Rapid degradation of cyclooxygenase-1 and hematopoietic prostaglandin D synthase through ubiquitin–proteasome system in response to intracellular calcium level (original) (raw)
Cyclooxygenase-1 and hematopoietic prostaglandin D synthase proteins were rapidly degraded within 30 and 60 min, respectively, after an increase in the intracellular calcium level in human megakaryocytic MEG-01 cells.
Abstract
Cyclooxygenase (COX)-1 and hematopoietic prostaglandin (PG) D synthase (H-PGDS) proteins, which are both involved in the arachidonate cascade, were stable in human megakaryocytic MEG-01 cells. In contrast, once the intracellular calcium level was increased by treatment with a calcium ionophore, both protein levels rapidly decreased with a half-life of less than 30 and 120 min for COX-1 and H-PGDS, respectively. In the presence of a proteasome inhibitor, COX-1 and H-PGDS proteins accumulated within 10 and 30 min, respectively, and concurrently appeared as the high-molecular-mass ubiquitinated proteins within 30 and 60 min, respectively, after an increase in the intracellular calcium level. The ubiquitination of these proteins was also observed when ADP, instead of a calcium ionophore, was used as an inducer to elevate the intracellular calcium level. When the entry of calcium ion into the cells was inhibited by ethylene glycol tetraacetic acid (EGTA), the ubiquitination of COX-1 and H-PGDS was clearly suppressed; and the addition of CaCl2 to the medium cleared the EGTA-mediated suppression of the ubiquitination. These results indicate that COX-1 and H-PGDS were rapidly ubiquitinated and degraded through the ubiquitin–proteasome system in response to the elevation of the intracellular calcium level.
INTRODUCTION
The production of prostaglandins (PGs) is restricted to certain areas, where they function as local hormones with short half-lives and play a number of physiological roles. PGs are synthesized through the following three enzymatic steps: 1) cytosolic phospholipase A2 (cPLA2) releases arachidonic acid from membrane phospholipids; 2) the arachidonic acid is then converted to PGH2 by the action of either cyclooxygenase (COX)-1 or COX-2; and 3) PGH2, the common precursor of all prostanoids, is converted to various PGs, including PGD2, by the specific terminal PG synthases (Urade and Hayaishi, 2000a, 2000b; Helliwell et al., 2004; Ueno et al., 2005).
COXs are the rate-limiting enzymes in the arachidonate cascade (Smith et al., 2000; Kang et al., 2007). COX-1 and COX-2 are highly homologous, but their expression profiles are quite different. COX-1 is constitutively expressed in a variety of cells, and its expression in megakaryocytic cells is enhanced by 12-_O_-tetradecanoylphorbol-13-acetate (TPA). In contrast, COX-2 expression is inducible in response to various stimuli, such as proinflammatory cytokines (Smith et al., 2000; Kang et al., 2007). The transcriptional regulatory mechanisms of these COXs have been well investigated (Kang et al., 2007). Posttranscriptional regulation of COX-2, but not that of COX-1, has been well studied. COX-2 mRNA is rapidly degraded through AU-rich, sequence element–mediated degradation (Dixon, 2004). Moreover, the COX-2 protein is degraded through the ubiquitin–proteasome system via the endoplasmic reticulum (ER)-associated degradation system (Mbonye et al., 2006, 2008; Mbonye and Song, 2009).
PGD2 is produced in a variety of cells by two distinct types of PGD synthases (PGDS; EC 5.3.99.2; Urade and Hayaishi, 2000a). One is the lipocalin-type PGDS, and the other is hematopoietic PGDS (H-PGDS). H-PGDS is the 26-kDa cytosolic enzyme responsible for the glutathione-dependent biosynthesis of PGD2 and is the only vertebrate member of the sigma class of cytosolic glutathione _S_-transferases (Kanaoka and Urade, 2003). It is also called GSTS1 (Hayes et al., 2005). H-PGDS has been isolated from human, mouse, rat, and chicken sources, and its molecular properties have been extensively studied (Kanaoka et al., 1997; Thomson et al., 1998; Kanaoka et al., 2000). H-PGDS is expressed in antigen-presenting cells (Urade et al., 1989), megakaryocytes (Fujimori et al., 2000), type 2 helper T lymphocytes (Tanaka et al., 2000), microglia (Mohri et al., 2003), and dendritic cells (Shimura et al., 2010). Moreover, H-PGDS acts as a mediator of allergies (Yamamoto et al., 2009), suppresses intestinal adenomas in ApcMin/+ mice (Park et al., 2007), and shows enhanced expression and involvement in the progression of muscle necrosis (Mohri et al., 2009), which is inhibited by HQL-79, a potential inhibitor of H-PGDS (Tanaka et al., 2011). Thus H-PGDS inhibitors are considered to be useful in suppressing several diseases, including neurodegenerative diseases and muscular dystrophy (Mohri et al., 2006, 2009). The H-PGDS mRNA level is increased through the Oct-1 and AP-2 elements by TPA in megakaryocytes (Fujimori et al., 2000). Moreover, H-PGDS is translocated from the cytoplasm to the ER to produce PGD2 when the intracellular calcium level is increased (Ueno et al., 2001). However, the posttranslational regulation of COX-1 and H-PGDS proteins has never been identified.
In this study, we show the rapid degradation of COX-1 and H-PGDS proteins through the ubiquitin–proteasome system in response to the elevation of the intracellular calcium level in human megakaryocytic MEG-01 cells. This is the first identification of the ubiquitination-dependent degradation of COX-1 and H-PGDS as a terminal PG synthase involved in the arachidonate cascade.
RESULTS
Rapid decrease in COX-1 and H-PGDS protein levels in response to an increase in the intracellular calcium level
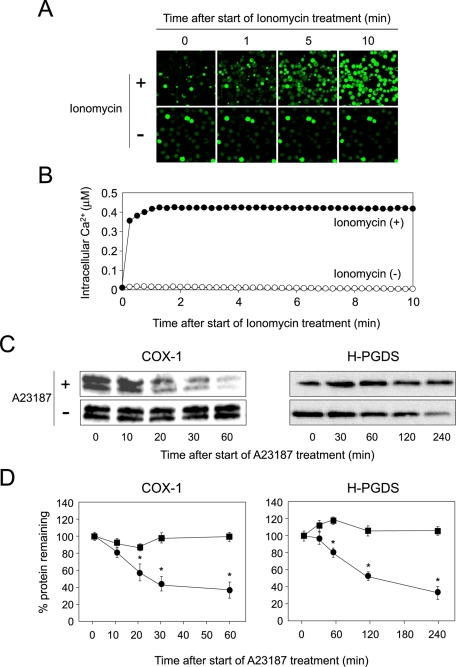
COX-1 is localized in the membranes of the ER and nucleus (Smith et al., 2000), while H-PGDS is translocated from the cytosol to the ER membranes by treatment with A23187 (Ueno et al., 2001), indicating that H-PGDS protein is translocated to the ER membranes in response to elevation of intracellular calcium level. We first measured the intracellular calcium level of human megakaryocytic MEG-01 cells after having treated them with ionomycin. We used ionomycin instead of A23187, because A23187 displays autofluorescence and is thus unsuitable for use in fluorescence analysis. MEG-01 cells were cultured with TPA for 16 h and then incubated with ionomycin and Fluo 4-AM, which binds Ca2+. Fluorescence microscopy was used to track the time course of Fluo 4-AM–derived fluorescence after the start of treatment with ionomycin. Intracellular calcium concentration quickly increased, and its level was further enhanced 10 min after the start of treatment with ionomycin (Figure 1, A and B). However, when cells were not treated with the ionophore, no apparent increase in the intracellular calcium level was detected (Figure 1, A and B).
