The SKP1-Cul1-F-box and Leucine-rich Repeat Protein 4 (SCF-FbxL4) Ubiquitin Ligase Regulates Lysine Demethylase 4A (KDM4A)/Jumonji Domain-containing 2A (JMJD2A) Protein (original) (raw)
Abstract
Chromatin-modifying enzymes play a fundamental role in regulating chromatin structure so that DNA replication is spatially and temporally coordinated. For example, the lysine demethylase 4A/Jumonji domain-containing 2A (KDM4A/JMJD2A) is tightly regulated during the cell cycle. Overexpression of JMJD2A leads to altered replication timing and faster S phase progression. In this study, we demonstrate that degradation of JMJD2A is regulated by the proteasome. JMJD2A turnover is coordinated through the SKP1-Cul1-F-box ubiquitin ligase complex that contains cullin 1 and the F-box and leucine-rich repeat protein 4 (FbxL4). This complex interacted with JMJD2A. Ubiquitin overexpression restored turnover and blocked the JMJD2A-dependent faster S phase progression in a cullin 1-dependent manner. Furthermore, increased ubiquitin levels decreased JMJD2A occupancy and BrdU incorporation at target sites. This study highlights a finely tuned mechanism for regulating histone demethylase levels and emphasizes the need to tightly regulate chromatin modifiers so that the cell cycle occurs properly.
Keywords: Chromatin, DNA Replication, E3 Ubiquitin Ligase, Ubiquitin, Ubiquitination, Histone Demethylase, Histone Tri-demethylase, JMJD2A, Jmjc, KDM4A
Introduction
In eukaryotes, the genome is packaged into chromatin, a highly ordered structure that contains DNA, RNA, histones, and other chromosomal proteins. Histones are targeted by several posttranslational modifications that are regulated by a number of modifying enzymes. Maintaining the proper balance of these posttranslational modifications by chromatin-modifying enzymes is critical for regulating gene expression, cell fate, cell cycle, chromosomal structure, and genomic stability (1–3). For example, balancing the histone methylation state (e.g. the degree of methylation: mono, di, and tri) within the genome is important for regulating expression of differentiation programs and coordinating DNA replication during the G1 and S phases of the cell cycle (4–6). Furthermore, aberrant expression of histone methyltransferases and demethylases results in cancer, developmental defects, and mental retardation (7–9). Understanding how these enzymes are regulated is required for comprehending their physiologic and pathological functions.
Ubiquitination is a key regulatory pathway for protein stability. Ubiquitin is transferred to lysine residues by a three-enzyme cascade involving E1 activating, E2 conjugating, and E3 ligase enzymes. The E3 ligases, such as the cullin family, provide substrate specificity by coordinating interactions between the E2 and specific substrates (10, 11). Recent studies have demonstrated the importance of the ubiquitin system in regulating chromatin modifiers. For example, the mammalian homolog of trithorax in Drosophila, myeloid/lymphoid or mixed lineage leukemia (MLL), is responsible for H3K4 methylation, and its tight regulation is necessary for proper cell cycle progression (12). Mixed lineage leukemia is regulated via bimodal ubiquitin-dependent degradation, which results in biphasic expression during the cell cycle. This regulation involves two complexes, SCFSkp2 during S phase, and the anaphase-promoting complex APCCdc20 in late M phase (12). Another methyltransferase regulated via the ubiquitin system during cell cycle is the Su(var)3-9, Enhancer of zeste, Trithorax (SET) domain containing lysine methyltransferase 8, Set8/PR-Set7/KMT5A, which catalyzes the monomethylation of H4K20. Set8/PR-Set7 is targeted by three different ubiquitination complexes, SCFSkp2 in G1 (13), APCCdh1 for proper mitotic progression (14), and the cullin 4A RING E3 ubiquitin ligase CRL4Cdt2 during S phase and after DNA damage. CRL4Cdt2 specifically targets Set8 on the chromatin through interaction with proliferating cell nuclear antigen (15–19). These examples illustrate the importance of regulating methyltransferases during the cell cycle and imply that specific regulation of histone demethylases is necessary as well. To date, only the yeast H3K4 demethylase Jhd2 has been shown to be ubiquitinated, which impacted its transcriptional activity (20).
The JMJD2/KDM43 histone demethylase family removes trimethylated H3K9, H3K36, and H1.4K26 (21–24). We previously demonstrated that JMJD2A regulates cell cycle progression by increasing chromatin accessibility and antagonizing the heterochromatin protein HP1γ occupancy (25). Interestingly, we observed that JMJD2A levels changed during the cell cycle, peaking in G1/S and becoming the lowest in G2/M, whereas the RNA levels were more stable (25). Here we demonstrate that the decrease in JMJD2A protein levels during the cell cycle was regulated by the proteasome. Cullin 1 interacted with JMJD2A and contributed to ubiquitination and turnover. We then demonstrated that the F-box protein FbxL4 interacted with JMJD2A and regulated protein turnover. Interestingly, ubiquitin overexpression increased JMJD2A protein turnover in cells stably overexpressing JMJD2A and blocked the ability of JMJD2A to promote S phase progression by modulating turnover, occupancy, and replication at target sites. Cullin 1 was critical for the ubiquitin-dependent suppression of the JMJD2A-dependent faster S phase progression. These data highlight the role of SCF-FbxL4 in regulating JMJD2A protein levels and highlight the need to modulate JMJD2A protein levels during the cell cycle.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Treatments
For tissue culture and generation of stables cell lines, see Black et al. (25). Cells were synchronized by treatment with 1 mm hydroxyurea (HU) (Sigma) for 24 h or 50 ng/ml nocodazole (Sigma) for 11 h. G2/M-arrested cells were collected by shakeoff from nocodazole arrest. Because HEK293T cells attach loosely, there are some G2 cells; therefore, we refer to this as G2/M phase (25). To release arrested cells, they were washed once with media and supplied with fresh media. Cycloheximide (Sigma) was used at 400 μm final concentration for HEK293T cells and 89 μm for HeLa cells. MG132 (Sigma) was used at 20 μm final concentration.
Plasmids, siRNAs, and Transfections
Plasmid transfections were done using the FuGENE 6 (Roche) or X-tremeGENE 9 DNA reagents (Roche) on 5 × 105 cells plated 12 h prior to transfection in 10-cm dishes or 0.8 × 105 cells in 6-well plates. The complexes were incubated with the cells in OptiMEM for 4 h or 8 h before fresh media was added. The transfected plasmids are as follows: pcDNA3-FLAG-DNCul1 and pcDNA3-FLAG-DNCul4a (26), pcDNA3–3×Myc-Cullin1, pEGFN-FLAG-FbxL4, pMSCV-GFP, and pMSCV-GFP-JMJD2A (25), pMSCV-FLAG-HA, pMSCV-FLAG-HA-JMJD2A, and pRK5-HA-Ub (Addgene).
siRNA transfections were done using the X-tremeGENE 9 siRNA (Roche) reagent on 0.8 × 105 HEK293T cells plated 12 h prior to transfection in 6-well dishes. The complexes were incubated in media for 48 h at a final concentration of 20 nm. HeLa cells were transfected at 30–40% confluence using the RNAi-Max (Invitrogen) reagent. The complexes were incubated in media for 72 h at a final concentration of 20 nm. The siRNA targeting the F-box genes were purchased from Dharmacon, whereas all others were purchased from Invitrogen. The siRNA control is an oligo duplex targeting the luciferase gene (si-GL2 (27)). The siRNA oligonucleotide sequences used are as follows: Cul1 (siCul1), 5′-GUUCAUAGCAGCCAGCCUGdTdT-3′ (siCul4 is a combination of oligos that target both Cul4a and Cul4b); Cul4a, 5′-GACAAUCCGAAUCAGUACCdTdT-3′; Cul4b, 5′-AGAUAAGGUUGACCAUAUAdTdT-3′; FbxW1 (siFbxW1), 5′-ACAGGAUCAUCGGAUUCCAdTdT-3′; FbxW2 (siFbxW2), 5′-CUCCUGAGAUAGCAAACUUdTdT-3′; FbxW11 (siFbxW11), 5′-GAUGUCUCCAGAUAAGUAAdTdT-3′; FbxW12 (siFbxW12), 5′-UUGCCUGACUUAGCUUUGAdTdT-3′; FbxL4 (siFbxL4), 5′-UGAUAGGAGCCAAGUGUAAdTdT-3′; FbxL13 (siFbxL13), 5-CUCCGGAAUUGAUGAUAAAdTdT-3′; FbxL17 (siFbxL17), 5′-UCACUGAACUGGAUAAUGAdTdT-3′; and FbxL19 (siFbxL19), 5′-CAAUACGGUUUGCUAUAAAdTdT-3′.
Western Blot Analyses
Western blot analyses were performed according to Whetstine et al. (24). Some of the Western blot analyses in this manuscript were spliced together from the same exposure and experiment so that controls (e.g. inputs) and experimental conditions were in the same figure.
Antibodies
The antibodies used were as follows: JMJD2A N154/32 mouse monoclonal (NeuroMab (25)), JMJD2A rabbit polyclonal (25), β-actin (Millipore, MAB1501), ubiquitin (Santa Cruz Biotechnology, Inc., sc-8017), cullin 1 (Santa Cruz Biotechnology, Inc., sc-17775), FbxL4 (Santa Cruz Biotechnology, Inc., sc-54489), Myc immunoprecipitation (Santa Cruz Biotechnology, Inc., sc-40), Myc western (Cell Signaling Technology, Inc., 2276), p-H3-Ser10 (Invitrogen, 441190G), tubulin (Sigma), FbxW2 (Abcam, Inc., ab83467), FLAG M2 (Sigma, F1807), HA 12CA5 (Roche, 11583816001), and BrdU (BD Biosciences, 347580).
Coimmunoprecipitation
Cells were lysed in IPH buffer (50 mm Tris (pH 8), 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40), sonicated and cleared by centrifugation. 1 mg was immunoprecipitated overnight in the presence of 25 μl of protein A or G magnetic Dynabeads (Invitrogen) and 100 μg/ml ethidium bromide. The beads were washed with IPH buffer and boiled for 10 min in 2× protein loading buffer. In denaturing conditions, beads were washed four times with IPH buffer supplemented with 4 m urea.
Flow Cytometry
Complete methods can be found in Black et al. (25).
ChIP
Complete methods can be found in Black et al. (25). A large batch of transfected cells was arrested and released from HU for 1 h before being split into three sets used for the triplicate of chromatin.
Analysis of single-stranded DNA during S Phase
Complete methods can be found in Black et al. (2010) (25).
EdU DNA Immunoprecipitation
Cells were incubated in presence of 10 μm of ethynyl deoxyuridine (EdU) for 1 h. The genomic DNA was purified as for the BrdU DNA immunoprecipitation (25). 100 μg of genomic DNA was biotinylated using click chemistry in 50 mm Tris (pH) 8.5, 10 μm biotin-azide, 1 mm CuSO4, and 1 mm ascorbic acid. The reaction was quenched with 2 mm EDTA and precipitated immediately. 2 μg of biotinylated DNA was affinity-purified with 30 μl of M280 streptavidin beads (Invitrogen). The DNA was eluted in 1% SDS and 10 μg of proteinase K for 1 h at 55 °C. The DNA was then column-purified (Promega) before analysis by quantitative PCR.
BrdU DNA Immunoprecipitation, JMJD2A ChIPs, and Chromosome 1 to 4 ChIP-chip
BrdU immunoprecipitations and JMJD2A ChIPs were performed as described in Black et al. (25). BrdU and JMJD2A immunoprecipitations were amplified with the WGA2 kit (Sigma). The amplified material was hybridized to chromosome 1–4 tiling arrays (Roche Nimblegen) according to the manufacturer's protocol. The microarrays were scanned on a Nimblegen MS200 at 2-μm resolution.
A total of 2408 JMJD2A peak locations in the JMJD2A-overexpressed cells were detected by using the MA2C software (p value cutoff = 1.0 E-3) (28). Raw BrdU ChIP intensity values in each channel were log-transformed (based 2) followed by quantile normalization (29). The normalization formula was determined on the basis of intensity values at the 12,900 control probes printed on the Nimblegen array and applied to adjust values at all other probes. After normalization, the replicates were averaged for each experiment. We averaged the normalized BrdU intensities around 2 MB regions centered at the JMJD2A peak locations. Because of the incomplete coverage of the Nimblegen array, we excluded those regions that were less than 50% covered by the microarray, corresponding to 380 peaks. The average BrdU intensities were smoothed using a moving average with a 50-kb window size in 1-kb increment step sizes. For comparison, we randomly selected the same number of regions from the genome and obtained an average profile using the same procedure. For each channel, the mean intensity level was subtracted to further remove any potential artifact. We used a one-sided paired t test to determine whether the difference was statistically significant.
Statistics
All error bars represent S.E. JMJD2A half-lives were calculated using a polynomial trend line. p values were determined with a two-tailed Student's t test.
RESULTS
Ubiquitination and Proteasomal Degradation Regulate JMJD2A Levels during the Cell Cycle
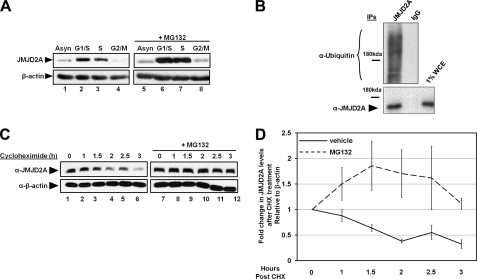
We demonstrated previously that JMJD2A protein levels are regulated during the cell cycle (25). This observation was confirmed in Fig. 1A (left panel, lanes 1–4). JMJD2A protein levels were highest in G1/S, decreased through S phase, and reached their lowest levels during G2/M (Fig. 1A, left panel) (25). Because JMJD2A RNA levels remained relatively stable (25), we reasoned that the decrease in JMJD2A protein levels was regulated by protein degradation. To test this hypothesis, we treated HEK293T cells synchronized in different cell cycle phases with the proteasomal inhibitor MG132. JMJD2A levels were stabilized in S and G2/M phases (Fig. 1A, right panel). However, we did not observe as substantial an increase in JMJD2A levels in G2/M-synchronized cells treated with MG132. This suggests that the majority of degradation occurred during S phase. The increased levels of JMJD2A upon proteasome inhibition suggested that JMJD2A could be regulated by an ubiquitin-dependent turnover mechanism. Consistent with this hypothesis, endogenous JMJD2A was ubiquitinated under denaturing conditions (Fig. 1B). Taken together, these data suggest that JMJD2A is ubiquitinated and degraded by the 26S proteasome.
FIGURE 1.
JMJD2A is ubiquitinated and degraded by the 26S proteasome. A, the proteasomal inhibitor MG132 prevents the decrease in JMJD2A levels during cell cycle. Asynchronous HEK293T cells (Asyn), cells arrested in G1/S after 1 mm HU for 24 h, cells released from HU for 2.5 h into S phase, or cells arrested in G2/M with nocodazole were treated or not treated with 20 μm MG132 for 2.5 h before lysis and immunoblotting. B, JMJD2A immunoprecipitated under denaturing conditions contains ubiquitin. Asynchronous HEK293T cells were treated with 20 μm MG132 for 2.5 h before lysis and then immunoprecipitated (IP) in the presence of 100 μg/ml of EtBr for JMJD2A, washed in denaturing buffer, and immunoblotted for ubiquitin. C, JMJD2A is degraded by the 26S proteasome. HEK293T cells were treated for the indicated times with 400 μm cycloheximide (CHX) in the presence of 20 μm MG132 or vehicle for 4 h and then lysed and immunoblotted. D, graphical representation of C. The y axis represents the ratio of JMJD2A relative to time 0, which was normalized to β-actin. The average of three independent experiments is shown. The errors bars represent the mean ± S.E. WCE, whole cell extract.
To confirm that the turnover of JMJD2A was mediated by proteasomal degradation, we inhibited protein synthesis with cycloheximide treatment and analyzed JMJD2A protein levels following treatment with vehicle or MG132. Endogenous JMJD2A had a half-life of 2 h. The turnover of JMJD2A was blocked by the addition of the proteasomal inhibitor MG132 (Fig. 1C and graphed in D), demonstrating that JMJD2A turnover is 26S proteasome-dependent.
The E3 Ligase Cullin 1 Regulates JMJD2A Turnover
To resolve the E3 ligase(s) that were responsible for ubiquitinating JMJD2A, cells were transfected with dominant negative cullin constructs and then synchronized with nocodazole and Western blotted for JMJD2A. Because JMJD2A levels decreased through S phase and are lowest in G2/M, we hypothesized that stabilizing JMJD2A would result in increased levels during G2/M. The dominant negative cullins that were used have a C-terminal truncation, which eliminates the ability to interact with Rbx-1 and the E2 ubiquitin-conjugating enzyme, blocking ubiquitination and turnover. However, the dominant negative cullins are still able to interact with their substrate. For example, dominant negative cullin 1 interacts with the substrate through the S-phase kinase-associated protein 1 (SKP1) and the F-box-containing protein but lacks the ability to promote ubiquitination and turnover (26).
