Distinct Functional Effects for Dynamin 3 During Megakaryocytopoiesis (original) (raw)
Abstract
Dynamin 3 (DNM3) is a member of a family of motor proteins that participate in a number of membrane rearrangements such as cytokinesis, budding of transport vesicles, phagocytosis, and cell motility. Recently, DNM3 was implicated as having a role in megakaryocyte (MK) development. To further investigate the functional role of DNM3 during megakaryocytopoiesis, we introduced sequence-specific short hairpin RNAs (shRNAs) into developing MKs. The results showed that knockdown of DNM3 inhibited a stage of MK development that involved progenitor amplification. This was evident by significant decreases in the number of colony forming unit-megakaryocytes, the total number of nucleated cells, and the number of CD41+ and CD61+ MKs produced in culture. Using a styrl membrane dye to quantify the demarcation membrane system (DMS) of terminally differentiated MKs, we found that DNM3 co-localized with the DMS and that DNM3 lentiviral shRNAs precluded the formation of the DMS. Knockdown of dynamin 3 in murine MKs also caused a decrease in the number of morphologically large MKs and the overall size of large MKs was decreased relative to controls. MK protein lysates were used in overlay blots to show that both DNM3 and actin bind to nonmuscle myosin IIA (MYH9). Consistent with these observations, immunofluorescence studies of MKs and proplatelet processes showed co-localization of DNM3 with MYH9. Overall, these studies demonstrate that DNM3 not only participates in MK progenitor amplification, but is also involved in cytoplasmic enlargement and the formation of the DMS.
Introduction
Dynamin 3 (DNM3) is one of 3 members of a superfamily of proteins (DNM1, DNM2, and DNM3). This family of proteins has mechanochemical properties [1] and each contains an amino-terminal GTPase domain, which hydrolyzes nucleotides to link cellular membranes to the actin cytoskeleton, a pleckstrin homology (PH) domain, and a carboxy-terminal proline/arginine-rich (PRD) domain. The PH domain of dynamin binds phosphatidylinositol lipids to link dynamin with membranes, whereas the PRD domain interacts with Src-homology-3-domains of many different actin-associated proteins [1–3]. Given the low affinity and weak specificity of the PH domain for negatively charged phosphatidylinositol lipids and the widespread use of dynamin in vesicle scission, the many binding partners of the PRD domain likely target dynamin to various sites of action. Together, these features allow the dynamins to participate in a number of membrane trafficking events, which include podosome formation [4,5], membrane vesiculation from the plasma membrane and _trans_-Golgi network [6], lamellipodial protrusion, phagocytosis [7], and cytokinesis [8].
Development of megakaryocytes (MKs) starts with commitment of hematopoietic stem cells to form MK progenitors that proliferate and differentiate to subsequently undergo a maturation process that involves endomitosis and cytoplasmic enlargement [9–12]. During MK differentiation, the promegakaryoblast (immature MK) increases its DNA content by a poorly understood process called endomitosis [13]. The resulting mature MK is a large polyploid cell with a highly elaborate demarcation membrane system (DMS) that contains cytoplasmic proteins and granules essential for the formation of platelets [14–16] and platelet function [17–19]. In anticipation of platelet release, membranes of the DMS acquire phosphatidylinositol 4, 5-bisphosphate (PIP2), which activates the WASp-Arp2/3 complex to induce actin polymerization in preparation for platelet formation [20].
Despite the discovery of platelets over a century ago, there is still controversy about the mechanisms involved in the actual formation of platelets. One model of platelet formation is based on the hypothesis that the DMS defines plasma membrane territories corresponding to future platelets that are liberated via cytoplasmic fragmentation [21,22]. However, in a more highly favored model, the DMS serves as a reservoir of cytoplasmic membrane that evaginates to form long processes, termed proplatelets, which release platelets along their length or only at their tip [23–25]. During the final stages of MK development, the proplatelets ultimately yield individual platelets [26].
We previously reported that DNM3 is upregulated during MK development [27], and that there is an amplification of MK clonogenic progenitors when DNM3 is overexpressed in human umbilical cord blood (UCB) CD34+ cells [28]. Here we use a functional genomics approach to introduce short hairpin RNAs (shRNAs) into developing MKs. This study confirms that DNM3 is involved in progenitor amplification, and also presents new evidence that DNM3 is involved in the maturation of MKs.
Materials and Methods
Lentiviral shRNA clone sets
Sequence-verified shRNA lentiviral plasmid (pLKO.1-puro) sets, each containing a different shRNA sequence-directed against human DNM3 (NM_015569) or murine dynamin 3 (NM_172646), were obtained from Sigma-Aldrich. Human dynamin 3 shRNA lentiviral plasmids are TRCN0000051404 (shRNA51404), TRCN0000051405 (shRNA51405), TRCN0000051406 (shRNA51406), and TRCN0000051407 (shRNA51407) (Fig. 1A). Negative control plasmids, pLKO.1 empty and nonspecific shRNA and a TurboGFP positive control plasmid, were purchased from Addgene and Sigma-Aldrich, respectively. Murine dynamin 3 shRNA lentiviral plasmids are TRCN0000091644 (shRNA91644, sequence: CCGGCCCACTATAATCCGTCCACTACTCGAGTAGTGGACGGATTATAGTGGGTTTTTG) and TRCN0000091645 (shRNA91645, sequence: CCGGCGTGTTAAATCTAACGCTAATCTCGAGATTAGCGTTAGATTTAACACGTTTTTG).
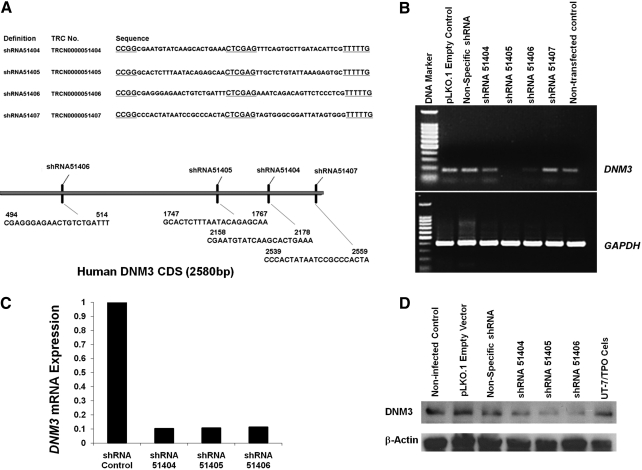
FIG. 1.
In vitro knockdown validation of _DNM3_-specific shRNAs. (A) Human _DNM3_-specific lentiviral shRNA plasmid sequences. Schematic that represents the coding sequence for human DNM3 and the shRNA targeting sites. (B) RT-PCR was performed to determine the down-regulation levels of DNM3 after UT-7/TPO cells were transfected with the indicated plasmid constructs. (C) Quantitative RT-PCR was performed to confirm the knockdown of DNM3 in culture-derived MKs produced from human UCB CD34+ cells that had been transduced with the indicated shRNA lentiviral vectors. Expression levels are expressed as ratios relative to the shRNA control, which is set to 1. (D) Western blot showed that all 3 lentiviral shRNA constructs knocked down DNM3 protein expression levels. All experiments were performed using cells infected with the pLKO.1 empty vector as a negative control. Three independent experiments were performed. DNM3, dynamin 3; MK, megakaryocyte; shRNA, short hairpin RNA; TPO, thrombopoietin; UCB, umbilical cord blood; PCR, polymerase chain reaction.
Validation of knockdown efficiency
Nucleofection of UT-7/TPO cells, a human megakaryoblastic cell line (kindly provided by Dr. Norio Komatsu [29,30]) was performed by culturing cells in Iscove's modified Dulbecco's medium (IMDM; American Type Culture Collection, ATCC) supplemented with 10% fetal bovine serum (FBS; Hyclone), and 10 ng/mL of recombinant human thrombopoietin (TPO; PeproTech). About 1×106 cells were electroporated with 1 μg of plasmid DNA containing DNM3 shRNA using the Amaxa Nucleofector System. Forty-eight hours after transfection, cells were harvested for reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Isolation of human UCB CD34+ cells
After obtaining parental informed consent, human UCB units were collected from placentas of healthy full-term donor pregnancies. UCB was drawn into blood bags containing acid-citrate-dextrose/adenine (50 mL/bag; Citra Anticoagulant, Inc.) and transported to the Puget Sound Blood Center for processing. UCB was processed by using a modification of a previously published procedure [31]. Briefly, UCB was diluted 1:2 with phosphate-buffered saline (PBS) containing 2% bovine serum, 6% Hetastarch (Abbott) was added at a 1:5 dilution, and cells were allowed to sediment for 1 h. The leukocyte-rich supernatant was removed and centrifuged at 400 g for 10 min, and the leukocyte-poor plasma was removed. The remaining cell pellet was washed twice with PBS containing 2% bovine serum followed by an enrichment for CD34+ cell.
CD34+ cells were isolated from the UCB-nucleated cell fraction by using an AutoMACS CD34 Microbead Kit and an autoMACS Separator (Miltenyi Biotec Inc.) according to the manufacturer's instructions. After UCB-nucleated cell fractions were passed twice over columns, cells were enumerated after staining with trypan blue (Sigma-Aldrich). Purity levels were determined by flow cytometry (FACSCalibur; Becton-Dickinson). Cells were stained with CD34 phycoerythrin (PE) and CD45 fluoresceine isothiocyanate-conjugated monoclonal antibodies (Becton-Dickinson). Greater than 90% of the cells were CD34+ as confirmed by flow analysis.
Lentivirus production, transduction, and analysis
Lentiviral particles were packaged by co-transfecting 293T cells (ATCC) with the following: (1) shRNA plasmid transfer vector, (2) envelope coding plasmid pMD2.G (Addgene), and (3) a second-generation packaging plasmid psPAX2 (Addgene). First, 293T cells were cultured in T25 flasks containing Dulbecco's modified Eagle's medium supplemented with 10% Cosmic Calf Serum (Hyclone). After reaching 80%–90% confluence, 293T cells were co-transfected with the 3 plasmids. Twenty-four hours after transfection, supernatant containing lentiviral particles was harvested 3 times at 12-h intervals. Combined supernatants were filtered through a 0.22-μm low-protein-binding filter, and concentrated ∼350–400-fold by ultracentrifugation at 7,200 g for 19 h at 4°C. Supernatant was aspirated; the viral pellet was re-suspended in complete X-vivo10 (BioWhittaker, Inc.), and 100 μL of aliquots was stored at −80°C. Lentiviral titers were determined using an HIV-1 p24 antigen ELISA Kit (ZeptoMatrix) according to the manufacturer's instruction. p24 protein levels were determined and converted to transfection units of virus (TU/mL) according to a procedure defined by the Trono laboratory (http://tronolab.epfl.ch/webdav/site/tronolab/shared/protocols/TUvsp24.html) [32].
For transduction of human UCB CD34+ cells, flat-bottomed 96-well nontreated tissue culture plates were coated with 100 μL of 50 μg/mL Retronectin (Takara Bio Inc.). Human UCB CD34+ cells were seeded at 4×104 cells/mL onto Retronectin (Takara Bio Inc.)-coated plates and cultured in serum-free X-vivo 10 medium supplemented with Interleukin 3 (IL-3; 10 ng/mL), Interleukin 6 (IL-6; 10 ng/mL), stem cell factor (SCF; 10 ng/mL), and TPO (50 ng/mL). Using a previously published method [33], CD34+ cells were infected at 24, 48, and 72 h with lentiviral shRNAs at a multiplicity of infection of 40 and were cultured for up to 10 days. At day 4, an aliquot of cells was collected from both experimental and control groups to perform colony forming unit-megakaryocytes (CFU-MKs) assays according to the manufacturer's instructions (Stemcell Technologies, Inc). After a 10-day culture period total nucleated cell (TNC) counts and immunophenotyping by flow cytometry were performed.
For transduction of murine bone marrow cells, TNCs were harvested from bone marrow of C57BL/6 mice (Jackson Laboratory). All animal work was approved by the University of Washington Institutional Animal Care and Use Committee. Single nucleated murine cell suspensions were treated with ACK lysing buffer (Invitrogen), passed through a 70-μm cell strainer (BD Biosciences), and seeded at 8×104 cells/mL onto Retronectin coated plates. The cells were cultured overnight in 250 μL of IMDM with 1×Nutridoma-SP serum-free media supplement (Roche Applied Science) with IL-3 (10 ng/mL), IL-6 (10 ng/mL), and SCF (10 ng/mL). Cells were infected with murine lentiviral shRNAs at 24 and 48 h. At day 4, 100 μL of medium was removed and 100 μL of fresh IMDM supplemented with 1×Nutridoma-SP serum-free media containing TPO at a final concentration of 16 ng/mL and 5 μg/mL puromycin (Sigma-Aldrich). At day 7, cells were examined using a Leica DMIL microscope equipped with a Retiga digital CCD camera and QCapture 2.94 software (QImaging). Feret's diameter was calculated using the NIH ImageJ software program (http://rsb.info.nih.gov/ij/). Absolute numbers of total CD41+ MKs were determined by flow cytometry and using TruCount beads (Becton Dickinson). Cells were stained with PE-conjugated anti-mouse CD41 (eBioscience). Data were acquired by BD LSR II flow cytometer and analyzed by BD FACSDiva software (BD Biosciences).
For transduction of UT-7/TPO cells, 5×104 UT-7/TPO cells were seeded into 24-well culture plates with IMDM supplemented with 10% FBS plus 10 ng/mL of TPO. Cells were treated with 2.5 μM SU6656 (Invitrogen), an Src family kinase inhibitor [34]. At 24 and 48 h the cells were infected with lentiviral shRNA viruses. At day 4, culture supernatant was examined for the presence of platelets and the remaining cells were enumerated and examined for ploidy and DMS.
Ploidy and DMS analyses
To perform ploidy analyses, cells were harvested and stained with 500 μL of 4 mM sodium citrate buffer containing 50 μg/mL of propidium iodide (Invitrogen), 0.1% Triton X-100, and 0.2 mg/mL RNase A (Invitrogen) for 30 min. Ploidy analysis was conducted using flow cytometry and the software program CellQuest software (BD Biosciences).
DMS was analyzed using a modification of a previously published procedure [35]. Briefly, UT-7/TPO cells were cytocentrifuged at 2,000 rpm for 2 min onto glass slides. Cells were fixed with 2.5% glutaradehyde (Sigma-Aldrich), stained with 10 μM of the styrl membrane dye, 4-(2-[6-(Dioctylamino)-2-naphthalenyl]ethenyl)-1-(3-sulfopropyl) pyridinium (di-8-ANEPPS; Invitrogen) in PBS, and mounted using SlowFade Gold antifade reagent (Invitrogen). Fluorescence images were viewed and captured using an MRC-1024 confocal laser scanning imaging microscope system (Bio-Rad Laboratories, Inc.). Three-dimensional (3D) images were acquired and analyzed by the Nikon NIS-Elements AR software under the Nikon ECLIPSE Ti inverted microscope (Nikon). To obtain intact 3D images, the di-8-ANEPPS-stained UT-7/TPO cells were viewed and captured in suspension in a 24-well glass-bottom sensoplate (greiner bio-one North America, Inc.).
To quantify DMS staining, 1×103 transduced UT-7/TPO were suspended in 200 μL PBS in a single well of a 96-well plate and stained with di-8-ANEPPS at a final concentration of 10 μM. As a background control, 1×103 cells were suspended in 200 μL PBS without di-8-ANEPPS. The fluorescent intensity of cells was measured 60 min after adding di-8-ANEPPS and measured with an Flx800 fluorescence plate reader (Bio-Tek) using an excitation wavelength filter of 485±20 nm and a fluorescence wavelength emission filter of 620±40 nm.