FIGURE 1:
Calcium-dependent degradation of COX-1 and H-PGDS proteins. (A) Increasing the intracellular calcium level in MEG-01 cells. Data are representative of three independent experiments. (B) Measurement of intracellular calcium level in MEG-01 cells. Ionomycin: closed circles; vehicle: open circles. Data are representative of three independent experiments. (C) Stability of COX-1 and H-PGDS proteins in MEG-01 cells. Cells were cultured with CHX (10 μM; Sigma) for 15 min prior to addition of A23187 and CHX. Data are representative of three independent experiments. (D) The intensities of immunoreactive bands. The value at 0 min was shown as 100% of protein remaining. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with A23187(−). A23187(−): closed squares; A23187(+): closed circles.
To examine the stability of COX-1 and H-PGDS proteins in MEG-01 cells, we cultured cells for 16 h in medium containing TPA and then incubated them for various times with cycloheximide (CHX) in the presence or absence of A23187. The levels of both proteins were determined by immunoprecipitation and Western blot analysis. The COX-1 protein in MEG-01 cells was stable for at least 60 min (Figure 1, C and D, left). However, once the intracellular calcium level became elevated, the COX-1 protein level rapidly decreased, with a half-life of <30 min (Figure 1, C and D, left).
The H-PGDS protein in the cells was stable for at least 240 min (Figure 1, C and D, right). However, when the intracellular calcium level was increased by treatment with A23187, the level of H-PGDS protein rapidly decreased, with a half-life of <120 min (Figure 1, C and D, right). These results, taken together, indicate that both COX-1 and H-PGDS proteins were stable at the normal intracellular calcium level, but were rapidly degraded once the intracellular calcium level had increased in the MEG-01 cells.
Accumulation and ubiquitination of COX-1 and H-PGDS in response to the increase in the intracellular calcium level
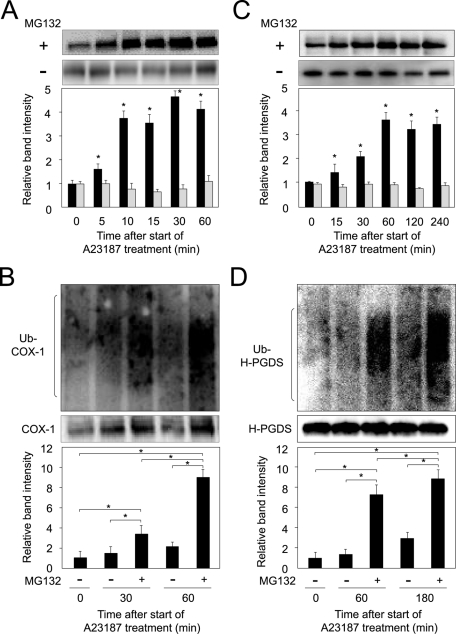
To investigate whether the COX-1 and H-PGDS protein levels were decreased due to proteasomal degradation, we incubated MEG-01 cells with A23187 for 60 or 240 min with or without MG132, a proteasome inhibitor that irreversibly inhibits the 20S proteasome and causes the accumulation of ubiquitinated proteins normally degraded by the proteasome system (Welchman et al., 2005).
Accumulated COX-1 protein was detected at 10 min after initiation of treatment with A23187 and MG132, and its level increased in a time-dependent manner (Figure 2A), suggesting that COX-1 is a likely substrate of the ubiquitin–proteasome complex, whereas COX-1 did not accumulate when cells were cultured in medium containing A23187 alone (Figure 2A); these results are consistent with previous results (Moraitis and Giguere, 2003; Mishra et al., 2009). To further investigate COX-1 degradation through the ubiquitin–proteasome system, we examined the time dependence of the effect of MG132 on the ubiquitination of the COX-1 protein in the cells. When cells were incubated with MG132 and A23187 for 0, 30, or 60 min, a high-molecular-weight smeared mass of the ubiquitinated COX-1 protein was detected at 30 min, and it increased in a time-dependent manner after an increase in the intracellular calcium level (Figure 2B). No high-molecular-weight smeared mass of ubiquitinated COX-1 protein was detected at 15 min after the elevation of the intracellular calcium level (unpublished data).
FIGURE 2:
Rapid ubiquitination of COX-1 and H-PGDS. (A) Accumulation of COX-1 in response to an increase in the intracellular calcium level was dependent upon the incubation time with MG132. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with before treatment (0 min). MG132(−): gray columns; MG132(+): black columns. (B) Ubiquitin-dependent degradation of COX-1 (Ub-COX-1). High-molecular-weight smeared signals were measured. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets. (C) Accumulation of H-PGDS was dependent on the incubation time with MG132. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with before treatment (0 min). MG132(−): gray columns; MG132(+): black columns. (D) Time dependence of the change in the ubiquitinated H-PGDS (Ub-H-PGDS) level in MEG-01 cells. High-molecular-weight smeared signals were measured. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets.
H-PGDS protein accumulated at 30 min after the intracellular calcium level was increased, and its level was enhanced in an incubation time–dependent manner (Figure 2C), revealing that H-PGDS becomes a substrate of the ubiquitin–proteasome system. In contrast, the H-PGDS level was not changed when the cells were cultured in medium containing A23187 alone (Figure 2C). Moreover, when MG132 was present, a high-molecular-weight smeared mass of the ubiquitinated H-PGDS protein was detected at 60 min after the elevation of the intracellular calcium level, and the level of the ubiquitinated H-PGDS protein increased in a time-dependent manner (Figure 2D). No high-molecular-weight smeared mass of ubiquitinated H-PGDS protein was observed at 30 min after the enhancement of the intracellular calcium level (unpublished data).
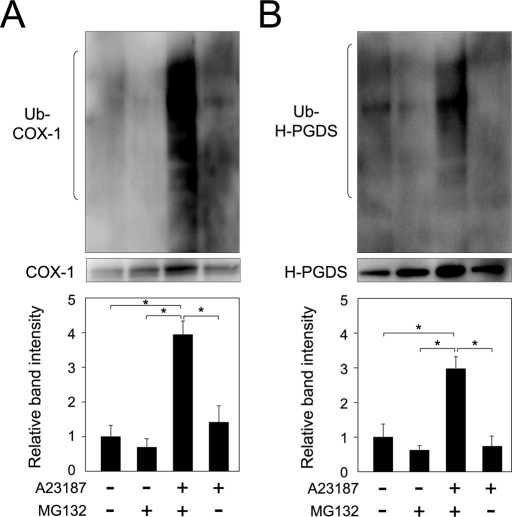
Furthermore, ubiquitinated COX-1 and H-PGDS proteins were not detected when the cells were cultured in medium containing either MG132 or A23187, although a high-molecular-weight smeared mass of ubiquitinated COX-1 and H-PGDS was detected in both A23187- and MG132-treated cells (Figure 3, A and B). These results, taken together, reveal that ubiquitin-dependent degradation of COX-1 and H-PGDS proteins was initiated within 10 and 30 min, respectively, after a rise in the intracellular calcium level in MEG-01 cells.
FIGURE 3:
Ubiquitination of COX-1 and H-PGDS in the presence of MG132. (A) MG-132–dependent ubiquitination of COX-1 in MEG-01 cells. The cells were incubated in medium with TPA for 16 h and then cultured for 30 min in medium containing A23187 in the presence or absence of MG132. High-molecular-weight smeared signals were measured. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets. (B) Ubiquitination of H-PGDS is dependent on MG132. The cells were cultured for 60 min, as described in the legend of (A). High-molecular-weight smeared signals were measured. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets.
Substrate and product-independent ubiquitination of COX-1 and H-PGDS
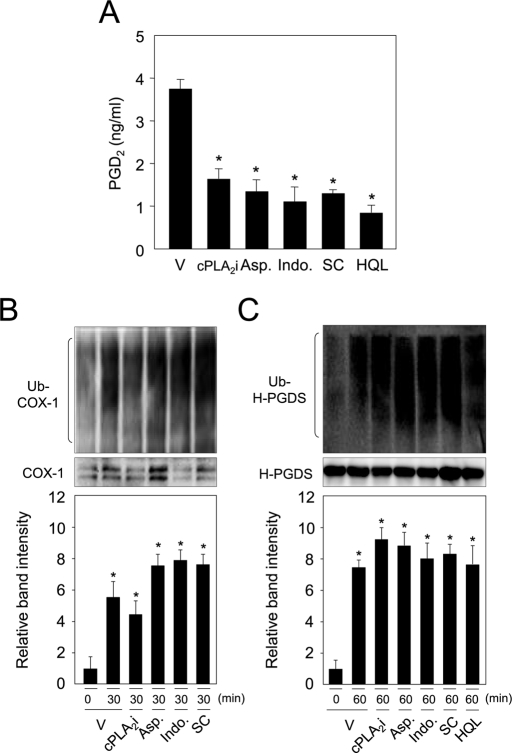
To study the roles of substrates and products of COX-1 and H-PGDS on the ubiquitin-dependent degradation of both enzymes, ubiquitination of both proteins was examined under the influence of inhibitors for cPLA2, COX, or H-PGDS in MEG-01 cells. PGD2 production was inhibited with cPLA2 inhibitor (cPLA2i); aspirin; indomethacin, COX inhibitors; SC-560 (SC), a COX-1 inhibitor; and HQL-79 (HQL), an H-PGDS inhibitor in MEG-01 cells (Figure 4A). Cells were treated with cPLA2, COX, or H-PGDS inhibitors and then incubated in medium containing A23187 and MG132 for 60 min, and ubiquitination of both proteins was examined. COX-1 was ubiquitinated by treatment with A23187, and this ubiquitination was not inhibited, even in the presence of these four inhibitors (Figure 4B). In addition, COX-1 protein level was decreased to the same extent, even in the presence of each inhibitor in both A23187- and CHX-treated cells, as compared with the vehicle-treated cells (Supplemental Figure S1A, left). Moreover, when cells were cultured under the same conditions without stimulation with ionophore, the COX-1 level was not changed (Figure S1A, right).
FIGURE 4:
Ubiquitination of COX-1 and H-PGDS independent of their substrates and products in MEG-01 cells. (A) PGD2 production in MEG-01 cells was inhibited by cPLA2i (10 μM); Aspirin (Asp.; 10 μM); indomethacin (Indo.; 2 μM), COX inhibitors; SC-560 (SC; 5 μM), a COX-1 inhibitor; and HQL-79 (HQL; 3 μM), an H-PGDS inhibitor. MEG-01 cells were incubated in medium containing TPA for 16 h and then incubated with A23187 in the presence or absence of each inhibitor for 30 min. Data represent the mean ± SD (*p < 0.01) from three independent experiments, as compared with the vehicle. (B) Ubiquitination of COX-1 independent of the inhibitors. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with the vehicle. (C) H-PGDS ubiquitination under the existence of either inhibitor. MEG-01 cells were incubated for 60 min, as described in the legend of (B), together with A23187 and MG132. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with the vehicle.
Ubiquitination of H-PGDS was detected in A23187-treated MEG-01 cells, and the level of this ubiquitination was not altered by cotreatment with any of the five inhibitors (Figure 4C). Moreover, the protein level of H-PGDS was eliminated by treatment of cells with A23187 and CHX, and its decreased level was not altered, even when the cells were cultured separately in medium containing each inhibitor (Figure S1B, left). Furthermore, the H-PGDS level was not altered when the cells were cultured in the same conditions without A23187 stimulation (Figure S1B, right). These results indicate that the calcium ion–mediated ubiquitination of COX-1 and H-PGDS was independent on the levels of their substrates and their products in MEG-01 cells.
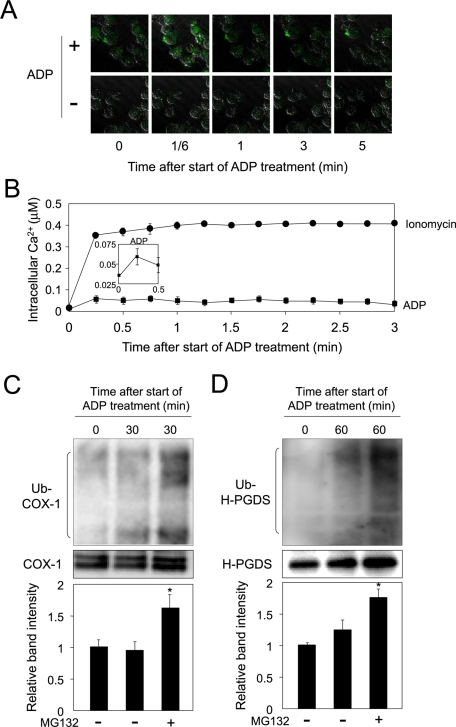
Ubiquitin-dependent degradation of COX-1 and H-PGDS in ADP-treated MEG-01 cells
The entry of calcium ions into cells treated with a calcium ionophore may not represent physiological conditions. As ADP is known to enhance the intracellular calcium level in MEG-01 cells (Nakata et al., 2005), we next measured the intracellular calcium level in MEG-01 cells incubated with ADP or ionomycin. Cells were cultured with TPA for 16 h and then incubated with Fluo 4-AM in the presence or absence of ADP or ionomycin for 5 min, after a 15-min preincubation with Fluo 4-AM. Intracellular calcium level increased slightly at 10 s, then decreased, and became low level by 3 min after initiation of treatment with ADP (Figure 5, A and B; note Figure 5B, inset), indicating that ADP weakly enhanced the intracellular calcium level in MEG-01 cells. The enhancement of the intracellular calcium level by ADP was ∼21% of the ionomycin-mediated elevation at 10 s in MEG-01 cells (Figure 5B, inset).
FIGURE 5:
ADP enhances intracellular calcium level and concurrently degrades ubiquitinated COX-1 and H-PGDS proteins. (A) Elevation of the intracellular calcium level in ADP-treated MEG-01 cells. ADP (0.5 μM): closed squares; ionomycin (1 μM): closed circles. (B) Measurement of intracellular calcium level in MEG-01 cells. Data are representative of three independent experiments and represent the mean ± SD. (C) COX-1 protein accumulated in the ADP-treated MEG-01 cells. The cells were incubated in medium containing TPA for 16 h and then incubated with A23187 in the presence or absence of MG132 for 30 min. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with MG132(−) at 30 min. (D) Ubiquitination of H-PGDS protein in the ADP-treated MEG-01 cells. The cells were incubated for 60 min, as described in the legend of (B), together with A23187 and/or MG132. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with MG132(−) at 60 min.
We next examined the ubiquitination-dependent degradation of COX-1 and H-PGDS proteins in the ADP-treated MEG-01 cells. When the cells were cultured in medium containing ADP with or without MG132 for 30 min, ubiquitination of COX-1 protein was observed, as compared with the levels of ubiquitination in the pretreated cells or only MG132-treated cells (Figure 5C). In addition, COX-1 protein level was decreased in both ADP- and CHX-treated cells, as compared with the vehicle- or CHX-treated cells (Figure S2A). Moreover, ubiquitinated H-PGDS protein was detected at 60 min after initiation of ADP treatment, as compared with the level of ubiquitinated protein in the MG132-treated cells (Figure 5D). Furthermore, the protein level of H-PGDS was reduced by treatment of cells with both ADP and CHX, as compared with the vehicle- or CHX-treated cells (Figure S2B). These results reveal that the calcium-dependent degradation of COX-1 and H-PGDS proteins was also observed in MEG-01 cells treated with ADP, a natural inducer that causes an increase in intracellular calcium level. Thus the degradation of both enzymes was initiated by the treatment with ADP as well as with ionophore.
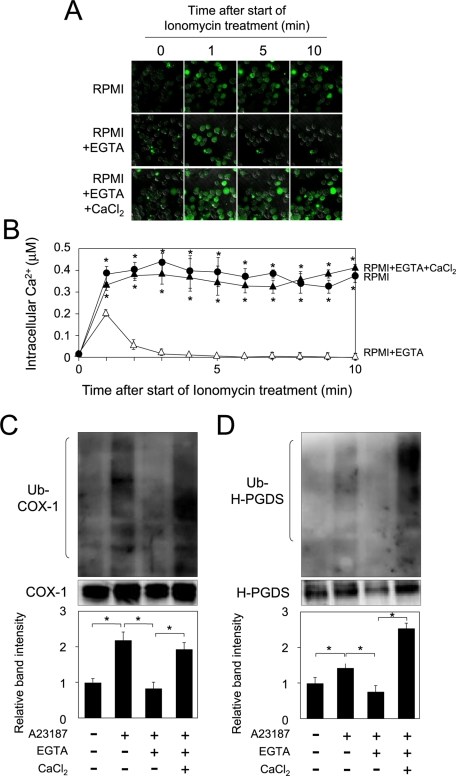
Essential roles of the entry of calcium ion into cells upon ubiquitination-dependent degradation of COX-1 and H-PGDS
To investigate the molecular mechanism of the calcium-dependent degradation of COX-1 and H-PGDS proteins, we investigated the roles of intracellular and extracellular calcium ions in the ubiquitination-dependent degradation of COX-1 and H-PGDS proteins. MEG-01 cells were first incubated with TPA for 16 h, and then for an additional 10 min in RPMI1640 containing ionomycin and Fluo 4-AM with ethylene glycol tetraacetic acid (EGTA) and/or CaCl2, after preincubation with Fluo 4-AM for 15 min. The intracellular calcium level was increased by treatment with ionomycin (Figure 6, A, top, and B). On the contrary, when cells were cultured in medium with ionomycin and EGTA, the intracellular calcium level was transiently elevated at 1 min after initiation of treatment and then quickly decreased (Figure 6A, middle, and B). Moreover, when cells were incubated in medium containing ionomycin, EGTA, and CaCl2, the entry of calcium ions into the cells was greater (Figure 6, A, bottom, and B) than that seen in cells cultured in RPMI1640 containing ionomycin and EGTA (Figure 6, A, top, and B).
FIGURE 6:
Essential roles of calcium entry into cells for ubiquitination of COX-1 and H-PGDS. (A) Suppression of the entry of extracellular calcium ion by EGTA. MEG-01 cells were incubated with TPA for 16 h, washed with PBS, and then pretreated with EGTA (5 mM) for 15 min. They were subsequently incubated for 10 min with Fluo 4-AM (1 μM) and ionomycin (1 μM) in the presence of EGTA (5 mM) with or without CaCl2 (2 mM). (B) Measurement of intracellular calcium level in MEG-01 cells. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with that for culture in medium containing EGTA. (C) Roles of extracellular calcium ions in the ubiquitination of COX-1 protein in MEG-01 cells. The cells were incubated for 30 min in RPMI1640 containing the various indicated combinations of EGTA, CaCl2, and A23187, together with MG132. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets. (D) Essential role of the increased intracellular calcium in the ubiquitination of H-PGDS. The cells were cultured for 60 min, as described in the legend of (C). Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets.
We next investigated the ubiquitination of COX-1 and H-PGDS proteins in RPMI1640 with or without EGTA and/or CaCl2. When MEG-01 cells were cultured in medium containing A23187, the ubiquitination of COX-1 protein was enhanced, as compared with that in the vehicle control (Figure 6C). Moreover, when cells were incubated in medium containing A23187 and EGTA, the ubiquitination of COX-1 protein was clearly decreased, as compared with that in A23187-treated cells (Figure 6C). Conversely, when cells were cultured in medium containing A23187 and EGTA together with CaCl2, the ubiquitination level of COX-1 protein was elevated, as compared with that for cells cultured in medium with A23187 and EGTA (Figure 6C). In addition, the protein level of COX-1 was reduced by the treatment with A23187 and CHX, and this reduction was restored by cotreatment with A23187, CHX, and EGTA (Figure S3A). Moreover, when cells were cultured in medium containing A23187, CHX, and EGTA together with CaCl2, the COX-1 protein level was clearly reduced (Figure S3A).
The ubiquitination level of H-PGDS protein was increased by incubation with A23187, as compared with that in the vehicle control (Figure 6D). Furthermore, when cells were incubated in the presence of A23187 and EGTA, ubiquitination of H-PGDS protein was clearly decreased, as compared with that for cells cultured in medium with A23187 alone (Figure 6D). Conversely, when MEG-01 cells were cultured in medium containing A23187 and EGTA together with CaCl2, the ubiquitination level of H-PGDS protein was enhanced, as compared with that for cells cultured in medium containing A23187 and EGTA (Figure 6D). Furthermore, the H-PGDS protein level was decreased by treatment with A23187 and CHX, as compared with that of the vehicle-treated cells (Figure S3B). This decrease was restored by cotreatment with A23187, CHX, and EGTA (Figure S3B). Moreover, when cells were cultured in medium containing A23187, CHX, and EGTA together with CaCl2, the H-PGDS protein level was reduced (Figure S3B).
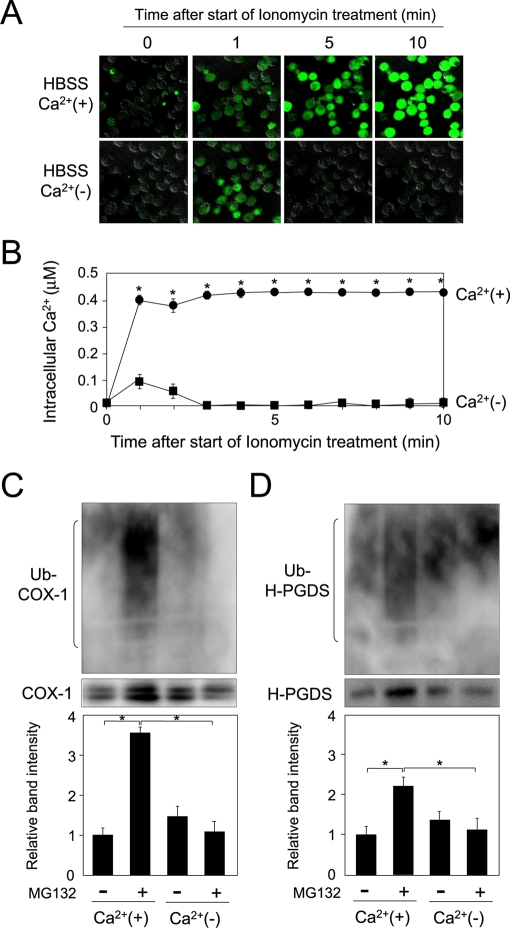
To confirm the requirement of the entry of calcium ion for the ubiquitination of the COX-1 and H-PGDS proteins, we cultured MEG-01 cells in Hank's balanced salt solution (HBSS; containing 1.3 mM CaCl2) or Ca2+-free HBSS and detected the ubiquitinated COX-1 and H-PGDS proteins by immunoprecipitation and Western blot analysis. Ionomycin-induced entry of the extracellular calcium ion into the cells was observed with the cells in HBSS (Figure 7, A, top, and B). In contrast, the intracellular calcium level was transiently increased at 1 min in Ca2+-free HBSS, even when the cells were incubated with ionomycin (Figure 7, A, bottom, and B).
FIGURE 7:
Calcium-dependent ubiquitination of COX-1 and H-PGDS. (A) Determination of the intracellular calcium level in the cells cultured in HBSS or Ca2+-free HBSS. MEG-01 cells were incubated with TPA for 16 h, and then for 10 min in HBSS or Ca2+-free HBSS containing Fluo 4-AM and ionomycin after the preincubation with Fluo 4-AM for 15 min. (B) Measurement of intracellular calcium level in MEG-01 cells. HBSS: closed circles; Ca2+-free HBSS: closed squares. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as compared with that for culture in Ca2+-free medium. (C) Ubiquitination of COX-1 in MEG-01 cells cultured in HBSS or Ca2+-free HBSS with or without MG132. The cells were incubated in RPMI1640 with TPA for 16 h, washed twice with Ca2+-free HBSS, and then preincubated in Ca2+-free HBSS with 5 mM EGTA for 15 min. The cells were further incubated in HBSS or Ca2+-free HBSS with A23187 in the presence or absence of MG132 for 30 min. Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets. (D) Ubiquitination of H-PGDS in MEG-01 cells cultured in HBSS or Ca2+-free HBSS with or without MG132. The cells were cultured for 60 min, as described in the legend of (C). Data are representative of three independent experiments and represent the mean ± SD (*p < 0.01), as indicated by the brackets.
Finally, we investigated the requirement of extracellular calcium ions on the ubiquitination of COX-1 and H-PGDS proteins in MEG-01 cells. When the cells were cultured in HBSS containing A23187 and MG132, the ubiquitination level of COX-1 protein was enhanced, as compared with that in the A23187-treated cells (Figure 7C), while the level of accumulated COX-1 protein was not altered when the cells were incubated in Ca2+-free HBSS, even in the presence of A23187 and MG132 (Figure 7C). Protein level of COX-1 was decreased by treatment with A23187 and CHX in the presence of Ca2+, as compared with that cultured in medium containing the various indicated combinations of CaCl2 and A23187 together with CHX (Figure S4A).
H-PGDS protein level was also increased in cells cultured in HBSS containing A23187 and MG132, as compared with the A23187-treated cells (Figure 7D), whereas the level of ubiquitinated H-PGDS protein was not changed when the cells were incubated in Ca2+-free HBSS with A23187 and MG132 (Figure 7D). In addition, the protein level of H-PGDS was decreased in medium containing A23187, CHX, and CaCl2, as compared with the vehicle- treated cells or either A23187 or CaCl2-treated cells (Figure S4B).
Additionally, the ubiquitination of COX-1 and H-PGDS proteins was dependent upon extracellular Ca2+ concentration (Figure S5, A and B), indicating that both COX-1 and H-PGDS protein degraded through the ubiquitination-mediated proteasome system in response to the calcium ions. These results, taken together, reveal that the entry of calcium ions into the cells was essential for the initiation of ubiquitination-mediated degradation of COX-1 and H-PGDS proteins in MEG-01 cells.