Consistent with our hypothesis, JMJD2A levels increased with dominant negative cullin 1 overexpression (Fig. 2A, DN-1 or DN-Cul1). This stabilization of JMJD2A was specific as dominant negative cullin 4a (DN-4a), an E3 ligase that regulates proteins associated with proliferating cell nuclear antigen during S phase and failed to stabilize JMJD2A protein levels (Fig. 2A, compare lane 1 to lanes 2, 3, and 4) (30). DN-Cul1 was also able to regulate exogenously expressed JMJD2A (Fig. 2A, compare lane 5 to lanes 6, 7, and 8). Importantly, overexpression of these dominant negative cullins did not alter the asynchronous cell cycle or the ability to synchronize cells with nocodazole (data not shown). We also observed the same increase in JMJD2A protein levels when cullin 1 was depleted with siRNA in HeLa cells (Fig. 3A).
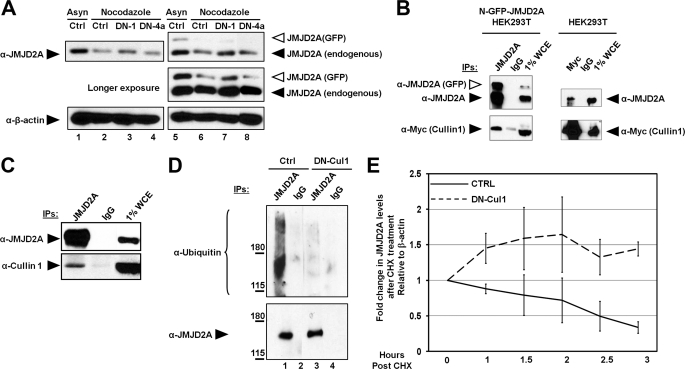
FIGURE 2.
The SCF complex regulates the degradation of JMJD2A. A, dominant negative cullin 1 prevents the degradation of JMJD2A. HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with the dominant negative version of cullin 1 (DN-1) or cullin 4a (DN-4a) and arrested in G2/M with nocodazole. Cells were then lysed and immunoblotted. Asyn, asynchronous. B and C, JMJD2A interacts with cullin 1. B, HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with Myc-cullin 1 and then lysed, immunoprecipitated (IP) with either JMJD2A or Myc in the presence of 100 μg/ml of EtBr, and immunoblotted. WCE, whole cell extract. C, HEK293T cells were treated with 20 μm MG132 2.5 h before lysis and then immunoprecipitated in presence of 100 μg/ml of EtBr and immunoblotted. D, dominant negative cullin 1 decreased the ubiquitination of JMJD2A. HEK293T cells overexpressing HA-ubiquitin and DN-Cul1 or a control vector were arrested in S phase in the presence of 20 μm MG132 and 25 mm _N_-ethylmaleimide, lysed, immunoprecipitated, and immunoblotted. E, dominant negative cullin 1 prevents the proteasomal degradation of JMJD2A. HEK293T cells overexpressing DN-Cul1 or a control vector were treated for the indicated times with 400 μm of cycloheximide (CHX) and then lysed and immunoblotted. The y axis represents the ratio of JMJD2A relative to time 0, which was normalized to β-actin. The average of two independent experiments is shown. The errors bars represent the mean ± S.E.
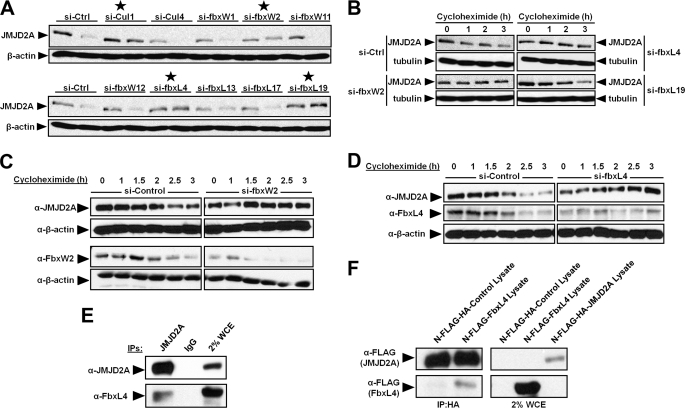
FIGURE 3.
The F-box protein FbxL4 interacts with JMJD2A and contributes to JMJD2A degradation. A, screen of F-box proteins involved in the degradation of JMJD2A. HeLa cells were transfected with siRNAs targeting F-box proteins. For each siRNA, lysates from both asynchronous cells and cells arrested in G2/M by nocodazole and a shake off were immunoblotted for JMJD2A or β-actin. The asynchronous lysates are the first lanes followed by G2/M lysates for each siRNA treatment. The stars denote siRNA treatments that exhibited increased JMJD2A protein levels after nocodazole treatment when compared to si-Ctrl. B, depletion of FbxL4 and FbxW2 prevent the proteasomal degradation of JMJD2A in HeLa cells. HeLa cells transfected with an siRNA targeting FbxL4, FbxW2, FbxL19, or a control siRNA were treated for the indicated times with 89 μm of cycloheximide and then lysed and immunoblotted. C, depletion of FbxW2 prevents the proteasomal degradation of JMJD2A in HEK293T cells. HEK293T cells transfected with an siRNA targeting FbxW2 or a control siRNA were treated for the indicated times with 400 μm of cycloheximide and then lysed and immunoblotted. D, depletion of FbxL4 prevents the proteasomal degradation of JMJD2A in HEK293T cells. HEK293T cells transfected with an siRNA targeting FbxL4 or a control siRNA were treated for the indicated times with 400 μm of cycloheximide and then lysed and immunoblotted. E and F, JMJD2A interacts with FbxL4. E, HEK293T cells were treated with 20 μm MG132 2.5 h before lysis and then immunoprecipitated (IP) in the presence of 100 μg/ml of EtBr and immunoblotted. F, HA-immobilized FLAG-HA-JMJD2A was incubated with HEK293T lysates overexpressing FLAG-FbxL4 in the presence of 100 μg/ml of EtBr and immunoblotted. WCE, whole cell extract.
To test if this functional interaction was direct, we conducted a series of coexpression experiments to demonstrate that JMJD2A and cullin 1 could coimmunoprecipitate. Immunoprecipitating either protein resulted in detectable association with either JMJD2A or cullin 1, respectively (Fig. 2B). These associations were also validated for endogenous proteins because cullin 1 was immunoblotted in endogenous JMJD2A immunoprecipitations (Fig. 2C). We next asked if cullin 1 contributed to the ubiquitination of JMJD2A. Consistent with the observed interactions, overexpression of DN-Cul1 decreased the amount of ubiquitination in the JMJD2A immunoprecipitation (Fig. 2D). Loss of ubiquitinated JMJD2A coincided with a complete block in the turnover of endogenous JMJD2A when DN-Cul1 was overexpressed (Fig. 2E). These data are consistent with the idea that one or more cullin 1-containing E3 ligase complex(es) contribute(s) to ubiquitination and turnover of JMJD2A.
The F-box and Leucine-rich Repeat Protein 4 (FbxL4) Interacts with JMJD2A and Regulates Protein Turnover
Cullin 1 is a shared subunit of the large family of SCF ubiquitin ligases. SCF family ligases also contain the linker protein SKP1 and one of a large number of F-box protein family members that act as substrate recognition subunits of SCF ligases (11). To identify the corresponding F-box protein(s) that interact with JMJD2A and modulate protein turnover, we performed an siRNA screen in HeLa cells for F-box proteins that effect JMJD2A protein levels. The cells were synchronized with nocodazole and immunoblotted for JMJD2A (the same method used to identify cullin 1). The depleted F-box proteins that appeared to increase JMJD2A levels were rescreened and tested for their impact on turnover. A total of eight siRNAs were rescreened, and only depletion of FbxL4, FbxL19, and the F-box and WD repeat domain-containing 2 (FbxW2) increased JMJD2A levels (Fig. 3A, stars). We then determined whether these F-box siRNAs affected protein turnover in HeLa cells. Only depletion of FbxL4 and FbxW2 blocked the turnover of JMJD2A (Fig. 3B). This observation was confirmed in HEK293T cells (Fig. 3, C and D).
To determine whether FbxL4 or FbxW2 interacted with JMJD2A, we performed coimmunoprecipitation experiments. Endogenous FbxL4 coimmunoprecipitated with JMJD2A, suggesting that the two proteins interact (Fig. 3E). We were not able to demonstrate endogenous interactions between JMJD2A and FbxW2. Therefore, we were not able to definitively address whether FbxW2 directly regulated JMJD2A turnover.
We next determined whether immunopurified JMJD2A could interact with lysate containing overexpressed FbxL4 in vitro. JMJD2A was immobilized via its HA tag on magnetic beads coated with anti-HA antibodies. The JMJD2A bound on the beads was incubated with extracts from mock-transfected cells or cells expressing FLAG-FbxL4. Unbound proteins were removed by washing, and FbxL4 association was assayed by Western blot analysis. The immunopurified JMJD2A was able to interact with FbxL4 in vitro (Fig. 3F). Taken together, our results are consistent with FbxL4 interacting with JMJD2A and promoting turnover.
Ubiquitin Overexpression Increases Exogenous JMJD2A Turnover and Prevents JMJD2A-dependent Faster S Phase Progression
We have demonstrated previously that increased JMJD2A levels result in faster S phase progression, which was accompanied by increased chromatin accessibility and more replication forks (25). These effects were dependent on the catalytic activity of JMJD2A. Our previous data also demonstrated that overexpressed JMJD2A was not properly regulated over the cell cycle. Therefore, we determined whether overexpressing ubiquitin would restore the turnover kinetics of exogenous JMJD2A. Indeed, JMJD2A overexpressing cells had increased turnover of the exogenous JMJD2A following overexpression of ubiquitin (Fig. 4A, the half-life went from 2 h 22 min to 1 h 25 min). Interestingly the exogenous ubiquitin did not affect turnover of endogenous JMJD2A when exogenous JMJD2A was present (Fig. 4B). The increased turnover observed for exogenous JMJD2A as well as the turnover of endogenous JMJD2A was blocked completely with coexpression of dominant negative cullin 1 (Fig. 4, A and B, DN-1, black dotted line). These results demonstrate that increased ubiquitin levels in the presence of functional cullin 1 are able to modulate JMJD2A protein turnover.
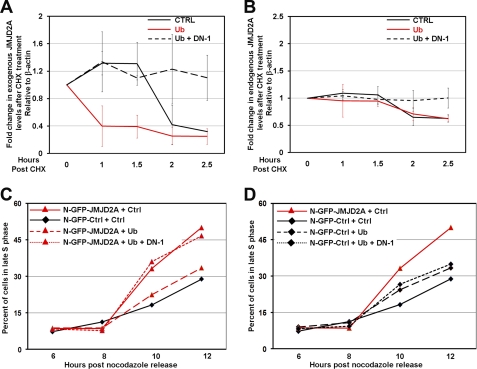
FIGURE 4.
Cullin 1-dependent ubiquitination regulates the JMJD2A-dependent faster S phase progression. A and B, the overexpression of ubiquitin (Ub) leads to a faster degradation of exogenous JMJD2A (A) while not altering endogenous JMJD2A turnover (B). Dominant negative cullin 1 (DN-1) prevented the turnover of both endogenous and exogenous JMJD2A. HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with HA-ubiquitin and DN-1 or control vector. Cells were then treated for the indicated times with 400 μm of cycloheximide (CHX), lysed, and immunoblotted. The y axis represents the ratio of JMJD2A relative to time 0, which was normalized to β-actin. The average of two independent experiments is shown. The errors bars represent the mean ± S.E. C and D, overexpression of ubiquitin prevents the faster S phase because of JMJD2A overexpression, which was blocked by dominant negative cullin 1. HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with HA-ubiquitin and DN-1 or a control vector, arrested in G2/M with nocodazole, and released for the indicated times prior to collection for FACS analysis. A graphical representation of the average of three independent experiments is shown.
On the basis of these observations, we determined whether increasing turnover of exogenous JMJD2A by overexpressing ubiquitin would rescue the faster S phase progression observed in JMJD2A-overexpressing cells (25). When ubiquitin was overexpressed in cells expressing exogenous JMJD2A, the faster S phase progression was ameliorated (Fig. 4C, compare the solid red line to both black and dashed red lines). Because cullin 1 interacts with multiple F-box proteins, contributes to the ubiquitination of JMJD2A, and affects both exogenous and endogenous protein turnover, we tested the impact dominant negative cullin 1 (DN-1) would have on the ubiquitin-based rescue of JMJD2A-dependent faster S phase progression.
When DN-1 was overexpressed with ubiquitin, JMJD2A overexpressing cells did not respond to ubiquitin overexpression (Fig. 4C, compare the solid red line to the dotted red line). The overexpression of ubiquitin and/or dominant negative cullin 1 had a subtle effect on cell cycle progression in control cells (Fig. 4D, compare the solid black line to the dotted black line). These results demonstrate that proper regulation of JMJD2A by cullin 1 and ubiquitin is critical for regulating JMJD2A-regulated S phase progression.
Overexpression of Ubiquitin Decreased JMJD2A Occupancy and BrdU Incorporation at JMJD2A Target Sites
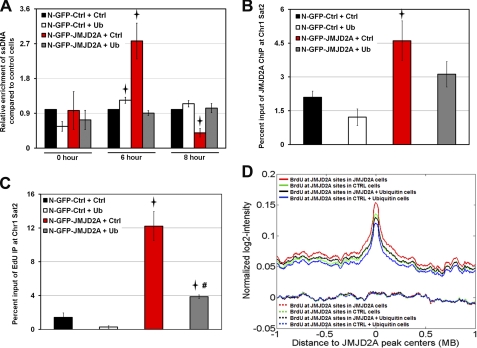
Our previous studies demonstrated that the JMJD2A-dependent faster S phase progression was accompanied by an increased number of replication forks, as determined by the increased presence of single-stranded DNA (ssDNA) (25). Therefore, we determined whether increased ubiquitin expression would alter the amount of replication forks in cells overexpressing JMJD2A. Consistent with our previous findings, we observed an increase in the amount of ssDNA at 6 h post-nocodazole with JMJD2A overexpression (Fig. 5A, red bars, p < 0.05). This increase in ssDNA was completely reversed upon overexpression of ubiquitin (Fig. 5A, gray bars). Six hours after nocodazole release, the control cells overexpressing ubiquitin showed a slight but significant increase in the presence of ssDNA (Fig. 5A, white bars, p < 0.05), which was consistent with the slightly faster progression in S phase (Fig. 4D). We also observed that ubiquitin overexpression rescued the decreased ssDNA observed in JMJD2A overexpressing cells 8 h after nocodazole release, whereas no change occurred in control cells. These observations coupled to the rescued S phase progression suggest that increased ubiquitination of JMJD2A through cullin 1 is able to antagonize the faster S phase phenotype.
FIGURE 5.
JMJD2A occupancy and BrdU incorporation are regulated by ubiquitin. A, ubiquitin (Ub) overexpression prevents the increase of ssDNA (indicative of replication forks) in JMJD2A-overexpressing cells (25, 35). HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with HA-ubiquitin or a control vector and then treated with BrdU for 24 h prior to arrest in G2/M with nocodazole. Cells were released for the indicated times prior to DNA collection. Samples were purified and slot-blotted, and then immunoblotted for BrdU. The y axis represents the ratio of native/denaturated BrdU signal normalized to the level in control cells at each time point. The average of two independent experiments is shown. The errors bars represent the mean ± S.E. Stars, p < 0.05. B, ubiquitin overexpression decreases the enrichment of JMJD2A at Chr1 sat2 (25). HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with HA-ubiquitin or a control vector and released 1 h after HU treatment before performing the ChIP experiment. JMJD2A cells were statistically different from control cells (p = 0.02), whereas JMJD2A cells overexpressing ubiquitin were not (p = 0.1). C, ubiquitin overexpression prevents the increased EdU incorporation at Chr1 sat2 in JMJD2A-overexpressing cells (25). HEK293T cells overexpressing control N-GFP or N-GFP-JMJD2A were transfected with HA-ubiquitin or a control vector and then treated with EdU for 1 h prior to DNA collection. The DNA was then biotinylated and replicated DNA was enriched with streptavidin beads prior to analysis by quantitative PCR. Stars, p < 0.05 relative to control cells treated with control vector; #, p < 0.05 relative to JMJD2A cells + control vector. D, JMJD2A overexpression leads to increased BrdU incorporation at JMJD2A binding sites, which was ameliorated upon overexpression of ubiquitin. JMJD2A binding sites in JMJD2A-overexpressing cells were identified by ChIP-chip and analysis with MA2C software (28). BrdU ChIP-chip data were then analyzed by meta-analysis at these JMJD2A-bound genomic locations in control and JMJD2A-overexpressing cells with and without exogenous ubiquitin. The graph depicts the average BrdU ChIP profile around JMJD2A binding sites (solid curves) compared with random regions (dashed curves). Data are centered on the JMJD2A binding sites (0 on x axis). The average BrdU intensities were smoothed using a moving average with a 50-kb window in 1-kb increment step sizes.
On the basis of these observations, we evaluated JMJD2A occupancy and EdU and BrdU incorporation at JMJD2A targets upon overexpression of ubiquitin. At chromosome 1 satellite 2 (Chr1 sat2, a known JMJD2A target) (25), JMJD2A occupancy was significantly increased in JMJD2A-overexpressing cells, which was consistent with our previous observations (25). Upon ubiquitin overexpression, JMJD2A occupancy was no longer statistically different from control cells (Fig. 5B). In agreement with our previous results, increased JMJD2A occupancy was accompanied by an increase in EdU incorporation at Chr1 sat2 (25). However, this EdU increase was significantly reduced with ubiquitin overexpression (Fig. 5C, p < 0.05).
To determine whether the ubiquitin-dependent changes observed at Chr1 sat 2 were generalizable, we evaluated the increased BrdU incorporation at JMJD2A target regions on chromosomes 1–4. We first identified JMJD2A binding sites by performing JMJD2A ChIP-chips on Nimblegen HD2.1 microarrays for chromosomes 1–4. We identified 2408 binding sites in JMJD2A-overexpressing cells using MA2C (28). We next asked if replication of these loci was affected by JMJD2A overexpression. Asynchronously growing control and JMJD2A-overexpressing cells were treated with BrdU for 1 h. Genomic DNA was extracted from these cells, sheared, and used in BrdU immunoprecipitations. The BrdU-enriched DNA, or replicated DNA, was amplified, labeled, and hybridized to the same Nimblegen HD2.1 microarrays for chromosomes 1–4.
To determine how replication was affected at the JMJD2A-bound loci, we compared BrdU ChIP-chip profiles from control and JMJD2A overexpressing cells with or without ubiquitin overexpression. We performed a meta-analysis of the BrdU profile surrounding all JMJD2A binding sites in overexpressing cells. The graph in Fig. 5D is centered on the “average” JMJD2A binding site and depicts the BrdU incorporation surrounding this site. Similar to Chr1 sat2, BrdU incorporation increased at and flanking JMJD2A binding sites in JMJD2A overexpressing cells (Fig. 5D, red line compared with green line, p < 2.97 E-18). Upon overexpressing ubiquitin in JMJD2A overexpressing cells, the increased BrdU incorporation that was observed at and flanking JMJD2A binding sites was significantly reduced (p < 3.22 E-10). In fact, the decrease in BrdU incorporation resulted in JMJD2A overexpressing cells having no statistical difference when compared to BrdU incorporation in control cells (Fig. 5D, green line compared with black line, p = 0.57). The altered BrdU incorporation at JMJD2A binding sites was specific because the same number of random regions within chromosomes 1–4 did not show any enrichment for BrdU and displayed no difference compared with either control or control cells overexpressing ubiquitin (Fig. 5D, dashed lines on the bottom). Taken together, these data highlight an unappreciated role for cullin 1 and ubiquitin in regulating JMJD2A protein levels as well as the impact JMJD2A has on S phase progression and replication of JMJD2A target regions within the genome.
DISCUSSION
Here we demonstrate that ubiquitin-dependent proteasomal degradation regulates JMJD2A protein levels over the cell cycle. Our data is consistent with the SCF-FbxL4 complex mediating turnover and ubiquitination of JMJD2A. Our work as well as the recent advances in the cell cycle-dependent regulation of mixed lineage leukemia (MLL) and Set8/PR-Set7 underscores the importance of regulating chromatin modifiers during the cell cycle (12–19). These data also highlight the need to better understand the role that chromatin-modifying enzymes play outside of the traditional asynchronous cell populations. It is likely that many, if not most, chromatin regulators could exhibit cell cycle-dependent functions and regulation that may be masked in standard asynchronous populations.
We observed that inhibiting proteasomal degradation in G2/M arrested cells was not able to restore JMJD2A protein levels to that observed in G1/S arrested cells. Consistent with this observation, interfering with the ubiquitination machinery by siRNA depletion or dominant negative overexpression of cullin-1 or FbxL4 only increased JMJD2A levels in G2/M to that of asynchronous levels while not achieving the levels of expression observed in G1/S or S phase. This data is consistent with the majority of JMJD2A regulation occurring during S-phase. Because our data demonstrated that degradation occurs during G2/M, it is possible that we did not observe a higher up-regulation of JMJD2A in G2/M cells because the degradation had already occurred during the nocodazole treatment, as the cells were not treated with MG132 until the final two and a half hours of mitotic arrest. It is also possible that JMJD2A turnover is regulated at multiple points in the cell cycle by different F-box-containing SCF ubiquitin ligase complexes, which is supported by the multiple F-box proteins regulating JMJD2A protein levels. Nonetheless, these data highlight the regulation of JMJD2A levels during the cell cycle.
Recently, Braun et al. (31) showed that the boundary factor Epe1, the yeast homolog of the plant homeodomain finger protein 2 (PHF2), is specifically targeted by Cul4-Ddb1Cdt2. This pathway controls the distribution of this antisilencing factor within heterochromatin and restricts Epe1 to heterochromatic boundaries, which prevents position effect variegation. Importantly, the authors showed that this heterochromatin-sculpting function of Cul4-Ddb1Cdt2 is sufficient to explain its requirement for silencing (31). Here, we demonstrate that JMJD2A is ubiquitinated by the SCF-FbxL4 complex. By linking cullin 1/SCF-dependent ubiquitination to a histone demethylase and chromatin dynamics, we have identified another mechanism to modulate heterochromatin formation and S phase progression. Because cullin 4 is known to function in S phase through interaction with PCNA and ubiquitination of targets on chromatin (30), it will be interesting to determine whether cullin 1-dependent regulation is occurring on chromatin and which other chromatin modifying enzymes cullin 1/SCF targets.
The SCF complex plays well established roles in cell growth control pathways. However, many aspects of its functions remain unknown (32). In this manuscript, we demonstrate that the protein degradation of JMJD2A involves at least two F-box proteins, FbxL4 and FbxW2. The regulation by multiple F-box proteins underscores the importance of down-regulating JMJD2A. Little is known about the function of FbxL4 and FbxW2. In Drosophila, FbxL4 overexpression rescued defects in photo response that are caused by the mutation of dCAMTA, the homolog of the candidate human brain tumor suppressor gene CAMTA1 (33). The only target described to date for FbxW2 is GCMa, a key factor in the formation of the human syncytiothrophoblast layer (34). Our work describes a new substrate for these proteins and suggests that they may have roles in chromatin states and cell cycle dynamics.
Most proteins regulated by post-translational modifications involve the coordinated action of multiple modifications. For example, the Cdk1/CyclinB complex phosphorylates the methyltransferase Set8/PR-Set7 during early anaphase, leading to the removal of Set8/PR-Set7 from mitotic chromosomes. This phosphorylation was followed by Cdc14 dephosphorylation. Set8/PR-Set7 was then ubiquitinated and degraded (14). In addition to ubiquitin, JMJD2A and other family members are likely targeted by other modifications. Identification of these additional modifications and resolving their interrelationship will prove vital to understanding the biological functions of histone demethylases. The signaling cascades mediating modification of histone demethylases may be amenable as targets for cancer therapeutics.
Acknowledgments
We thank Cristina Pineda, Rachel Lee, and Christopher Guenard for technical assistance and Grace Gill, Cyril Benes, and Nicholas Dyson for helpful discussions and comments on the manuscript.
*
This work was supported, in whole or in part, by the Ellison Medical Foundation, Howard Goodman Fellowship, and Grant CA059267 (to J. R. W.). This work was also supported by National Institutes of Health Grants R01CA60499 (to A. D.) and R01HG005085 (to G. C. Y.). This work has also been aided by grants from The Jane Coffin Childs Memorial Fund for Medical Research (to J. C. B.) and from NCI National Institutes of Health Grant KCA140774A (to T. A.).
3
The abbreviations used are:
JMJD2
Jumonji domain-containing 2
KDM4
lysine demethylase 4
HU
hydroxyurea
EdU
ethynyl deoxyuridine
DN-Cul1
dominant negative cullin 1
SCF
SKP1-Cul1-F-box
DN-1
dominant negative cullin 1
ssDNA
single-stranded DNA
Chr1 sat2
chromosome 1 satellite 2.
REFERENCES
- 1.Bannister A. J., Kouzarides T. (2011) Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J. S., Smith E., Shilatifard A. (2010) Cell 142, 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Negrini S., Gorgoulis V. G., Halazonetis T. D. (2010) Nat. Rev. Mol. Cell Biol. 11, 220–228 [DOI] [PubMed] [Google Scholar]
- 4.Corpet A., Almouzni G. (2009) Trends Cell Biol. 19, 29–41 [DOI] [PubMed] [Google Scholar]
- 5.Bernstein B. E., Meissner A., Lander E. S. (2007) Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 6.Méndez J. (2009) Crit. Rev. Biochem. Mol. Biol. 44, 343–351 [DOI] [PubMed] [Google Scholar]
- 7.Varier R. A., Timmers H. T. (2011) Biochim. Biophys. Acta 1815, 75–89 [DOI] [PubMed] [Google Scholar]
- 8.Pedersen M. T., Helin K. (2010) Trends Cell Biol. 20, 662–671 [DOI] [PubMed] [Google Scholar]
- 9.Shi Y., Whetstine J. R. (2007) Mol. Cell 25, 1–14 [DOI] [PubMed] [Google Scholar]
- 10.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 11.Petroski M. D., Deshaies R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Cheng E. H., Hsieh J. J. (2007) Genes Dev. 21, 2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Y., Yu V. C., Zhu G., Chang D. C. (2008) Cell Cycle 7, 1423–1432 [DOI] [PubMed] [Google Scholar]
- 14.Wu S., Wang W., Kong X., Congdon L. M., Yokomori K., Kirschner M. W., Rice J. C. (2010) Genes Dev. 24, 2531–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbas T., Shibata E., Park J., Jha S., Karnani N., Dutta A. (2010) Mol. Cell 40, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., Zou L. (2010) Mol. Cell 40, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda H., Hübner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., Reinberg D. (2010) Mol. Cell 40, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., Julien E. (2010) Nat. Cell Biol. 12, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen S., Eskildsen M., Fugger K., Hansen L., Larsen M. S., Kousholt A. N., Syljuåsen R. G., Trelle M. B., Jensen O. N., Helin K., Sørensen C. S. (2011) J. Cell Biol. 192, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mersman D. P., Du H. N., Fingerman I. M., South P. F., Briggs S. D. (2009) Genes Dev. 23, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fodor B. D., Kubicek S., Yonezawa M., O'Sullivan R. J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. (2006) Genes Dev. 20, 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 23.Trojer P., Zhang J., Yonezawa M., Schmidt A., Zheng H., Jenuwein T., Reinberg D. (2009) J. Biol. Chem. 284, 8395–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. (2006) Cell 125, 467–481 [DOI] [PubMed] [Google Scholar]
- 25.Black J. C., Allen A., Van Rechem C., Forbes E., Longworth M., Tschöp K., Rinehart C., Quiton J., Walsh R., Smallwood A., Dyson N. J., Whetstine J. R. (2010) Mol. Cell 40, 736–748 [DOI] [PubMed] [Google Scholar]
- 26.Jin J., Ang X. L., Shirogane T., Wade Harper J. (2005) Methods Enzymol. 399, 287–309 [DOI] [PubMed] [Google Scholar]
- 27.Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song J. S., Johnson W. E., Zhu X., Zhang X., Li W., Manrai A. K., Liu J. S., Chen R., Liu X. S. (2007) Genome Biol. 8, R178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 30.Jackson S., Xiong Y. (2009) Trends Biochem. Sci. 34, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun S., Garcia J. F., Rowley M., Rougemaille M., Shankar S., Madhani H. D. (2011) Cell 144, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skaar J. R., Pagano M. (2009) Curr. Opin. Cell Biol. 21, 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J., Gong P., Reddig K., Mitra M., Guo P., Li H. S. (2006) Cell 127, 847–858 [DOI] [PubMed] [Google Scholar]
- 34.Yang C. S., Yu C., Chuang H. C., Chang C. W., Chang G. D., Yao T. P., Chen H. (2005) J. Biol. Chem. 280, 10083–10090 [DOI] [PubMed] [Google Scholar]
- 35.Jasencakova Z., Scharf A. N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. (2010) Mol. Cell 37, 736–743 [DOI] [PubMed] [Google Scholar]