Quantitative RT-PCR
Total RNA was isolated using the Protein and RNA Isolation System (PARIS) kit (Applied Biosystems/Ambion). For conventional RT-PCR, primers for DNM3 and housekeeping genes were designed using the software program Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). DNM3 forward primer: 5′-TCGCAACCTCGTAGACTCCT-3′, reverse primer: 5′-GCTCAGCAGATTCCTCCATC-3′. GAPDH forward primer: 5′-GTCAGTGGTGGACCTGACCT-3′, reverse primer: 5′-AGGGGAGATTCAGTGTGGTG-3′. Using the ThermoScript RT-PCR System (Invitrogen), RNA was reverse transcribed into cDNA according to the kit protocol, and amplified for 40 cycles.
For real-time quantitative RT-PCR, the gene specific primers and the Universal ProbeLibrary probes for DNM3 and housekeeping G6PDH were designed by the Assay Design Center (www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=UP030000). DNM3 was amplified using left primer: 5′-CTCCTGGATACAGCACTCTCG-3′, forward primer: 5′-TTAGTGTTGGCCTCCTTTGG-3′ and the Universal ProbeLibrary probe #25 from Roche Applied Science. G6PDH was amplified using reverse primer: 5′-AACAGAGTGAGCCCTTCTTCA-3′, forward primer: 5′-GGAGGCTGCATCATCGTACT-3′, and the Universal ProbeLibrary probe #5 (Roche Applied Science). The LightCycler TaqMan Master kit was used to amplify DNM3 or G6PDH with the LightCycler 2.0 instrument (Roche Applied Science) under the following conditions: a preincubation step for 15 min at 95°C followed by 45 cycles for 10 s at 95°C, 10 s at 60°C, and 15 s at 72°C. Data were analyzed with LightCycler Software 4.0 (Roche Applied Science). mRNA levels were normalized versus the amount of the housekeeping gene transcript G6PDH using the comparative CT method [36]. All experiments were performed using cells infected with the pLKO.1 empty vector as a negative control. Expression levels are expressed as ratios relative to the shRNA control, which is set to 1.
Western blotting
Cells were lysed in Cell Disruption Buffer using the PARIS kit. Protein concentrations were determined using the Pierce Coomassie Plus-The Better Bradford Assay Kit (Thermo Fisher Scientific). Protein (20 μg) from each sample was mixed in Laemmili Sample Buffer (Bio-Rad Laboratories), heated to 95°C for 5 min, loaded onto a 4%–15% Ready Gel Tris-HCI Gel (Bio-Rad Laboratories), and electrophoresed. Separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad Laboratories), probed with anti-DNM3 antibody diluted 1/1,000 (GeneTex Inc.), followed by a goat anti-rabbit IgG (H+L)-HRP conjugate diluted 1/2,500 (Bio-Rad Laboratories). Immunoreactive bands were observed by Amersham ECL Western Blotting Detection Reagents (GE Healthcare). As a control, the membrane was also probed with mouse beta actin antibody diluted 1/1,000 (Abcam Inc.) followed by a rabbit polyclonal antibody to mouse IgG H&L-HRP conjugate diluted 1/2,500 (Abcam Inc.).
Mass spectrometry
Cell lysates were incubated with purified DNM3 antibody that was attached to beads. The beads were then washed and immunoprecipitated proteins were eluted. After running a portion of the eluted fractions on a sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS-PAGE) gel, bands were excised, and remaining eluates were prepared for mass spectrometry analysis by reducing each sample with dithiothreitol, alkylating with iodoacetamide and digesting with trypsin according to in-house protocols (www.psbc.org/research/mass_spec_protocols.htm). The tryptic peptides were analyzed by mass spectrometry. Spectra were searched against the Human International Protein Index database, using Proteome Discoverer 1.0 software (Thermo Scientific).
Immunoprecipitation and overlay blot assay
Approximately 2.6×106 UT7/TPO cells were harvested and washed 2 times with PBS. Cell pellets were treated with 100 μL cold lysis buffer (50 mM Tris pH 7.5 containing 1% Triton X-100, 150 mM NaCl, 2 mM EDTA, and 1×Halt Proteases and Phosphatase Inhibitor Cocktail; Pierce Biotechnology, Inc.). Cell pellets were gently vortexed and after 5–10 min on ice, and 500 μL of cold lysis buffer was added. Cells were kept on ice for 10 min and centrifuged at 4°C, 16,000 g for 10 min. The resulting supernatant was loaded onto a micro-column (Miltenyi Biotec) and prewashed twice with lysis buffer, and the precleared lysate was used for immunoprecipitation assays. An aliquot of lysate containing about 2 mg of total protein was mixed with 10 μg of nonmuscle Myosin IIA antibody (Novus Biological) and incubated overnight at 4°C. The antibody–antigen mixture was labeled by adding 100 μL of Protein G microbeads (Miltenyi Biotec), mixed well, and incubated on ice for 60 min. A micro-column was placed in a magnetic separator (Miltenyi Biotec) and washed twice with lysis buffer. The magnetically labeled antibody–antigen mixture was applied onto the column and flow through lysate was collected as a nonbound fraction. The column was washed 5 times with 400 μL of lysis buffer without the inhibitor cocktail and once with 50 mM Tris pH 7.5, 250 mM NaCl, and 2 mM EDTA. The column was then washed twice with 50 mM NaCl in Tris pH 7.5. SDS sample buffer was diluted 1:1 and heated to 95°C. First, 10 μL of heated elution buffer was applied and collected as a waste. The immunoprecipitated fraction was collected after adding an additional 90 μL of heated elution buffer, boiled with 10 μL of 4× loading buffer for 3 min, and loaded onto a 4%–15% SDS-PAGE gel (BioRad). After electrophoresis, proteins were transferred onto PVDF membranes (Millipore, Immobilon P) for immunoblotting. Membranes were blocked with tris-buffered saline (TBS; BioRad) containing 0.05% Tween 20 and 0.2% I-block reagent (Applied Biosystems) for 4 h at room temperature. Each protein lane from the PVDF membrane was cut into a strip. An aliquot of 750 μL of fresh UT-7/TPO lysate containing 1 mg total protein was applied to each test membrane strip and incubated overnight at 4°C. Control lane membrane strips remained in blocking buffer. After membrane strips were washed with TBS-0.05% Triton, each individual strip was immunostained with the following antibodies: anti-dynamin 3 (GeneTex), anti-nonmuscle myosin IIA (Novus Biologicals), or anti-actin-SC1616 (Santa Cruz). Protein A horseradish preoxidase (HRP) conjugate (BioRad) was used to bind the primary antibodies, and gels were observed using an ECL substrate (Perkin Elmer Inc.), followed by exposure with X-ray film (CL-XPosure; ThermoScientific).
Immunofluorescence staining and laser-scanning confocal microscopy analysis
UT-7/TPO cells were transfected with a custom-made DNM3_mCherry fusion construct (GeneCopoeia), cultured in IMEM supplemented with 10 ng/mL TPO and 2.5 μM SU6656 for 3 days. At day 4, the transfected cells were fixed by 3.7% IC Fixation Buffer (eBioscience) for 15 min, then washed with PBS, and cytocentrifuged onto poly-L-lysine-coated slides (Vescor, Inc.). Transfected cells were stained with 10 μM di-8-ANEPPS and 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) and mounted with Prolong Gold Antifade Reagent (Molecular Probes).
