and are associated with complex chromosomal abnormalities. Blood. 1995;85:2189–93. [PubMed] [Google Scholar]
- 17.Kita-Sasai Y, Horiike S, Misawa S, et al. International prognostic scoring system and TP53 mutations are independent prognostic indicators for patients with myelodysplastic syndrome. Br J Haematol. 2001;115:309–12. doi: 10.1046/j.1365-2141.2001.03073.x. [DOI] [PubMed] [Google Scholar]
- 18.Rocquain J, Carbuccia N, Trouplin V, et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. [Errata, PLoS ONE 2010;5(9).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badalian-Very G, Vergilio J-A, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116:1919–23. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 22.Sekeres MA. The epidemiology of myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24:287–94. doi: 10.1016/j.hoc.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Sekeres MA, Schoonen WM, Kantarjian H, et al. Characteristics of US patients with myelodysplastic syndromes: results of six cross-sectional physician surveys. J Natl Cancer Inst. 2008;100:1542–51. doi: 10.1093/jnci/djn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rollison DE, Howlader N, Smith MT, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001–2004, using data from the NAACCR and SEER programs. Blood. 2008;112:45–52. doi: 10.1182/blood-2008-01-134858. [DOI] [PubMed] [Google Scholar]
- 25.Grand FH, Hidalgo-Curtis CE, Ernst T, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–92. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 26.Sanada M, Suzuki T, Shih L-Y, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–8. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 27.Odero MD, Vizmanos JL, Román JP, et al. A novel gene, MDS2, is fused to ETV6/ TEL in a t(1;12)(p36. 1; p13) in a patient with myelodysplastic syndrome. Genes Chromosomes Cancer. 2002;35:11–9. doi: 10.1002/gcc.10090. [DOI] [PubMed] [Google Scholar]
- 28.Silva FPG, Morolli B, Storlazzi CT, et al. ETV6 mutations and loss in AML-M0. Leukemia. 2008;22:1639–43. doi: 10.1038/leu.2008.34. [DOI] [PubMed] [Google Scholar]
- 29.Suarez HG, du Villard JA, Caillou B, Schlumberger M, Parmentier C, Monier R. gsp Mutations in human thyroid tumours. Oncogene. 1991;6:677–9. [PubMed] [Google Scholar]
- 30.Kalfa N, Lumbroso S, Boulle N, et al. Activating mutations of Gsα in kidney cancer. J Urol. 2006;176:891–5. doi: 10.1016/j.juro.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 32.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 33.Smith AE, Mohamedali AM, Kulasekararaj A, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–32. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 34.Fisher CL, Pineault N, Brookes C, et al. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115:38–46. doi: 10.1182/blood-2009-07-230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyl-transferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement1