Immune-related MicroRNAs are Abundant in Breast Milk Exosomes (original) (raw)
Abstract
Breast milk is a complex liquid rich in immunological components that affect the development of the infant's immune system. Exosomes are membranous vesicles of endocytic origin that are found in various body fluids and that can mediate intercellular communication. MicroRNAs (miRNAs), a well-defined group of non-coding small RNAs, are packaged inside exosomes in human breast milk. Here, we identified 602 unique miRNAs originating from 452 miRNA precursors (pre-miRNAs) in human breast milk exosomes using deep sequencing technology. We found that, out of 87 well-characterized immune-related pre-miRNAs, 59 (67.82%) are presented and enriched in breast milk exosomes (P < 10-16, _χ_2 test). In addition, compared with exogenous synthetic miRNAs, these endogenous immune-related miRNAs are more resistant to relatively harsh conditions. It is, therefore, tempting to speculate that these exosomal miRNAs are transferred from the mother's milk to the infant via the digestive tract, and that they play a critical role in the development of the infant immune system.
Keywords: breast milk, exosome, immune-related miRNAs, deep sequencing.
Introduction
The past decade has seen a large increase in the list of immunological components detected in human breast milk 1. Large quantities of secretory immunoglobulins (particularly IgA), measurable levels of leukocytes, as well as antimicrobial factors (such as lysozyme, lactoferrin and oligosaccharides) are abundant in breast milk 2. These immune-related factors not only provide passive immunity to infants, but could also prompt and modulate the development of the infant's own immune system.
Exosomes are tiny endosome-derived membrane vesicles (~30-100 nm in diameter) that are released into the extracellular environment from a variety of different cells 3, 4. Exosomes are present in various body fluids (such as amniotic fluid, blood, malignant ascites fluid, milk, saliva and urine) and contain distinct subsets of microRNAs (miRNAs) 5. miRNAs, a large family of ~22 nucleotide (nt) non-coding small RNAs, are derived from ~70 nt long stem-loop precursors (pre-miRNAs), have emerged as key post-transcriptional regulators of gene expression and play important roles in a wide range of physiological and pathological processes 6. Circulating miRNA levels have been found to be elevated in diseased states, such as ovarian cancer, lung cancer and melanoma 7-9. The finding that miRNAs are present in exosomes triggered the hypothesis that extracellular miRNAs may function in cell-cell communication 4, 10.
In the present study, we investigated the miRNA transcriptome of exosomes in human breast milk using deep sequencing technology to elucidate the distribution and expression profile of immune-related miRNAs. Particular emphasis was placed on the resistance and stability of breast milk miRNAs under different harsh conditions.
Materials and Methods
Ethics statement
All women involved in the study gave their signed informed consent to participate. The study was approved by the ethics committees of Ya'an Vocational College and the College of Animal Science and Technology, Sichuan Agricultural University.
Sample collection
Human breast milk samples (20-50 ml) were collected into sterile tubes using a manual breast pump from four healthy women (30 ± 0.9 years old, primiparity) when their infants were aged 60 days. Smoking or medication during pregnancy, known autoimmune disorders, diabetes, or allergic asthma excluded women from participation in this study. All samples were stored at -80 °C until analyzed. All mothers had full term vaginal deliveries of healthy, normal birth-weight infants.
Enrichment of exosomal RNA in breast milk
Raw milk (1.5 ml) was centrifuged at 2,000 × g for 10 min to remove fat globules. The supernatant was centrifuged at 12,000 × g for 30 min and further filtered through a 0.45 μm PVDF filter to eliminate cells and cellular debris. Approximately 1 ml of supernatant was mixed with 500 µl of ExoQuick Exosome Precipitation Solution (SBI, CA, USA) and incubated at 4 °C for 12 hours. The ExoQuick/supernatant mixture was centrifuged at 1,500 × g for 30 min to obtain a beige exosome pellet, which was then re-suspended in 250 µl nuclease-free water. Total RNA from exosomes was extracted using TRIzol-LS (Invitrogen, CA, USA) following the manufacturer's instructions. Small RNAs were analyzed with the Agilent Bioanalyzer 2100 and the RNA 6000 Nano LabChip Kit (Agilent, CA, USA).
Small RNA sequencing
Total RNA isolated from each woman's milk was used for library construction and subjected to single-end sequencing in 36 bp reads using an Illumina Genome Analyzer II. The bioinformatics pipeline for miRNA discovery and profiling was carried out as previously described 11, 12. In brief, the raw reads that passed through a series of filters (such as the length and sequence comparison) were called “mappable reads”. All mappable reads were counted and the identical reads were combined into a single kind, and then were mapped to the 1,424 human pre-miRNAs registered in miRBase 17.0 (April 2011) with no more than one mismatch. As our previous reports 11, 12, the most abundant mature variant of a given miRNA was chosen as a reference sequence, which provides the most robust approach for evaluation of expression level. The small RNA sequence data from this study have been submitted to the NCBI Gene Expression Omnibus under accession no. GSE32253.
Harsh treatments of miRNAs and quantitative PCR (q-PCR)
Three exogenous miRNAs (ath-miR-159a-3p of A. thaliana, cel-lin-4-5p and cel-miR-2-3p of C. elegans), which have no sequence similarity to human miRNAs, were chemically synthesized. To determine the resistance and stability of exogenous miRNAs, ath-miR-159a-3p was added directly to raw milk and subjected to four types of treatment: (1) incubated at 26 °C for 0.5, 1, 2, 4, 8 and 24 hours; (2) subjected to six freeze-thaw cycles at -20 °C; (3) treated with RNase A (0.16 µg·µl-1) and RNase T1 (0.4 U·µl-1) (Fermentas, Shenzhen, China) for 60 min at 37 °C; and (4) incubated at 100 °C for 10 min. After these treatments, in order to normalize inter-sample variation in the RNA isolation step, cel-lin-4-5p and cel-miR-2-3p were added to the raw milk after the addition of TRIzol-LS (Invitrogen, CA, USA), which contains RNase inhibitors.
EvaGreen-based q-PCR was performed with a High-Specificity miRNA qRT-PCR Detection Kit (Stratagene, CA, USA) on the CFX96™ Real-Time PCR Detection System (Bio-Rad, CA, USA). The forward primers of the three exogenous and ten endogenous miRNAs were identical in sequence and length to the miRNAs themselves. The ΔΔCt method was used to determine expression level differences between surveyed samples. Each miRNA of each sample was analyzed in triplicate.
Results and Discussion
An overview of exosomal miRNAs in milk
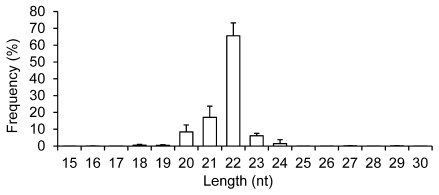
Four exosomal small RNA libraries were generated from the milk of four women and each library was individually sequenced. This generated ~21.59 million (M) 36 nt reads. The four libraries produced ~86.37 M raw reads and of these, 96.70% (~83.52 M) passed through filters for sequence patterns (simple sequence compositions) of length, copy number and other known RNA classes, and are referred to as “mappable reads” 11, 12. Similar to previous studies on different tissue types 11, 12, the majority of the small RNAs in breast milk exosomes are 20-24 nt in size, with the predominant species being 22 nt in length (65.56 ± 7.65%), which is typical of Dicer-processed products (Fig. 1).
Figure 1.
Length distribution and frequency percent of mappable reads. Values are means ± s.d.
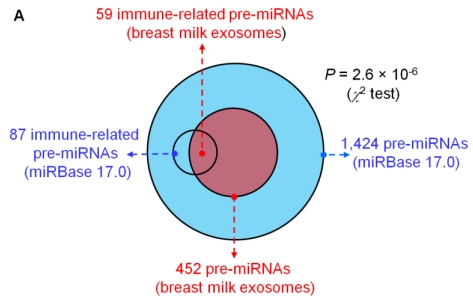
Analysis of the mapped human pre-miRNAs indicates that 452 of 1,424 (~31.74%) known human pre-miRNAs deposited in the latest version of miRBase 17.0 were detected in the four exosomal small RNA libraries (Fig. 2A). Predictably, we detected identical mature miRNA sequences that originate from distinct pre-miRNAs; 639 mature miRNAs originating from 452 pre-miRNAs were identified, corresponding to 602 unique miRNAs.
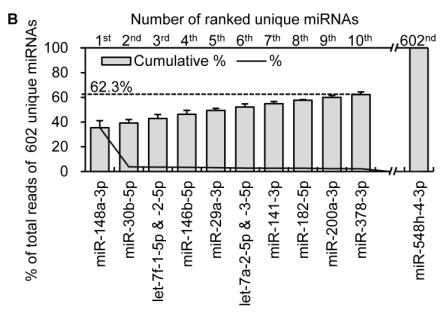
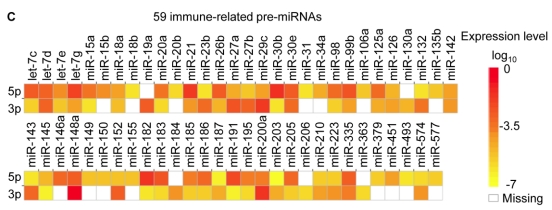
Figure 2.
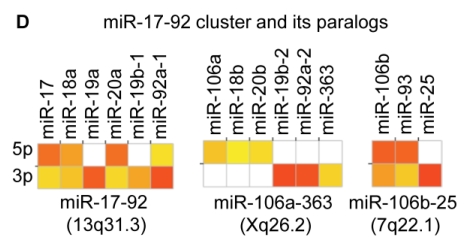
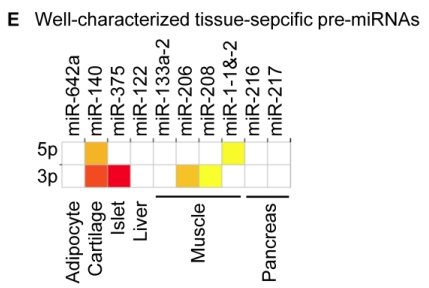
Distribution and characterization of miRNAs in breast milk exosomes. (A) Distribution of pre-miRNAs. Out of 1,424 pre-miRNAs deposited in miRBase 17.0 (blue circle), 87 (6.11%) pre-miRNAs have been designated as immune-related pre-miRNAs based on annotation in the Pathway Central database (SABiosciences, MD, USA). Immune-related pre-miRNAs (59 of 452, 13.05%) are enriched in breast milk exosomes (red circle). _χ_2 test: Number of immune-related miRNAs and others detected in breast milk exosomes compared with the total entries in miRBase 17.0. (B) Top ten most highly expressed unique miRNAs. Plot of the unique miRNAs (starting from the miRNA with the highest counts, x-axis) versus their cumulative % of the total counts from 602 unique miRNAs detected in breast milk exosomes for each small RNA library. Values are means ± s.d. The dashed horizontal line at 62.3% represents the level of the top ten unique miRNAs and the % of individual miRNAs is marked by the gray line. (_C_-E) The expression profile of selected miRNAs in breast milk exosomes. (C) 59 immune-related pre-miRNAs; (D) miR-17-92 cluster and paralogous clusters, and (E) ten well-characterized tissue-specific pre-miRNAs.
The biological replicates were highly correlated with each other (average r = 0.94) (Table 1), which suggested experimental reliability and further highlighted the low variation in exosomal miRNA profiles in breast milk across different healthy individuals.
Table 1.
Pearson's correlation of the counts of 602 unique miRNAs among four biological replicates. (**: P < 10-16)
| Sample No. | 2 | 3 | 4 |
|---|---|---|---|
| 1 | 0.93** | 0.95** | 0.89** |
| 2 | 0.98** | 0.94** | |
| 3 | 0.96** |
Immune-related miRNAs are enriched in breast milk exosomes
The miRNA transcriptome in breast milk exosomes consists of unevenly distributed counts of reads, varying from millions of reads for the highest expression to single count miRNAs. The ten most highly expressed unique miRNAs account for 62.3 % of the total counts for all 602 unique miRNAs (Fig. 2B).
Notably, four of the top ten miRNAs (1st: miR-148a-3p; 2nd: miR-30b-5p; 8th: miR-182-5p; and 9th: miR-200a-3p) are designated as immune-related pre-miRNAs based on the annotation of the Pathway Central database (SABiosciences, MD, USA). miR-30b-5p promotes cellular invasion and immunosuppression 13; miR-182-5p is induced by IL-2 (interleukin-2) and promotes T cell-mediated immune responses 14; and miR-200a-3p is associated with Hodgkin lymphoma 15 and oral cancers 16. Specifically, miR-148a-3p was the most highly expressed miRNA over all four libraries and, on average, accounts for ~35.45 % of the total counts of all 602 unique miRNAs. This is consistent with a previous report showing that miR-148a-3p was highly expressed in bovine milk and was identified as a potential biomarker for the quality control of raw milk and other milk-related products 17. Intriguingly, direct protein coding gene targets of miR-148a-3p so far reported include TGIF2 (TGFB-induced factor homeobox 2), which is overexpressed in ovarian cancer 18 and PXR (pregnane X receptor), a major transcription factor regulating the inducible expression of a variety of transporters and drug-metabolizing enzymes 19. Interestingly, DNMT3B (DNA methyltransferase 3b), which encodes a de novo methyltransferase that sets up DNA methylation patterns early in development, is another known target of miR-148a 20. A recent study showed that the exogenous plant miRNAs in food are present in the sera and tissues of various animals, and can regulate the expression of target genes in mammals 21. Therefore, we tentatively speculate that the enrichment of these highly expressed miRNAs in human breast milk lead to a post-transcriptional repression of their target genes in cells of the infant digestive tract.
Recent reports indicate that other miRNAs in the top ten are also related to various pathological responses. For example, miR-29a-3p suppresses immune responses to intracellular pathogens by targeting interferon-γ 22; plasma miR-141-3p was identified as a potential biomarker for colon cancers 23; miR-378-3p acts as a molecular switch involved in breast cancer cell metabolism via interference with a well-integrated bioenergetics transcriptional pathway 24, and miR-146b-5p targets NF-κB signaling in innate immune responses 25.
In addition, two of the top ten miRNAs are from the ubiquitous let-7-family of miRNAs (let-7f-1-5p & -2-5p, and let-7a-2-5p & -3-5p), which are key miRNA regulators in development that are present in abundance across various species, including mammals, flies, worms and plants 26.
The distribution of pre-miRNAs in breast milk exosomes shows that more than half of the immune-related pre-miRNAs (59 of 87, 67.82%) are present and enriched in breast milk exosomes (P < 10-16, _χ_2 test) (Fig. 2A, C), which is consistent with previous studies 27. Interestingly, six pre-miRNAs from the miR-17-92 cluster and nine pre-miRNAs from paralogous clusters were both abundant in breast milk exosomes (Fig. 2D). In addition to being well-known oncogenes, recent studies establish essential roles for the miR-17-92 and paralogous clusters in development of the immune system 28. In contrast, tissue-specific miRNAs were generally less abundant in breast milk exosomes (Fig. 2E).
Previous reports suggested that exosomes play a significant role in intercellular communication by transferring miRNAs to neighboring cells 4, 10. Although the molecular mechanism underlying this phenomenon is still unclear, it is intriguing to speculate that the these immune-related miRNAs in milk exosomes may be transferred into the infant body via the digestive tract, and may play a critical role in the development of the immune system in infants.
Resistance to degradation and stability of breast milk miRNAs
We next sought to investigate the stability of miRNAs in breast milk, given that it is an important prerequisite for our hypothesis that these exosomal miRNAs are transferable genetic material from mother to infant.