Staining for DNM3, MYH9, and the DMS of primary MKs was performed by first sorting for culture-derived CD41+ MKs produced from human CD34+ cells. The MKs were fixed with 3.7% paraformadehyde, cytocentrifuged onto poly-L-lysine-coated slides, and permeabilized with 0.25% Triton X-100 for 10 min. Cellular preparations were incubated overnight with a mixture of nonmuscle Myosin IIA mouse monoclonal antibody (Abcam) at 1:500 and DNM3 rabbit polyclonal antibody (Abcam) at 1:1,000. After 3 washes, the slides were incubated with a mixture of an Alexa Fluor 488 goat anti-mouse IgG and an Alexa Fluor 633 goat anti-rabbit IgG (Invitrogen) at 1:1,000 for 1 h at room temperature. The slides were mounted using SlowFade Gold antifade reagent containing DAPI for nuclear staining (Invitrogen). For proplatelet formation, 7-day sorted culture-derived CD41+ cells were transferred to glass coated slides with PDMS (50:1) bound with 5 μg of vWF (Heamatologic Technologies Inc.) and cultured for an additional 3 days until proplatelet formation was observed. Proplatelets were stained using the same methods described above.
Single confocal optical sections were acquired on a Zeiss LSM 510 META laser scanning confocal microscope. All images were acquired with a Zeiss Plan-Neofluar 40×/1.3 oil immersion objective. Alexa 488 fluorescence was excited with a 488 nm Argon laser, and detected through a 500–550 nm bandpass filter. Alexa 633 fluorescence was excited with a 633 nm HeNe laser, and detected through a 650–710 nm bandpass filter. DAPI fluorescence was excited in 2-photon mode with a pulsed laser tuned to 780 nm, and emission was detected through a 435–485 nm bandpass filter. DAPI, Alexa 488, and Alexa 633 images were pseudocolored blue, green, and red, respectively. All images were processed and analyzed by Zeiss LSM Image Brower and NIH ImageJ software.
Statistical analysis
Results are expressed as mean±standard error of the mean. Statistical significance was determined using the _t_-test and a repeated measures analysis of variance. Significance was accepted as the P<0.05.
Results
Selection of lentiviral shRNA clones
Four shRNA clones directed against human DNM3 were tested to validate their ability to knockdown the gene (Fig. 1A). RT-PCR results showed that shRNA51405 and shRNA51406 were the most effective at precluding the expression of DNM3 mRNA followed by shRNA51404. This is evident by comparing PCR products for DNM3 in cells transfected with DNM3 shRNA plasmids to cells treated with empty vector and nonspecific shRNA plasmid controls (Fig. 1B). The shRNA51407 plasmid also reduced the expression of DNM3 mRNA, but the reduction in DNM3 expression was not as profound as was found for the other 3 plasmids. Consequently, shRNA51404, shRNA51405, and shRNA51406 were selected for lentiviral packaging for use in subsequent transduction experiments.
Down-regulation of DNM3 decreases progenitor cell proliferation
The functional role of DNM3 at different stages of MK development was investigated by infecting human UCB CD34+ cells with _DNM3_-specific shRNA lentiviral vectors and culturing the cells along the MK lineage. After a 10-day culture period, quantitative RT-PCR, TNC counts, immunophenotyping, and clonogenic activities were determined.
Quantitative RT-PCR confirmed that all 3 lentiviral DNM3 shRNA constructs knocked down DNM3 mRNA relative to MKs transduced with empty vector shRNA controls (Fig. 1C). Western blot analysis also confirmed that all 3 DNM3 shRNA constructs visibly knocked down DNM3 protein levels (Fig. 1D).
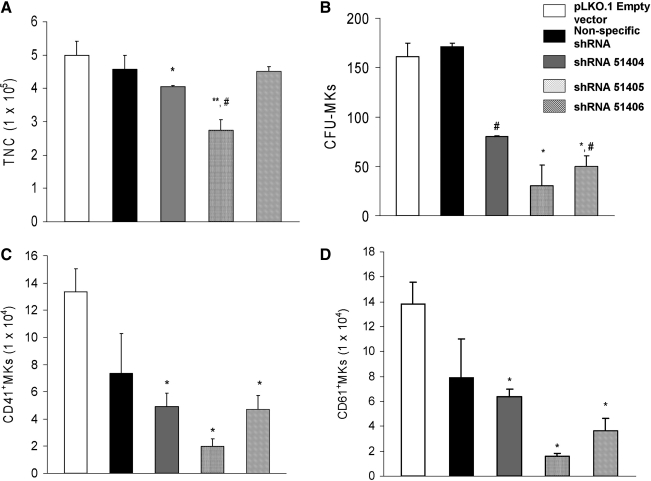
The total number of nucleated cells generated in culture from UCB CD34+ cells infected with shRNA51404 and shRNA51405 was significantly reduced relative to cells infected with negative shRNA control vectors (Fig. 2A). CFU-MK clonogenic assays also showed that there were significantly fewer CFU-MK colonies generated from UCB CD34+ treated with lentiviral DNM3 shRNAs than from cells treated with negative shRNA control vectors (Fig. 2B). Additionally, when expression levels for CD41 and CD61 were assessed on day 10 of culture, there was a significant decrease in the number of CD41+ and CD61+ MKs produced from UCB CD34+ cells treated with lentiviral DNM3 shRNAs (Fig. 2C, D).
FIG. 2.
DNM3 knockdown inhibits the proliferation of MK progenitors. Human UCB CD34+ cells were cultured with IL-3, IL-6, stem cell factor, and TPO and transduced with _DNM3_-specific shRNA lentiviral vectors at 24, 48, and 72 h of culture. (A) Total nucleated cell counts after cells were maintained in liquid culture for 10 days. (B) On day 4 of liquid culture, cells were transferred to MegaCult C collagen-based medium and maintained in culture for an additional 10 days. Colonies were stained mouse anti-human GPIIb/IIIa antibody and the total number of colony forming unit-megakaryocyte colonies was determined. (C and D) Total number of CD41+ and CD61+ MKs that is present at day 10 of liquid culture. Data represent the mean±SEM of 4 experiments. Each experiment was conducted in duplicate. *P<0.05, compared to pLKO.1 Empty vector control; **P<0.01, compared to pLKO.1 Empty vector control; #P<0.05, compared to nonspecific shRNA control. IL, interleukin; SEM, standard error of the mean.
Suppression of DNM3 precludes the maturation of MKs
Late stages of MK development are characterized by polyploidization, cellular enlargement, and the formation of the DMS. Consequently, to investigate the role of DNM3 during late stages of MK development, _DNM3_-specific shRNA lentiviral particles were used to identify changes in ploidy levels, cell size, and DMS formation in treated vs. untreated MKs.
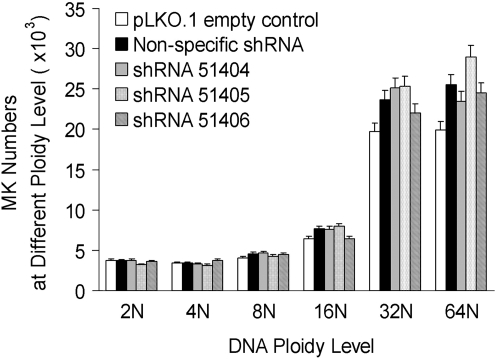
One approach to assess the functional role of DNM3 during late stages of MK development was to use the megakaryocytic cell line UT-7/TPO treated. Cells were treated with a Src-kinase inhibitor SU6656, which induces polyploidization and promotes the release of platelet-sized particles from these cells [34]. Using SU6656-treated UT-7/TPO cells, we found no significant changes in DNA ploidy levels with the knockdown of DNM3 (Fig. 3). However, evidence presented in the following paragraphs implicates DNM3 as having roles in cellular enlargement and DMS formation.
FIG. 3.
DNA ploidy analysis. UT-7/TPO cells treated with SU6656 were transfected with the indicated plasmids as described in the Materials and Methods section. Ploidy level of UT-7/TPO cells treated with SU6656 and transduced with control or DNM3 shRNA vectors. Data represent the mean±SEM of 4 experiments performed in duplicate. No significant changes in DNA ploidy levels were observed with the knockdown of DNM3.
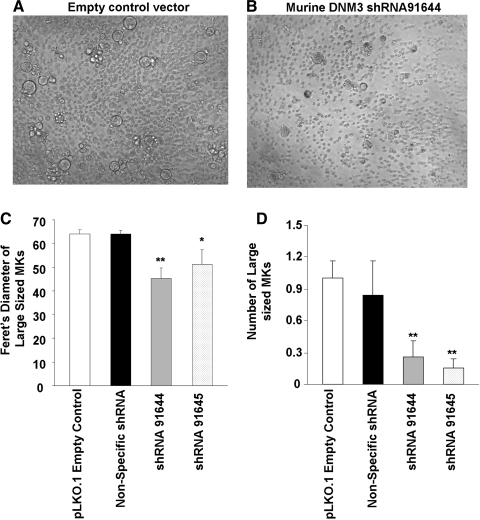
For the next experiment, shRNA clones directed against murine dynamin 3 were first tested to validate their ability to knockdown dynamin 3 in mouse MKs. RT-PCR results showed that both shRNA91644 and shRNA91645 effectively precluded dynamin 3 expression in mouse MKs (data not shown). Use of the murine dynamin 3-specific shRNA lentiviral particles to infect murine bone marrow showed that a knockdown of DNM3 resulted in fewer MKs with large size phenotypes relative to control cultures (Fig. 4A, B). This was evident by visually comparing microscopic images and using the ImageJ software program to quantify cell sizes. As shown in Fig. 4C, the Feret's diameter of cells treated with both murine shRNA91644 and shRNA91645 was significantly lower than for controls. Next, to determine the number of the largest sized MKs present in cultures, we used flow cytometry to gate on cells with the highest forward scatter, a measure of cell size. As shown in Fig. 4D, cultures treated with dynamin 3-specific shRNAs showed a significant decrease in the number of CD41+ cells with a large size phenotype relative to controls (Fig. 4D).
FIG. 4.
Suppression of dynamin 3 reduces the number of large MKs. Murine BM cells were treated with lentiviral vectors that drive expression of shRNA sequences targeted to murine dynamin 3. On day 4, puromycin (5 μg/mL) was added to cultures to select for transduced cells and on day 7 the cells were observed microscopically. (A) Cells treated with an empty vector control. (B) Cells treated with lentiviral construct driving the expression of dynamin 3-specific shRNA. (C) Overall size of cells as determined by using the “Analyze Particles” command in ImageJ to measure the Feret's diameter. (D) Total number of large cells as determined by flow cytometry. Data represent the mean±SEM of 3 experiments. Each experiment was conducted in duplicate. *P<0.05, compared to the controls; **P<0.01, compared to the controls.
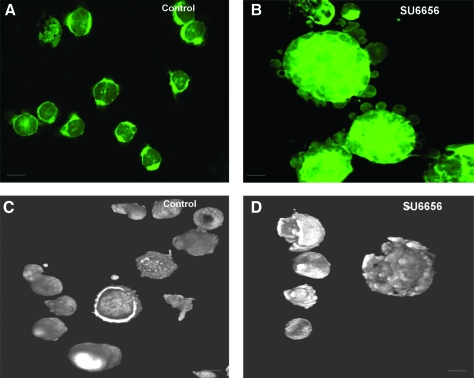
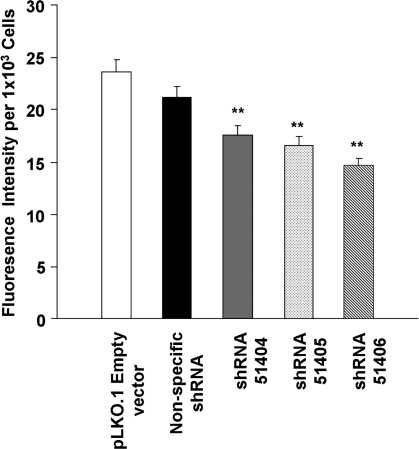
To monitor the formation of the DMS, we used the styrl membrane fluorescence dye, di-8-ANEPPS, which has been successfully shown by others to image and quantify the DMS of rat MKs [35]. Using this dye, we observed that in the absence of SU6656 treatment, UT-7/TPO cells show a peripheral staining pattern that is indicative of cells that do not have extensive plasma membrane invaginations (Fig. 5A, C). In contrast, the staining pattern of UT-7/TPO cells when treated with SU6656 clearly shows a well-developed plasma membrane system with invaginations (Fig. 5B, D). There also appears to be staining of bleb-like structures on the surface of cells that have been treated with SU6656. Since UT-7/TPO cells treated with SU6656 showed a well-developed DMS, these cells were transduced with control and human _DNM3_-specific shRNA constructs and the degree of DMS formation was determine by quantifying the fluorescent intensity of the cells. The results showed that when DNM3 was silenced in SU6656-treated UT-7/TPO cells, there was a significant reduction in di-8-ANEPPS membrane staining relative to empty vector and nonspecific shRNA negative controls (Fig. 6B).
FIG. 5.
UT-7/TPO cells when treated with SU6656 show a well-developed and invaginated plasma membrane system. Representative confocal and three-dimensional images of UT-7/TPO cells that were treated with and without SU6656 and stained with the styrl membrane dye, di-8-ANEPPS. (A and C) Cells not treated with SU6656. (B and D) Cells treated with SU6656. Scale bar represents 10 μm.
FIG. 6.
Functional knockdown of DNM3 precludes the formation of the MK demarcation membrane system. Relative fluorescence intensity of MKs after staining with di-8-ANEPPS. Data represent the mean±SEM of 2 experiments (_n_=6). **P<0.01 when compared to the controls.
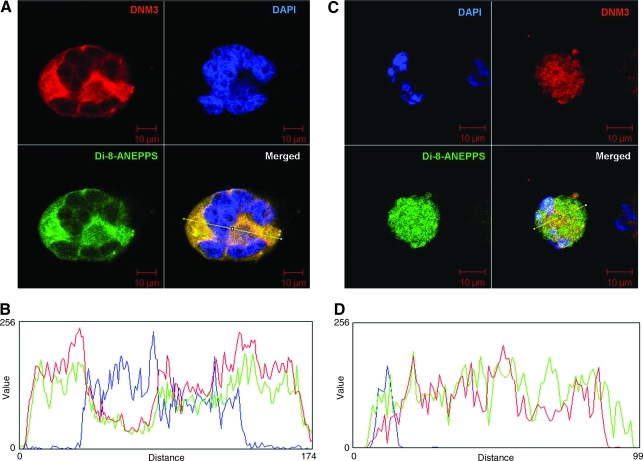
To observe the location of DNM3 relative to the DMS in MKs, we used a DNM3_mCherry fusion construct packaged as a lentiviral vector. Observation of cells infected with DNM3_mCherry vector and stained with di-8-ANEPPS showed a staining pattern that indicated that there is co-localization of the DMS with DNM3 expression in SU6656-treated UT-7/TPO cells (Fig. 7A, B). This staining pattern was also observed in primary culture-derived human MKs (Fig. 7C, D). Analysis with Image J RGB profiler also graphically supports a observation that DNM3 is closely associated with the distribution of DMS in MKs (Fig. 7B, D).
FIG. 7.
DNM3 is closely associated with the distribution of the DMS in MKs. (A) UT-7/TPO cells treated with SU6656 and transfected with a DNM3_mCherry lentiviral fusion vector were stained with DAPI, and the styrl membrane dye, di-8-ANEPPS. (B) UCB CD34+ cells were cultured in MK induction media as described in the Materials and Methods section. On day 7, CD41+ cell were isolated by flow cytometry and the CD41+ cells were cultured for an additional 3 days prior to staining with DAPI and di-8-ANEPPS. (C and D) Graphic depiction of cross-sectional line on merged images. Red, DNM3; green, DMS stained by di-8-AMEPPS; and blue, DAPI. Three independent experiments were performed. DMS, demarcation membrane system.
DNM3 is closely associated with actin and MYH9 in MKs and proplatelets
To identify binding partners of DNM3 during MK development, lysates from UT-7/TPO cells were immunoprecipitated with anti-DNM3 antibody. After running a portion of the eluted fractions on an SDS-polyacrylamide gel, we observed 2 major bands with estimated molecular weights of 200 and 44 kDa (data not shown). The bands were excised and remaining eluates were analyzed by mass spectrometry. We found that the 200 and 40 kDa bands corresponded to MYH9 and actin, respectively.
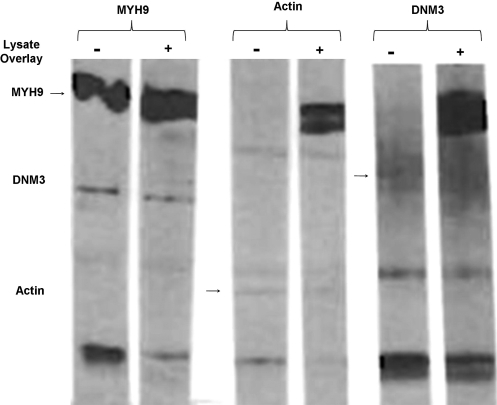
A blot overlay method was used to further investigate a possible association of DNM3 with MYH9. Lysates from UT7-/TPO cells were immunoprecipitated with anti-MYH9 antibody and the immunoprecipitated fraction was resolved by SDS-PAGE electrophoresis, transferred onto PVDF membrane, and each protein lane was cut into individual lanes for subsequent testing. As expected, strong bands were observed for both control and test lanes when membrane strips were treated with anti-MYH9 antibody (Fig. 8). Immunoreactivity with anti-MYH9 in the control lane in which no lysate overlay was performed was because the SDS-PAGE gel was loaded with the MYH-9 immunoprecipitated fraction. On the other hand, when strips were incubated with antibodies to DNM3 and actin, no band was observed for control lanes. Strong immunoreactivity was seen with test lanes in which the strips had been overlaid with lysates from UT7-TPO cells and incubated with anti-DNM3 or anti-actin. Also of interest is that control and test lanes in the overlay blot assay revealed a light band at 44 kDa with anti-actin antibody and a diffuse positive area at 100 kDa when stained with anti-DNM3 antibody (Fig. 8).
FIG. 8.
DNM3 and actin bind to immunoprecipitated MYH9 in UT-7/TPO cells. UT-7/TPO protein lysates were immunoprecipitated with anti-myosin IIA antibody. Eluted fractions were first separated on sodium dodecyl sulfate– polyacrylamide gel electrophoresis followed by transfer of the protein onto polyvinylidene fluoride membrane. After blocking, the protein lanes were cut into strips. For each antibody used in the experiment one lane remained in blocking solution (control), and a second strip for each antibody was incubated in fresh UT-7/TPO lysate (O/L=overlay). All strips were washed with TBS+0.05% Triton and immunostained with indicated antibodies.
Immunostaining was performed to evaluate the cellular localization of DNM3 relative to MYH9. UT-7/TPO cells treated with SU6656 and culture-derived human MKs were co-stained with antibodies to DNM3 and MYH9. Laser confocal microscopy showed that DNM3 is closely associated with the distribution of MYH9 in both UT-7/TPO cells treated with SU6656 and primary human MKs (Fig. 9). Interestingly, the strongest fluorescent intensities for DNM3 and MYH9 staining appear in bud-like structures displayed by the UT-7/TPO cells (Fig. 9A, B). Also, DNM3 and MYH9 expression patterns appear to be co-localized in proplatelet processes displayed by culture-derived MKs from UCB (Fig. 9E, F).
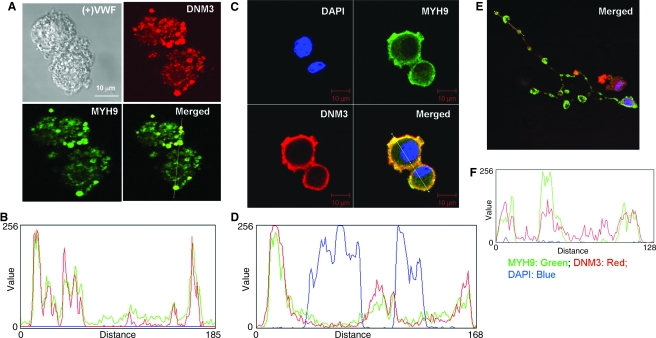
FIG. 9.
Co-localization of DNM3 and MYH9. (A) UT-7/TPO cells treated with SU6656. (B) Histogram of cross-sectional line depicted on merged image of UT-7/TPO cells. (C) UCB CD34+ cells cultured in MK induction media for 7 days, sorted for CD41+ cells, and cultured for an additional 3 days. (D) Histogram of cross-sectional line depicted on merged image of primary cultured-derived MKs. (E) Proplatelet processes extending from primary human cultured-derived MKs. (F) Histogram of cross-sectional line depicted on merged image of proplatelet processes. Red, DNM3; green, MYH9.
Discussion
Genes not previously associated with MK development were identified by comparative microarray expression analyses [27]. Among this list of genes was DNM3, which we verified in a previous study to be up-regulated during MK development [27]. We now report in this study that DNM3 functions at distinct stages of MK development, during the amplification of MK progenitors and during the maturation of MKs.
Evidence that DNM3 functions during a stage of MK development at which time progenitors are amplified is supported by our observations that knockdown of DNM3 in UCB CD34+ cells results in a decrease in the number of CFU-MK colonies as well as a decrease in TNC counts and a decrease in the number of cells that express markers of MK development. These results are consistent with a previous study of ours, in which we report that overexpression of DNM3 in human UCB CD34+ cells causes an increase in the number of CFU-MKs and TNCs [27]. Based on these findings, we suspect that the amplification of MK progenitors is due to DNM3 functioning during the cell division of MK progenitors, specifically during cytokinesis. Precedent for dynamin and dynamin-related protein participation during cell division are available in the literature. Dynamin is likely involved in the regulation of microtubule dynamics during cell division, but the mechanism is yet to be elucidated [37]. Dynamin localizes to cleavage furrow membranes and is required for cytokinesis in a number of organisms [8,38,39] and likely participates in membrane fission and fusion events that facilitate the final separation of dividing cells [37]. Dynamin has also been identified in numerous other RNAi screens to function during cell division [40–42], also suggesting that dynamin may indeed play a key role in coordinating membrane remodeling events that occur during cytokinesis [43].
During late stages of megakaryocytopoiesis before a release of platelets, MKs undergo a terminal maturation process that involves cellular enlargement. As MKs become relatively large (40–100 μm diameter), a massive expansion of their plasma membrane occurs that is estimated to increase the MK plasma membrane surface area 2,600% over a 72 h period [14]. In this study, we observed that not only did a dynamin 3 knockdown cause a significant decrease in the number of large MKs found in cultures, but also the overall size of the large MKs present in the cultures was decreased. A reduction in the total number of large MKs produced in culture is probably due to attenuation in the proliferation of CFU-MKs progenitors. While an explanation for why a dynamin 3 knockdown decreased the overall size of large MKs is not as evident and will require additional studies.