DISCUSSION
The ubiquitin–proteasome system is the principal mechanism responsible for the turnover of proteins in eukaryotic cells and is a fundamental mechanism for cellular controls (Clague and Urbe, 2010), such as signal transduction, transcription, and cell cycle progression (Wickliffe et al., 2009). Ubiquitination of a specific substrate is mainly regulated through the modulation of its degradation signal and through the control of the activity of its cognate E3 ubiquitin ligase (Ardley and Robinson, 2005; Welchman et al., 2005). Targeting of proteins to the proteasome generally requires the attachment of a multi-ubiquitin chain to the substrate (Komander, 2009). It is well known that many substrates of the ubiquitin–proteasome system can be ubiquitinated at multiple lysine residues and that multi-ubiquitination occurs with high-molecular-weight proteins, although a number of studies have shown that ubiquitin modification occurs at one or more specific lysine residues in the target protein (Kuhlbrodt et al., 2005; Miller and Gordon, 2005; Hurley et al., 2006).
PGs are known to act as local hormones that are rapidly synthesized in response to various stimuli; this synthesis is followed by quick termination of PG production. The half-lives of PGs are very short, indicating the existence of a critical regulation system for PG production. H-PGDS is localized in the cytosol, and is translocated to the ER membranes in response to elevation of the intracellular calcium level (Ueno et al., 2005). The results of our present study indicated that COX-1 and H-PGDS proteins in human MEG-01 cells were degraded through the ubiquitin–proteasome system in response to a rise in the intracellular calcium level. Treatment of cells with the proteasome inhibitor MG132 markedly increased the steady-state levels of COX-1 and H-PGDS proteins (Figure 2, A and C), demonstrating COX-1 and H-PGDS proteins to be direct substrates of the proteasome. This calcium-mediated ubiquitination of COX-1 and H-PGDS proteins was also observed in human embryonic kidney 293 cells expressing heterologous human COX-1 or H-PGDS protein (Figure S6), suggesting that the intracellular calcium concentration–dependent degradation of COX-1 and H-PGDS proteins is a common event in a variety of cells.
In the arachidonate cascade, COX is known to be a suicidal enzyme, because this enzyme is inactivated during catalysis within several minutes after treatment with endogenous or exogenous arachidonic acid (Smith et al., 2000; Smith and Song, 2002). COX-1 and COX-2 undergo irreversible suicide inactivation during COX catalysis under cell-free conditions (Smith and Song, 2002; Rouzer and Marnett, 2003). In vitro inactivation of COX is accompanied by significant changes in the structure of the enzyme (Mevkh et al., 1993). The inactivated COX-2 is degraded by the ubiquitin–proteasome system (Smith et al., 2000; Mbonye et al., 2006, 2008; Kang et al., 2007; Mbonye and Song, 2009; Wada et al., 2009) acting through the ER-associated degradation system (Hirsch et al., 2009; Mehnert et al., 2010), similar to other ER proteins that have recently been reported (Leichner et al., 2009; Wojcikiewicz et al., 2009; Hoseki et al., 2010). It has been shown that COX inhibitors do not inhibit the proteasome-dependent degradation of COX-2 (Figueiredo-Pereira et al., 2002), similar to our present results, which show that H-PGDS or COX inhibitor did not affect the proteasome-dependent degradation of COX-1 and H-PGDS (Figure 4, B and C). However, when cPLA2 inhibitor was used, the level of the ubiquitination of COX-1 was lowered, compared with levels seen with other inhibitors (Figure 4B). Moreover, the rate of degradation of COX-1 protein under the influence of cPLA2 inhibitor was lower than rates seen with other inhibitors (Figure S1A), suggesting the decrease of arachidonic acid levels by cPLA2 inhibitor might decrease the level of the ubiquitination of COX-1. The initiation of ubiquitination of COX-1 might be required for arachidonic acid–mediated inactivation of COX-1 protein (Mbonye and Song, 2009). Further analysis is needed to verify this hypothesis.
Moreover, when each of COX-1 and H-PGDS proteins was heterologously expressed in 293 cells that expressed negligible levels of COX-1 and H-PGDS, both of these proteins were degraded through the ubiquitin–proteasome system by the increase of the intracellular calcium level (Figure S6), indicating that each of the COX-1 and H-PGDS proteins is independently degraded through the ubiquitin–proteasome system by the increase of the intracellular calcium level.
In summary, the COX-1 and H-PGDS proteins were rapidly degraded through the ubiquitin–proteasome system in response to the increase in the intracellular calcium level. The overall regulation of PG synthesis, including the level of PG synthases, is carried out by the intracellular calcium level. This is the first report that COX-1 and H-PGDS, a terminal PG synthase involved in the arachidonate cascade, are ubiquitinated and concurrently degraded via the ubiquitin–proteasome system. In further study, the physiological meaning of the purpose of rapid degradation of COX-1 and H-PGDS through the ubiquitin–proteasome system should be elucidated. Furthermore, identification of the ubiquitin ligases for both enzymes will be essential for understanding the entire mechanism of the regulation of PGD2 synthesis in response to an increasing level of intracellular calcium.
MATERIALS AND METHODS
Cell culture
Human megakaryocytic MEG-01 cells (American Type Culture Collection, Manassas, VA) were cultured in RPMI1640 (Sigma-Aldrich, St. Louis, MO) supplemented with 10% (vol/vol) fetal calf serum and penicillin (10,000 U/ml) and streptomycin (10,000 U/ml) in a humidified incubator at 37°C and 5% CO2.
Calcium imaging
MEG-01 cells were seeded on 35-mm glass bottom culture dishes (MatTek, Ashland, MA) and were treated with TPA (20 nM) for 16 h. The cells were then washed with HBSS (Invitrogen, Carlsbad, CA) and preincubated with Fluo 4-AM (1 μM; Dojindo Laboratories, Kumamoto, Japan) for 15 min prior to treatment with ionomycin (1 μM; Calbiochem, Madison, WI) or ADP (0.5 μM; Sigma) in the presence of Fluo 4-AM (1 μM) for 10 min. Fluorescence was monitored with confocal laser scanning fluorescence microscopy (C-LSM510; Carl Zeiss, Jena, Germany) using 488-nm argon laser excitation with a 505- to 530-nm band-pass barrier filter. Fluorescence was recorded every 10–15 s after the initiation of every treatment. Known concentration of calcium solution was used for creation of a standard curve used to measure intracellular calcium levels.
Immunoprecipitation and Western blot analysis
MEG-01 cells were cultured with TPA (20 nM) for 16 h and then washed with phosphate-buffered saline (PBS). They were then incubated for various periods of time in fresh RPMI1640 containing A23187 (5 μM; Calbiochem) at 37°C in the presence or absence of MG132 (10 μM; Calbiochem). The cells were then lysed with RIPA buffer containing 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 0.5% (wt/vol) sodium deoxycholate, 0.1% (vol/vol) SDS, 1% (vol/vol) NP-40, 1 mM Na3VO4, 1 mM NaF, 50 μM Na2MoO4, and a protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). The cell lysates were then centrifuged at 12,000 × g for 30 min at 4°C to remove cell debris. Supernatants were incubated with anti–human H-PGDS monoclonal antibody (1E6; Cayman Chemicals, Ann Arbor, MI) overnight at 4°C with continuous agitation. Protein G Sepharose (GE Healthcare, Buckinghamshire, UK) was then added to the samples, and incubation was continued for 60 min at 4°C. Immunoprecipitates on the Sepharose resin were washed five times with ice-cold 50 mM Tris-Cl (pH 7.5) buffer containing 150 mM NaCl, 2 mM EDTA, and 0.1% (vol/vol) Triton X-100. Subsequently, the resin was suspended in Laemmli SDS-loading buffer containing 50 mM Tris-Cl (pH 6.8), 10% (vol/vol) glycerol, 2% (wt/vol) SDS, 0.005% (wt/vol) bromophenol blue, and 1% (vol/vol) 2-mercaptoethanol.