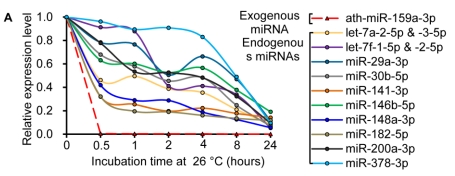
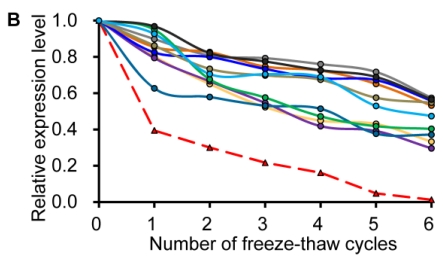
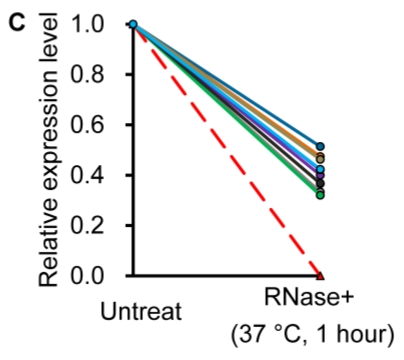
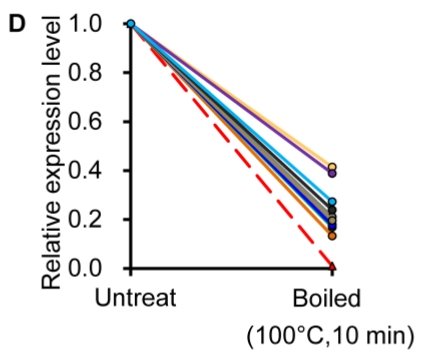
Similar to previous studies on blood 29, 30, the exogenous synthetic miRNAs were rapidly degraded, whereas endogenous miRNAs remain stable when milk is subjected to prolonged room temperature incubation (Fig. 3A) or freeze-thawed multiple times (Fig. 3B). Furthermore, compared with the complete degradation of exogenous miRNAs by treatment with RNase (Fig. 3C) or by incubation at 100°C for 10 min (Fig. 3D), the endogenous miRNAs are resistant to a certain extent to the harsh conditions.
Figure 3.
Characterization of breast milk miRNAs. The expression changes of a spiked-in C. elegans miRNA and of the top ten most highly expressed unique miRNAs from the small RNA libraries across various harsh conditions. Total RNA was extracted and then analyzed by q-PCR. Breast milk was (A) incubated at 26°C for 0.5, 1, 2, 4, 8 or 24 hours, (B) subjected to six freeze-thaw cycles at -20°C, (C) treated with RNase A and T1 for 60 min at 37°C, and (D) incubated at 100°C for 10 min.
Conclusion
The present study provides a close look at the immune-related miRNAs which are enriched in breast milk exosomes, and evidence that these exosomal miRNAs are resistant to general harsh conditions, such as prolonged room temperature incubation, multiple freeze-thaw, RNase digestion and even boiling. We tentatively propose that these exosomal miRNAs are transferable genetic material from mother to infant, and are essential for the development of the immune system in infants.
Acknowledgments
This work was supported by grants from the National Special Foundation for Transgenic Species of China (2009ZX08009-155B), the National Natural Science Foundation of China (30901024) and the International Science & Technology Cooperation Program of China (2011DFB30340). We thank Chris Heble for critically reading of the manuscript.
References
- 1.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R. et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 2.Petherick A. Development: mother's milk: a rich opportunity. Nature. 2010;468:S5–7. doi: 10.1038/468S5a. [DOI] [PubMed] [Google Scholar]
- 3.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 4.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH. et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Ba Y, Ma L, Cai X, Yin Y. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 8.Michael A, Bajracharya SD, Yuen PST, Zhou H, Star RA. et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N. et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Biochem. 2009;55:1977. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstr m K, Bossios A, Sj strand M, Lee JJ. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Liu Y, Wang T, Guan J, Luo Z. et al. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int J Biol Sci. 2011;7:1045–1055. doi: 10.7150/ijbs.7.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Xia Y, Gu Y, Zhang K, Lang Q. et al. MicroRNAome of porcine pre- and postnatal development. PLoS One. 2010;5:e11541. doi: 10.1371/journal.pone.0011541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaziel-Sovran A, Segura M, Di Micco R, Collins M, Hanniford D. et al. MiR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stittrich AB, Haftmann C, Sgouroudis E, Kühl AA, Hegazy AN. et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 15.Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A. et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 16.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA. et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Gao C, Li H, Huang L, Sun Q. et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- 18.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M. et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by microRNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 20.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Hou D, Chen X, Li D, Zhu L. et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2011;21:1–20. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma F, Xu S, Liu X, Zhang Q, Xu X. et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 23.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ. et al. Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M. et al. miR-378* mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D. et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]