Also, a hallmark feature of MK cytoplasmic maturation is the formation of the DMS, the direct source of platelets [20]. Studies of the DMS have depended upon electron microscopy of fixed specimens. Although these studies have provided significant information on MK ultrastructure, they do not allow studies of the real-time dynamics of morphological responses within the DMS or permit easy quantification of its development. Other studies have shown that styryl dyes such as di-8-ANEPPS and FM 2–10 can rapidly and extensively stain the DMS in unfixed rat MKs [35], which provides an important tool for studying membrane formation during megakaryocytopoiesis and its reorganization during thrombopoiesis. The extracellular-continuous channels formed by the DMS throughout the extranuclear volume of the MK account for the accessibility of demarcation membranes to styryl dyes [35]. Using the styryl membrane dye di-8-ANEPPS, the staining pattern of UT-7/TPO cells treated with SU6656 showed a well-developed DMS. Quantification of membrane staining showed that when knocking-down DNM3 expression that there was a decrease in fluorescent intensity. This suggests that by inhibiting DNM3 expression that the formation of the DMS is precluded.
The mechanism by which DNM3 may participate in MK cytoplasmic enlargement and DMS formation is unknown. However, it is known that PIP2, an important regulator of dynamin function [44–46], plays an instrumental role in facilitating DMS development and platelet biogenesis [20]. The DMS of MKs accumulates PIP2 late in differentiation. In anticipation of platelet release, membranes of the DMS harbor PIP2, and associate with WASp-Arp2/3 complex to induce actin polymerization in preparation for platelet formation [20]. Plasma membrane PIP2 is also essential for mediating the sequential recruitment of adaptor and accessory proteins to sites of clathrin-dependent endocytosis [47]. All 3 dynamin isoforms contain a PH domain that is able to bind PIP2. Binding of PIP2 to dynamin not only strongly increases the GTPase activity of dynamin but may also serve to target dynamin to the plasma membrane [48,49].
Our observation that DNM3 is associated with nonmuscle myosin 2A has important ramifications for the role of DNM3 in MK cytoskeleton rearrangements that involve not only MK development, but also proplatelet formation and platelet biogenesis. Defects in MYH9 are associated with 4 inherited macrothrombocytopenia disorders: the May-Hegglin anomaly, Fechtner syndrome, Sebastian syndrome, and Epstein syndrome [50]. Moreover, defects in MYH9 have been shown to affect different steps of MK development, including the maintenance of cell shape, formation of the DMS, anchorage to extracellular matix, and proplatelet formation [51,52]. Nonmuscle myosin II is a ubiquitous molecular motor that binds and contracts filamentous actin (F-actin) in an ATP-dependent manner [53,54]. The forces that are generated by contraction of the actin-myosin cytoskeleton contribute to a diverse array of cellular processes in a variety of cell types, including cytokinesis [55], platelet activation [56], cell rounding [57], and cell migration [58,59]. There are at least 2 distinct isoforms, IIA and IIB, which are encoded by genes located in different chromosomes [60]. Nonmuscle myosin IIA plays a fundamental role in basic cellular functions such as cell division, adhesion to extracellular matrices, and migration [61]. The myosin IIA heavy chain is encoded by the MYH9 gene and is the only isoform of nonmuscle myosin II present in blood platelets [62].
These studies clearly demonstrate that DNM3 participates at 2 distinct stages of MK development, during MK progenitor proliferation and during the maturation of MKs. These studies also implicate that DNM3 has a possible role in platelet biogenesis. We are currently in the process of developing a DNM3 knockout mouse, which should be instrumental in helping to unravel the mechanisms employed by DNM3 to regulate MK development and platelet biogenesis.
Acknowledgments
This study was supported in part by grants from the Puget Sound Blood Center and NIH grant P50HL081015. We would like to thank Ken Tsubata and Maria Nawrot for their technical assistance, and our biostatistician Doug Bolgiano for the assistance with data analysis. We would also like to thank Dr. Julio Vazquez and Dave Macdonald at the Fred Hutchinson Cancer Research Center for assistance with microscopy.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- 1.Orth JD. McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15:31–39. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 2.Lee E. De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci USA. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto PM. Herskovits JS. Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 4.McNiven MA. Kim L. Krueger EW. Orth JD. Cao H. Wong TW. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krutchen AE. McNiven MA. Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci. 2006;119(Pt 9):1683–1690. doi: 10.1242/jcs.02963. [DOI] [PubMed] [Google Scholar]
- 6.Perrais D. Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Gold ES. Underhill DM. Morrissette NS. Guo J. McNiven MA. Aderem A. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med. 1999;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson HM. Skop AR. Euteneuer U. Meyer BJ. McNiven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12:2111–2117. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman R. Regulation of megakaryocytopoiesis. Blood. 1989;74:1196–1212. [PubMed] [Google Scholar]
- 10.Long MW. Population heterogeneity among cells of the megakaryocyte lineage. Stem Cells. 1993;11:33–40. doi: 10.1002/stem.5530110107. [DOI] [PubMed] [Google Scholar]
- 11.Murphy MJ., Jr Megakaryocyte colony-stimulating factor and thrombopoiesis. Hematol Oncol Clin N Am. 1989;3:465–477. [PubMed] [Google Scholar]
- 12.Williams N. Stimulators of megakaryocyte development and platelet production. Prog Growth Factor Res. 1990;2:81–95. doi: 10.1016/0955-2235(90)90025-f. [DOI] [PubMed] [Google Scholar]
- 13.Vitrat N. Cohen-Solal K. Pique C. Le Couedic JP. Norol F. Larsen AK. Katz A. Vainchenker W. Debili N. Endomitosis of human megakaryocytes are due to abortive mitosis. Blood. 1998;91:3711–3723. [PubMed] [Google Scholar]
- 14.Behnke O. An electron microscope study of the megacaryocyte of the rat bone marrow. I. The development of the demarcation membrane system and the platelet surface coat. J Ultrastruct Res. 1968;24:412–433. doi: 10.1016/s0022-5320(68)80046-2. [DOI] [PubMed] [Google Scholar]
- 15.Cramer EM. Megakaryocyte structure and function. Curr Opin Hematol. 1999;6:354–361. doi: 10.1097/00062752-199909000-00014. [DOI] [PubMed] [Google Scholar]
- 16.Kaushansky K. Thrombopoietin and hematopoietic stem cell development. Ann NY Acad Sci. 1999;872:314–319. doi: 10.1111/j.1749-6632.1999.tb08475.x. [DOI] [PubMed] [Google Scholar]
- 17.Debili N. Hegyi E. Navarro S. Katz A. Mouthon MA. Breton-Gorius J. Vainchenker W. In vitro effects of hematopoietic growth factors on the proliferation, endoreplication, and maturation of human megakaryocytes. Blood. 1991;77:2326–2338. [PubMed] [Google Scholar]
- 18.Gewirtz AM. Human megakaryocytopoiesis. Semin Hematol. 1986;23:27–30. [PubMed] [Google Scholar]
- 19.Mazur EM. Megakaryocytopoiesis and platelet production: a review. Exp Hematol. 1987;15:348–357. [PubMed] [Google Scholar]
- 20.Schulze H. Korpal M. Hurov J. Kim SW. Zhang J. Cantley LC. Graf T. Shivdasani RA. Characterization of the megakaryocyte demarcation membrane system and its role in thrombopoiesis. Blood. 2006;107:3868–3875. doi: 10.1182/blood-2005-07-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulus JM. Bury J. Grosdent JC. Control of platelet territory development in megakaryocytes. Blood Cell. 1979;5:59–88. [PubMed] [Google Scholar]
- 22.Tavassoli M. Megakaryocyte–platelet axis and the process of platelet formation and release. Blood. 1980;55:537–545. [PubMed] [Google Scholar]
- 23.Italiano JE., Jr. Lecine P. Shivdasani RA. Hartwig JH. Blood platelets are assembled principally at the ends of proplatelet processes produced by differentiated megakaryocytes. J Cell Biol. 1999;147:1299–1312. doi: 10.1083/jcb.147.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radley JM. Scurfield G. The mechanism of platelet release. Blood. 1980;56:996–999. [PubMed] [Google Scholar]
- 25.Radley JM. Haller CJ. The demarcation membrane system of the megakaryocyte: a misnomer. Blood. 1982;60:213–219. [PubMed] [Google Scholar]
- 26.Italiano JE., Jr . The structure and production of blood platelets. In: Gresele P, editor; Fuster V, editor; Lopez JA, editor; Page CP, editor; Vermylen J, editor. Platelets in Hematologic and Cardiovascular Disorders: A Clinical Handbook. Cambridge University Press; New York, NY: 2008. pp. 1–10. [Google Scholar]
- 27.Shim MH. Hoover A. Blake N. Drachman JG. Reems JA. Gene expression profile of primary human CD34+CD38lo cells differentiating along the megakaryocyte lineage. Exp Hematol. 2004;32:638–648. doi: 10.1016/j.exphem.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Reems JA. Wang W. Tsubata K. Abdurrahman N. Sundell B. Tijssen MR. van der Schoot E. Di Summa F. Patel-Hett S. Italiano J., Jr. Gilligan DM. Dynamin 3 participates in the growth and development of megakaryocytes. Exp Hematol. 2008;36:1714–1727. doi: 10.1016/j.exphem.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu N. Kunitama M. Yamada M. Hagiwara T. Kato T. Miyazaki H. Eguchi M. Yamamoto M. Miura Y. Establishment and characterization of the thrombopoietin-dependent megakaryocytic cell line, UT-7/TPO. Blood. 1996;87:4552–4560. [PubMed] [Google Scholar]
- 30.Seiji M. Norio K. Jun M. Kouzoh K. Michio M. Yoichi S. Developmental expression of plasminogen activator inhibitor-1 associated with thrombopoietin-dependent megakaryocytic differentiation. Blood. 1999;94:475–482. [PubMed] [Google Scholar]
- 31.Rubinstein P. Dobrila L. Rosenfield RE. Adamson JW. Migliaccio G. Migliaccio AR. Taylor PE. Stevens CE. Processing and cryopreservation of placental/umbilical cord blood for unrelated bone marrow reconstitution. Proc Natl Acad Sci USA. 1995;92:10119–10122. doi: 10.1073/pnas.92.22.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini L. Blömer U. Gallay P. Ory D. Mulligan R. Gage FH. Verma IM. Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 33.O'Doherty U. Swiggard WJ. Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lannutti BJ. Blake N. Gandhi MJ. Reems JA. Drachman JG. Induction of polyploidization in leukemic cell lines and primary bone marrow by Src kinase inhibitor SU6656. Blood. 2005;105:3875–3878. doi: 10.1182/blood-2004-10-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahaut-Smith MP. Thomas D. Higham AB. Usher-Smith JA. Hussain JF. Martinez-Pinna J. Skepper JN. Mason MJ. Properties of the demarcation membrane system in living rat megakaryocytes. Biophys J. 2003;84:2646–2654. doi: 10.1016/S0006-3495(03)75070-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmittgen TD. Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 37.Konopka CA. Schleede JB. Skop AR. Bednarek SY. Dynamin and cytokinesis. Traffic. 2006;7:239–247. doi: 10.1111/j.1600-0854.2006.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wienke DC. Knetsch MLW. Neuhaus EM. Reedy MC. Manstein DJ. Disruption of a dynamin homologue affects endocytosis, organelle morphology and cytokinesis in Dictyostelium discoideum. Mol Biol Cell. 1999;10:225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng B. Schwarz H. Jesuthasan S. Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp Cell Res. 2002;279:14–20. doi: 10.1006/excr.2002.5579. [DOI] [PubMed] [Google Scholar]
- 40.Eggert US. Kiger AA. Richter C. Perlman ZE. Perrimon N. Mitchison TJ. Field CM. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:2135–2143. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Echard A. Hickson GR. Foley E. O'Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiger AA. Baum B. Jones S. Jones MR. Coulson A. Echeverri C. Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonner MK. Skop AR. Cell division screens and dynamin. Biochem Soc Trans. 2008;36(Pt 3):431–435. doi: 10.1042/BST0360431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Achiriloaie M. Barylko B. Albanesi JP. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol Cell Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barylko B. Binns D. Lin KM. Atkinson MA. Jameson DM. Yin HL. Albanesi JP. Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J Biol Chem. 1998;273:3791–3797. doi: 10.1074/jbc.273.6.3791. [DOI] [PubMed] [Google Scholar]
- 46.Klein DE. Lee A. Frank DW. Marks MS. Lemmon MA. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 47.Cremona O. de Camilli P. Phosphoinositides in membrane traffic at the synapse. J Cell Sci. 2001;114(Pt 6):1041–1052. doi: 10.1242/jcs.114.6.1041. [DOI] [PubMed] [Google Scholar]
- 48.Schmid SL. McNiven MA. De Camilli P. Dynamin and its partners: a progress report. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- 49.Hinshaw JE. Dynamin, and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seri M. Pecci A. Di Bari F. Cusano R. Savino M. Panza E. Nigro A. Noris P. Gangarossa S. Rocca B. Gresele P. Bizzaro N. Malatesta P. Koivisto PA. Longo I. Musso R. Pecoraro C. Iolascon A. Magrini U. Rodriguez Soriano J. Renieri A. Ghiggeri GM. Ravazzolo R. Balduini CL. Savoia A. MYH9-related disease: May-Hegglin anomaly, Sebastian syndrome, Fechtner syndrome, and Epstein syndrome are not distinct entities but represent a variable expression of a single illness. Medicine (Baltimore) 2003;82:203–215. doi: 10.1097/01.md.0000076006.64510.5c. [DOI] [PubMed] [Google Scholar]
- 51.Eckly A. Strassel C. Freund M. Cazenave JP. Lanza F. Gachet C. Léon C. Abnormal megakaryocyte morphology and proplatelet formation in mice with megakaryocyte-restricted MYH9 inactivation. Blood. 2009;113:3182–3189. doi: 10.1182/blood-2008-06-164061. [DOI] [PubMed] [Google Scholar]
- 52.Eckly A. Rinckel JY. Laeuffer P. Cazenave JP. Lanza F. Gachet C. Léon C. Proplatelet formation deficit and megakaryocyte death contribute to thrombocytopenia in Myh9 knockout mice. J Thromb Haemost. 2010;8:2243–2251. doi: 10.1111/j.1538-7836.2010.04009.x. [DOI] [PubMed] [Google Scholar]
- 53.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 54.Bresnick AR. Molecular mechanisms of nonmuscle myosin-II regulation. Curr Opin Cell Biol. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- 55.Satterwhite LL. Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- 56.Daniel JL. Molish IR. Rigmaiden M. Stewart G. Evidence for a role of myosin phosphorylation in the initiation of the platelet shape change response. J Biol Chem. 1984;259:9826–9831. [PubMed] [Google Scholar]
- 57.Majumdar M. Seasholtz TM. Goldstein D. de Lanerolle P. Brown JH. Requirement for Rho-mediated myosin light chain phosphorylation in thrombin-stimulated cell rounding and its dissociation from mitogenesis. J Biol Chem. 1998;273:10099–10106. doi: 10.1074/jbc.273.17.10099. [DOI] [PubMed] [Google Scholar]
- 58.Warrick HM. Spudich JA. Myosin structure and function in cell motility. Annu Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- 59.Wessels D. Soll DR. Knecht D. Loomis WF. De Lozanne A. Spudich J. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev Biol. 1988;128:164–177. doi: 10.1016/0012-1606(88)90279-5. [DOI] [PubMed] [Google Scholar]
- 60.Simons M. Wang M. McBride OW. Kawamoto S. Yamakawa K. Gdula D. Adelstein RS. Weir L. Human non-muscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69:530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- 61.Conti MA. Adelstein RS. Non-muscle myosin II moves in new directions. J Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- 62.Murakami N. Mehta P. Elzinga M. Studies on the distribution of cellular myosin with antibodies to isoform-specific synthetic peptides. FEBS Lett. 1991;288:247. [PubMed] [Google Scholar]