Protein samples were separated by SDS–PAGE and transferred onto polyvinylidene difluoride membranes (Immobilon P; Millipore, Bedford, MA). Blots were incubated with the specific primary antibodies, that is, anti-ubiquitin (Thermo Fisher Scientific, Rockford, IL), anti–COX-1 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA) or anti–human H-PGDS polyclonal antibody, washed, and then incubated with the appropriate second antibodies conjugated to horseradish peroxidase (GE Healthcare). Immunoreactive signals were detected by the use of ECL Plus Western blotting detection reagents (GE Healthcare) and a LAS-3000 imaging analyzer (Fujifilm, Tokyo, Japan). The band intensities were measured with Multi Gauge software (Fujifilm). Protein concentrations were measured with Pierce BCA protein assay reagent (Thermo Fisher Scientific).
Calcium-entry study
MEG-01 cells were incubated with TPA (20 nM) for 16 h, washed with PBS, and then pretreated with EGTA (5 mM) for 15 min. The cells were then cultured with 1 μM Fluo 4-AM in the presence of ionomycin (1 μM), ADP (0.5 μM; Sigma), EGTA (5 mM), and/or 2 mM CaCl2 for the desired times. Intracellular calcium was observed by using confocal laser scanning microscopy (C-LSM510) and measured using calcium solution as the standard.
Statistical analysis
Comparison of two groups was analyzed by Student's t test. For comparison of more than two groups with comparable variances, one-way analysis of variance and a Tukey's post hoc test were carried out. p < 0.05 was considered significant.
Supplementary Material
Supplemental Materials
Acknowledgments
We thank Fumio Amano (Osaka University of Pharmaceutical Sciences) for valuable discussions. This work was in part supported by a Grant-in-Aid for Scientific Research (21570151) and Scientific Research on Innovative Areas (23116516) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants from the Sumitomo Foundation, the Gushinkai Foundation, the Research Foundation for Pharmaceutical Sciences (to K.F.), Takeda Science Foundation (to Y.U. and K.F.), and the Japan Foundation for Applied Enzymology (to K.A. and K.F.).
Abbreviations used:
CHX
cycloheximide
COX
cyclooxygenase
cPLA2
cytosolic phospholipase A2
cPLA2i
cPLA2 inhibitor
EGTA
ethylene glycol tetraacetic acid
ER
endoplasmic reticulum
HBSS
Hank's balanced salt solution
H-PGDS
hematopoietic PGDS
PBS
phosphate-buffered saline
PG
prostaglandin
PGDS
prostaglandin D synthase
TPA
12-_O_-tetradecanoylphorbol-13-acetate
Footnotes
REFERENCES
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Dixon DA. Dysregulated post-transcriptional control of COX-2 gene expression in cancer. Curr Pharm Des. 2004;10:635–646. doi: 10.2174/1381612043453171. [DOI] [PubMed] [Google Scholar]
- Figueiredo-Pereira ME, Li Z, Jansen M, Rockwell P. N-acetylcysteine and celecoxib lessen cadmium cytotoxicity which is associated with cyclooxygenase-2 up-regulation in mouse neuronal cells. J Biol Chem. 2002;277:25283–25289. doi: 10.1074/jbc.M109145200. [DOI] [PubMed] [Google Scholar]
- Fujimori K, Kanaoka Y, Sakaguchi Y, Urade Y. Transcriptional activation of the human hematopoietic prostaglandin D synthase gene in megakaryoblastic cells. Roles of the Oct-1 element in the 5′-flanking region and the AP-2 element in the untranslated exon 1. J Biol Chem. 2000;275:40511–40516. doi: 10.1074/jbc.M007688200. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Helliwell RJ, Adams LF, Mitchell MD. Prostaglandin synthases: recent developments and a novel hypothesis. Prostaglandins Leukot Essent Fatty Acids. 2004;70:101–113. doi: 10.1016/j.plefa.2003.04.002. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Hoseki J, Ushioda R, Nagata K. Mechanism and components of endoplasmic reticulum-associated degradation. J Biochem. 2010;147:19–25. doi: 10.1093/jb/mvp194. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka Y, et al. Cloning and crystal structure of hematopoietic prostaglandin D synthase. Cell. 1997;90:1085–1095. doi: 10.1016/s0092-8674(00)80374-8. [DOI] [PubMed] [Google Scholar]
- Kanaoka Y, Fujimori K, Kikuno R, Sakaguchi Y, Urade Y, Hayaishi O. Structure and chromosomal localization of human and mouse genes for hematopoietic prostaglandin D synthase. Conservation of the ancestral genomic structure of sigma-class glutathione S-transferase. Eur J Biochem. 2000;267:3315–3322. doi: 10.1046/j.1432-1327.2000.01362.x. [DOI] [PubMed] [Google Scholar]
- Kanaoka Y, Urade Y. Hematopoietic prostaglandin D synthase. Prostaglandins Leukot Essent Fatty Acids. 2003;69:163–167. doi: 10.1016/s0952-3278(03)00077-2. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Mbonye UR, DeLong CJ, Wada M, Smith WL. Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog Lipid Res. 2007;46:108–125. doi: 10.1016/j.plipres.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Mouysset J, Hoppe T. Orchestra for assembly and fate of polyubiquitin chains. Essays Biochem. 2005;41:1–14. doi: 10.1042/EB0410001. [DOI] [PubMed] [Google Scholar]
- Leichner GS, Avner R, Harats D, Roitelman J. Dislocation of HMG-CoA reductase and Insig-1, two polytopic endoplasmic reticulum proteins, en route to proteasomal degradation. Mol Biol Cell. 2009;20:3330–3341. doi: 10.1091/mbc.E08-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye UR, Song I. Posttranscriptional and posttranslational determinants of cyclooxygenase expression. BMB Rep. 2009;42:552–560. doi: 10.5483/bmbrep.2009.42.9.552. [DOI] [PubMed] [Google Scholar]
- Mbonye UR, Wada M, Rieke CJ, Tang HY, Dewitt DL, Smith WL. The 19-amino acid cassette of cyclooxygenase-2 mediates entry of the protein into the endoplasmic reticulum-associated degradation system. J Biol Chem. 2006;281:35770–35778. doi: 10.1074/jbc.M608281200. [DOI] [PubMed] [Google Scholar]
- Mbonye UR, Yuan C, Harris CE, Sidhu RS, Song I, Arakawa T, Smith WL. Two distinct pathways for cyclooxygenase-2 protein degradation. J Biol Chem. 2008;283:8611–8623. doi: 10.1074/jbc.M710137200. [DOI] [PubMed] [Google Scholar]
- Mehnert M, Sommer T, Jarosch E. ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays. 2010;32:905–913. doi: 10.1002/bies.201000046. [DOI] [PubMed] [Google Scholar]
- Mevkh AT, Miroshnikov KA, Igumnova ND, Varfolomeev SD. Prostaglandin H synthase. Inactivation of the enzyme in the course of catalysis is accompanied by fast and dramatic changes in protein structure. FEBS Lett. 1993;321:205–208. doi: 10.1016/0014-5793(93)80109-8. [DOI] [PubMed] [Google Scholar]
- Miller J, Gordon C. The regulation of proteasome degradation by multi-ubiquitin chain binding proteins. FEBS Lett. 2005;579:3224–3230. doi: 10.1016/j.febslet.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Mishra A, Godavarthi SK, Maheshwari M, Goswami A, Jana NR. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J Biol Chem. 2009;284:10537–10545. doi: 10.1074/jbc.M806804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri I, Aritake K, Taniguchi H, Sato Y, Kamauchi S, Nagata N, Maruyama T, Taniike M, Urade Y. Inhibition of prostaglandin D synthase suppresses muscular necrosis. Am J Pathol. 2009;174:1735–1744. doi: 10.2353/ajpath.2009.080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri I, Eguchi N, Suzuki K, Urade Y, Taniike M. Hematopoietic prostaglandin D synthase is expressed in microglia in the developing postnatal mouse brain. Glia. 2003;42:263–274. doi: 10.1002/glia.10183. [DOI] [PubMed] [Google Scholar]
- Mohri I, et al. Prostaglandin D2-mediated microglia/astrocyte interaction enhances astrogliosis and demyelination in twitcher. J Neurosci. 2006;26:4383–4393. doi: 10.1523/JNEUROSCI.4531-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraitis AN, Giguere V. The co-repressor hairless protects RORα orphan nuclear receptor from proteasome-mediated degradation. J Biol Chem. 2003;278:52511–52518. doi: 10.1074/jbc.M308152200. [DOI] [PubMed] [Google Scholar]
- Nakata M, Maruyama I, Yada T. Leptin potentiates ADP-induced [Ca2+]i increase via JAK2 and tyrosine kinases in a megakaryoblast cell line. Diabetes Res Clin Pract. 2005;70:209–216. doi: 10.1016/j.diabres.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Park JM, et al. Hematopoietic prostaglandin D synthase suppresses intestinal adenomas in ApcMin/+mice. Cancer Res. 2007;67:881–889. doi: 10.1158/0008-5472.CAN-05-3767. [DOI] [PubMed] [Google Scholar]
- Rouzer CA, Marnett LJ. Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases. Chem Rev. 2003;103:2239–2304. doi: 10.1021/cr000068x. [DOI] [PubMed] [Google Scholar]
- Shimura C, Satoh T, Igawa K, Aritake K, Urade Y, Nakamura M, Yokozeki H. Dendritic cells express hematopoietic prostaglandin D synthase and function as a source of prostaglandin D2 in the skin. Am J Pathol. 2010;176:227–237. doi: 10.2353/ajpath.2010.090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- Smith WL, Song I. The enzymology of prostaglandin endoperoxide H synthases-1 and -2. Prostaglandins Other Lipid Mediat. 2002;68–69:115–128. doi: 10.1016/s0090-6980(02)00025-4. [DOI] [PubMed] [Google Scholar]
- Tanaka H, et al. Improvement in the quality of hematopoietic prostaglandin D synthase crystals in a microgravity environment. J Synchrotron Radiat. 2011;18:88–91. doi: 10.1107/S0909049510037076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Ogawa K, Sugamura K, Nakamura M, Takano S, Nagata K. Cutting edge: differential production of prostaglandin D2 by human helper T cell subsets. J Immunol. 2000;164:2277–2280. doi: 10.4049/jimmunol.164.5.2277. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Meyer DJ, Hayes JD. Sequence, catalytic properties and expression of chicken glutathione-dependent prostaglandin D2 synthase, a novel class Sigma glutathione S-transferase. Biochem J. 1998;333:317–325. doi: 10.1042/bj3330317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno N, Murakami M, Tanioka T, Fujimori K, Tanabe T, Urade Y, Kudo I. Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2. J Biol Chem. 2001;276:34918–34927. doi: 10.1074/jbc.M100429200. [DOI] [PubMed] [Google Scholar]
- Ueno N, Takegoshi Y, Kamei D, Kudo I, Murakami M. Coupling between cyclooxygenases and terminal prostanoid synthases. Biochem Biophys Res Commun. 2005;338:70–76. doi: 10.1016/j.bbrc.2005.08.152. [DOI] [PubMed] [Google Scholar]
- Urade Y, Hayaishi O. Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim Biophys Acta. 2000a;1482:259–271. doi: 10.1016/s0167-4838(00)00161-8. [DOI] [PubMed] [Google Scholar]
- Urade Y, Hayaishi O. Prostaglandin D synthase: structure and function. Vitam Horm. 2000b;58:89–120. doi: 10.1016/s0083-6729(00)58022-4. [DOI] [PubMed] [Google Scholar]
- Urade Y, Ujihara M, Horiguchi Y, Ikai K, Hayaishi O. The major source of endogenous prostaglandin D2 production is likely antigen-presenting cells. Localization of glutathione-requiring prostaglandin D synthetase in histiocytes, dendritic, and Kupffer cells in various rat tissues. J Immunol. 1989;143:2982–2989. [PubMed] [Google Scholar]
- Wada M, Saunders TL, Morrow J, Milne GL, Walker KP, Dey SK, Brock TG, Opp MR, Aronoff DM, Smith WL. Two pathways for cyclooxygenase-2 protein degradation in vivo. J Biol Chem. 2009;284:30742–30753. doi: 10.1074/jbc.M109.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wickliffe K, Williamson A, Jin L, Rape M. The multiple layers of ubiquitin-dependent cell cycle control. Chem Rev. 2009;109:1537–1548. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcikiewicz RJ, Pearce MM, Sliter DA, Wang Y. When worlds collide: IP(3) receptors and the ERAD pathway. Cell Calcium. 2009;46:147–153. doi: 10.1016/j.ceca.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, et al. Expression and characterization of PGD2 receptors in chronic rhinosinusitis: modulation of DP and CRTH2 by PGD2. Int Arch Allergy Immunol. 2009;148:127–136. doi: 10.1159/000155743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials