Incorporation of the Rpn12 Subunit Couples Completion of Proteasome Regulatory Particle Lid Assembly to Lid-Base Joining (original) (raw)
. Author manuscript; available in PMC: 2012 Dec 23.
Summary
The 26S proteasome, the central eukaryotic protease, comprises a core particle capped by a 19S regulatory particle (RP). The RP is divisible into base and lid subcomplexes. Lid biogenesis and incorporation into the RP remain poorly understood. We report several lid intermediates, including the free Rpn12 subunit and a lid particle (LP) containing the remaining eight subunits, LP2. Rpn12 binds LP2 in vitro, and each requires the other for assembly into 26S proteasomes. Stable Rpn12 incorporation depends on all other lid subunits, indicating Rpn12 distinguishes LP2 from smaller lid subcomplexes. The highly conserved C-terminus of Rpn12 bridges the lid and base, mediating both stable binding to LP2 and lid-base joining. Our data suggest a hierarchical assembly mechanism where Rpn12 binds LP2 only upon correct assembly of all other lid subunits, and the Rpn12 tail then helps drive lid-base joining. Rpn12 incorporation thus links proper lid assembly to subsequent assembly steps.
Introduction
Most regulatory and quality control protein degradation in eukaryotes is mediated by the ubiquitin-proteasome system (Finley, 2009; Ravid and Hochstrasser, 2008). This system is central to many cellular processes, including cell division, DNA repair, and adaptive immunity, and defects in it contribute to many human diseases. Typically, proteins to be degraded are tagged with a polyubiquitin chain that targets them to the 26S proteasome. The proteasome is a highly conserved 2.6 MDa protease complex consisting of a barrel-shaped 20S core particle (CP) that houses the proteolytic sites, and a 19S regulatory particle (RP) that binds to one or both ends of the CP. The RP confers ATP dependence on proteasomal proteolysis and mediates recognition and recycling of the polyubiquitin tag and unfolding and translocation of substrates into the CP.
The CP comprises four stacked heptameric rings of related subunits. Each outer ring contains seven distinct α subunits while each inner ring is composed of seven different β subunits. No atomic-resolution structure of the RP is available, and the positions of many individual subunits within it are uncertain. Our understanding of its structure to date is derived primarily from protein interaction analyses and medium-resolution electron microscopy (Bohn et al., 2010; da Fonseca and Morris, 2008).
The RP can be separated into two subcomplexes, the lid and base (Glickman et al., 1998). The base includes a ring of six related AAA+ ATPase (Rpt) subunits (Rpt1–6) that directly abuts the CP and three non-ATPase (Rpn) subunits, Rpn1, Rpn2, and Rpn13. Rpn13 and another RP subunit, Rpn10, are intrinsic polyubiquitin receptors that cooperate with mobile extrinsic ubiquitin receptors to promote ubiquitinated substrate binding to the proteasome.
The lid shares sequence and structural similarities with the COP9 signalosome (CSN) and the eIF3 translation initiation complex. Subunits of these three complexes each contain one of two characteristic motifs: PCI (Proteasome/COP9/Initiation complex) domains or MPN (Mpr1/Pad1, N-terminal) domains (Hofmann and Bucher, 1998). Both PCI and MPN domains are thought to be protein-protein interaction domains. The proteasome lid contains nine subunits: six PCI domain-containing subunits, Rpn3, Rpn5–7, Rpn9, and Rpn12; two MPN subunits, Rpn8 and Rpn11; and Sem1/Rpn15. Rpn11 bears a variant of the MPN domain, termed MPN+ or JAMM (Maytal-Kivity et al., 2002), which harbors an essential deubiquitylating activity that cleaves polyubiquitin chains from substrates (Finley, 2009).
Although the composition and functions of the proteasome are well documented, how this complex and highly abundant molecular machine is assembled from at least 33 different polypeptides is still poorly understood. Proteasome biogenesis is conserved across species and involves elements of stochastic self-assembly as well as chaperone-mediated subunit assembly. Much progress has been made recently in understanding both CP and RP-base assembly. CP biogenesis proceeds through the assembly of two half-proteasomes, which then dimerize, triggering the autocatalytic cleavage of β-subunit propeptides and yielding active CP (Chen and Hochstrasser, 1996). CP assembly is facilitated by three proteasome-specific assembly chaperones (Kusmierczyk and Hochstrasser, 2008). Base assembly is facilitated by four evolutionarily conserved chaperones; chaperone-bound intermediates associate to yield the assembled base precursor (Tomko and Hochstrasser, 2011).
In contrast to base assembly, very little information is available on lid biogenesis, although it appears to be able to form independently of the base in both yeast and mammals (Isono et al., 2007; Kaneko et al., 2009). The lid can be divided into two modules based on protein-protein interaction data (Sharon et al., 2006). Module 1 contains subunits Rpn5, Rpn6, Rpn8, Rpn9, and Rpn11, and module 2 consists of Rpn3, Rpn7, Rpn12, and Sem1 (see Fig. 4C). Several lid subcomplexes have been reported in vivo, including module 1 (Fukunaga et al., 2010), but the specific steps in lid assembly remain unclear. In yeast, fully formed base and lid complexes are present at low levels; no RP assembly intermediates containing subcomplexes of both the lid and base have been identified. Thus, lid and base probably join only upon complete assembly of each complex. By contrast, a complex consisting of fully formed lid bound to base subunits Rpt3, Rpt6, Rpn2, and the deubiquitylating enzyme Uch37 has been purified from mammals (Thompson et al., 2009). This suggests that in mammals either lid assembly occurs in conjunction with a subcomplex of base subunits or precedes association with base subunits, as in yeast. In short, neither the pathways and mediator(s) of lid assembly nor the means by which lid-base association is regulated during RP assembly are known.
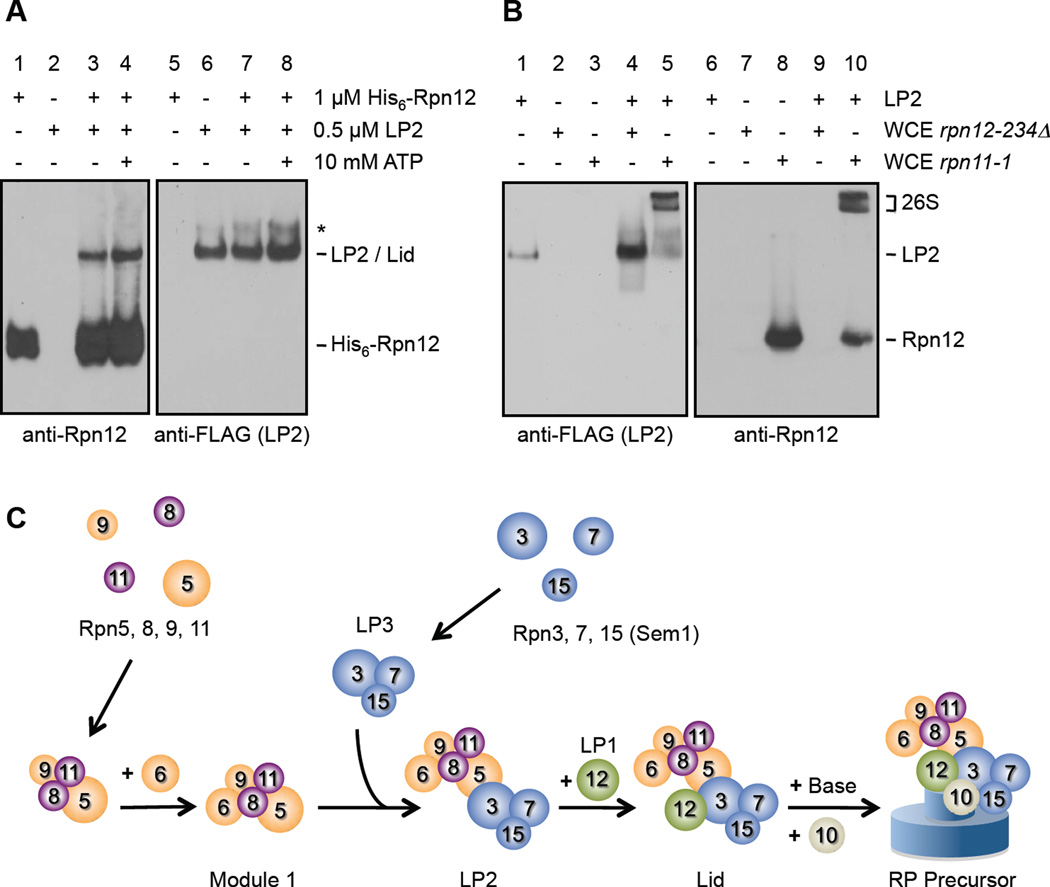
Figure 4. LP2 and Rpn12 are competent for assembly.
(A) Recombinant Rpn12 binds LP2 in vitro. Immunoblotting of nondenaturing gel-separated mixtures of the indicated species. *, the native PAGE-induced LP2 multimer.
(B) LP2 is incorporated into functional 26S proteasomes in vitro. Purified LP2 (containing Rpn5-3xFLAG) was added to WCEs from the indicated proteasome assembly-deficient strains, followed by nondenaturing PAGE and immunoblotting. Only the rpn11-1 WCE, which contains functional Rpn12, can support incorporation of LP2 into 26S proteasomes.
(C) A model of the lid biogenesis pathway. Module 1 subunits are either orange (PCI domain-containing) or purple (MPN domain-containing). Relative positions of subunits, particularly between the lid and base, are approximate.
Here, we describe a set of yeast lid intermediates, which led to the discovery of several novel features of RP assembly that help explain its high fidelity. Among the identified lid particles (LPs), a species containing only Rpn12 accumulates in all tested lid mutants, suggesting that stable Rpn12 binding to the assembling lid depends on prior incorporation of all other lid subunits. Yeast with rpn12 mutations accumulate a complementary lid intermediate, herein named LP2, containing all lid subunits except Rpn12. Rpn12 and LP2 associate in vitro to form a particle indistinguishable from the proteasomal lid, and neither of these particles can be efficiently incorporated into 26S proteasomes in the absence of the other. The highly conserved Rpn12 C-terminal tail is important both for stable Rpn12 binding to LP2 and for efficient lid-base association. Using site-directed protein crosslinking, we show that the Rpn12 tail contacts subunits in both the lid and base. Further, a C-terminal domain of Rpn12 is sufficient for proteasome formation and cell viability. Our data suggest a hierarchical lid assembly mechanism in which Rpn12 binds LP2 only upon correct assembly of all other lid subunits, and the conserved Rpn12 tail then helps drive lid-base association. Rpn12 incorporation thus acts as an assembly checkpoint linking proper lid assembly and subsequent RP assembly steps.
Results
A Sem1-containing lid intermediate
Identification of lid subcomplexes formed in vivo can provide information not only on the arrangement of subunits in the lid but also on potential assembly intermediates. Protein interaction data suggest the existence of two structural modules within the lid: module 1 contains Rpn5, Rpn6, Rpn8, Rpn9, and Rpn11 whereas module 2 contains Rpn3, Rpn7, Rpn12, and Sem1/Rpn15 (Sharon et al., 2006). Similarly, studies using yeast lid mutants identified module 1 as a stable subcomplex in the cell and a related complex bearing all module 1 subunits except Rpn6 (Isono et al., 2004; Isono et al., 2005). A module 2 subparticle containing Rpn3 and Rpn7 has been reported in strains harboring mutations in the RPN9 and RPN12 genes (Fukunaga et al., 2010), but the full module 2 has not been observed as a free-standing complex in vivo, nor have any module 2 subcomplexes bearing Sem1 or Rpn12. We therefore sought to identify lid intermediates containing Sem1 or Rpn12.
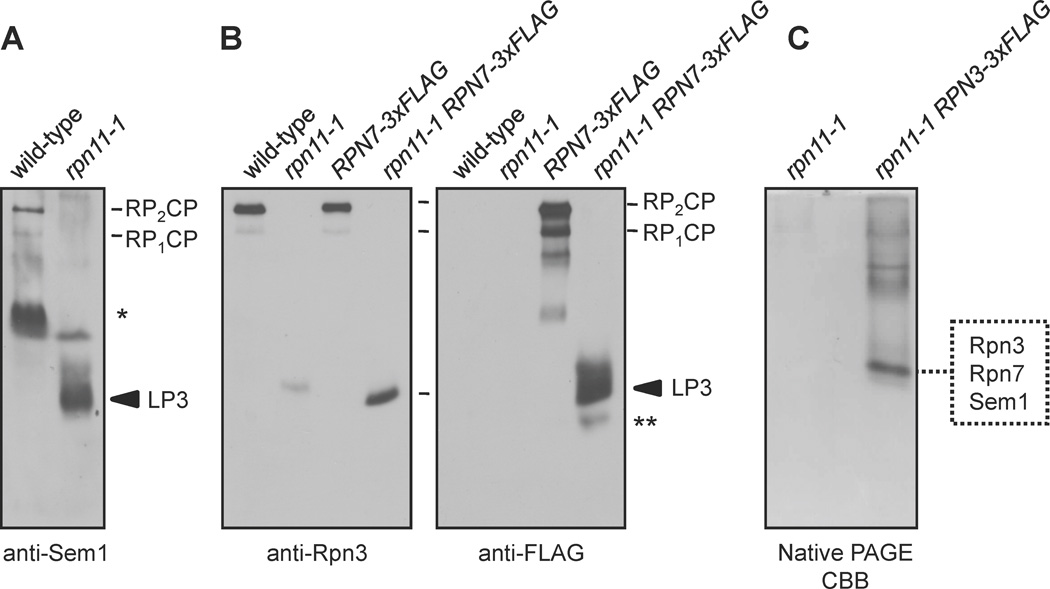
If lid assembly proceeds by formation and subsequent joining of modules 1 and 2, then disruption of module 1 might cause accumulation of module 2 intermediates. To disrupt module 1, we used the rpn11-1 mutant, which has a defect in lid assembly (Verma et al., 2002). Anti-Sem1 immunoblot analysis of yeast whole cell extracts (WCE) separated by nondenaturing polyacrylamide gel electrophoresis (native PAGE) was used to compare the profiles of Sem1-containing complexes from wild-type (WT) and rpn11-1 strains. In WT yeast, Sem1 existed primarily in 26S proteasomes and in a faster-migrating species that was unreactive with other proteasomal antibodies (Fig. 1A, asterisk and not shown), likely one of several known nonproteasomal Sem1-containing complexes (Wilmes et al., 2008). In rpn11-1 WCEs, Sem1 was not detected in 26S proteasomes but instead was found in a novel species, which we called lid particle 3 (LP3); LP3 was not seen in WT yeast (Fig. 1A).
Figure 1. Identification of lid intermediates containing Rpn3, Rpn7, and Sem1/Rpn15.
(A and B) Immunoblot analysis of native PAGE-separated proteins from the indicated strains. LP3 (lid particle 3) indicates a species that is enriched in rpn11-1 mutants. *, a species observed in anti-Sem1 blots that is not reactive with other proteasome antibodies; **, a species apparently containing only Rpn7.
(C) Native PAGE separation of FLAG affinity-purified proteins from rpn11-1 cells followed by Coomassie Brilliant Blue (CBB) staining. LC-MS/MS analysis of the excised species identified Rpn3, Rpn7, and Sem1/Rpn15.
LP3 was also observed in immunoblots using antibodies against Rpn3 and Rpn7 (Fig. 1B), but not in immunoblots with antibodies to components of the base or CP or to lid subunits Rpn5, Rpn8, or Rpn12 (not shown). We frequently observed a poorly-resolved species that trailed LP3 in Sem1 and FLAG (Rpn7) blots, but not in Rpn3 blots (Fig. 1A and B). This may indicate that Rpn7 and Sem1 form a less stable particle not containing Rpn3. Rpn3, Rpn7, and Sem1 also co-eluted from a gel filtration column at a position close to that predicted for the three-subunit complex (Supp. Fig. S1).
To purify LP3 and determine its composition, we modified the RPN3 locus with a sequence encoding a 3xFLAG epitope tag. Rpn3-3xFLAG-containing complexes were eluted from an anti-FLAG antibody resin using excess 3xFLAG peptide and resolved by native PAGE. The LP3 band (and the same region from an untagged RPN3 rpn11-1 control purification) was excised, digested with trypsin, and subjected to LC-MS/MS peptide sequencing (Fig. 1C). The analysis indicated that LP3 indeed consisted exclusively of Rpn3, Rpn7, and Sem1 (Fig.1C and Supp. Table S3). LP3 might thus represent an intermediate in the formation of module 2 or the full lid.
Rpn12 accumulates in a panel of lid mutants
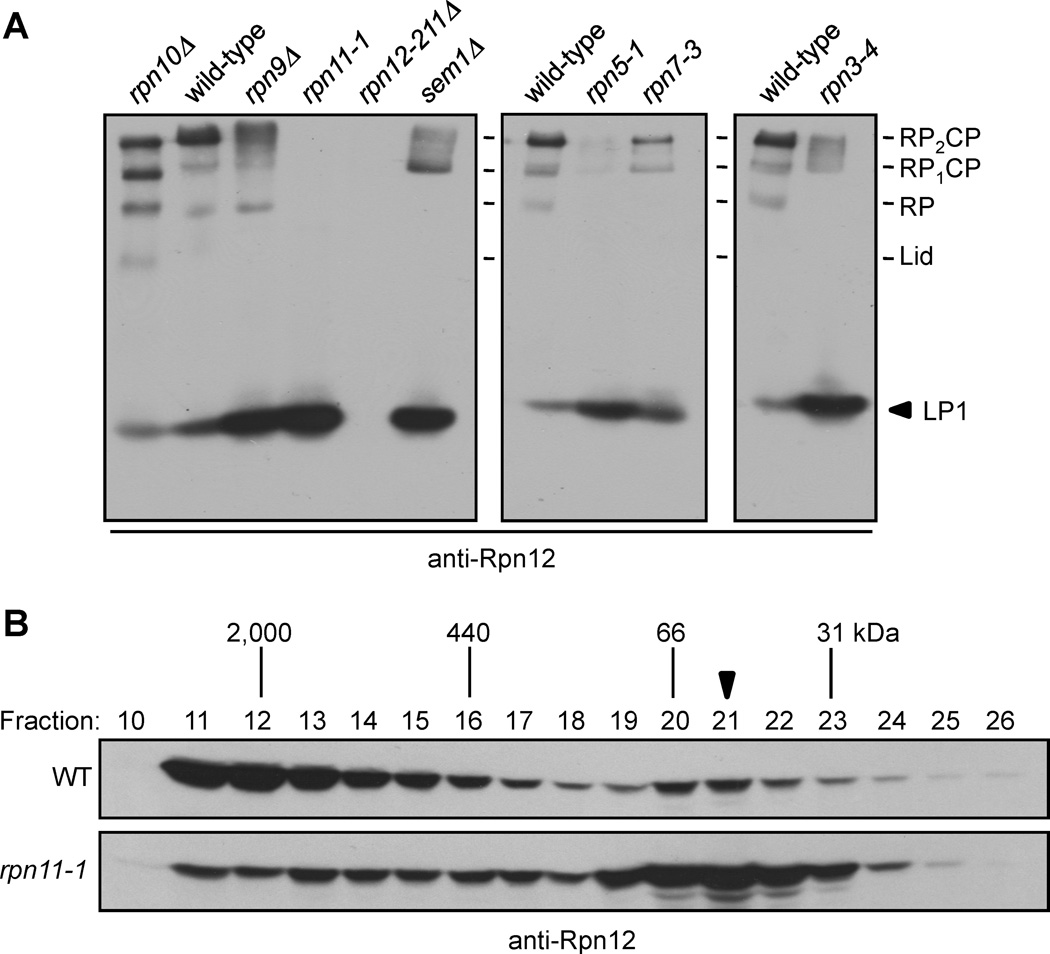
Although Rpn12 interacted with Rpn3 in yeast two-hybrid experiments (Fu et al., 2001), we did not detect Rpn12 in LP3. Free Rpn12 has been observed in rpn7-3 and rpn9Δ mutants (Fukunaga et al., 2010), but these strains expressed Rpn12 with a large C-terminal GFP-3xFLAG tag; this might have caused accumulation of free Rpn12 by impairing its incorporation into 26S proteasomes. We therefore searched for Rpn12-containing subparticles in strains expressing untagged WT Rpn12. We used native PAGE and anti-Rpn12 immunoblot analysis of a panel of lid mutants that encompassed nearly all lid subunits (Fig. 2A). In WT cells, Rpn12 was present mainly in doubly capped 26S proteasomes (RP2CP), with a small amount present as singly capped proteasomes (RP1CP) and free RP. A fast-migrating species, which we called LP1, was also present at low levels.
Figure 2. Rpn12 incorporation is a late step in lid assembly and depends upon the integrity of the assembling lid.
(A) Immunoblot analysis of native PAGE-separated proteins from whole-cell extracts (WCEs) of the indicated strains. The arrowhead indicates LP1, an Rpn12-containing species.
(B) Superose-12 fractionation of WCEs demonstrate a shift of Rpn12 from full proteasomes to a fast-migrating species in rpn11-1 cells (arrowhead).
Remarkably, in each lid mutant, the abundance of 26S proteasomes was decreased and was accompanied by an increase in LP1. (Rpn12 was not detected in the rpn12-211Δ mutant because the antibody epitope is deleted (Supp. Fig. S2A)). LP1 was unreactive to CP, base, Rpn3, Rpn5, Rpn8, or Sem1 antibodies, and was present in rpn9Δ and sem1Δ cells (Fig. 2A and not shown), indicating it also did not contain Rpn9 or Sem1. LP1 accumulation was not due to compensatory proteasome subunit overexpression because it still accumulated in cells lacking Rpn4, a transcription factor necessary for increased proteasome expression when proteasome activity is limiting (Supp. Fig. S2B). We also observed the accumulation of LP1, with an estimated size of ~40 kDa, in Superose 12-separated WCEs from rpn11-1 yeast (Fig. 2B, arrowhead), very close to the calculated mass of Rpn12 (32 kDa). These data implied that LP1 likely consisted of Rpn12 alone. This was confirmed by purification of LP1 from rpn11-1 cells and comparison to bacterially expressed recombinant Rpn12 (Supp. Fig. S2C).
We conclude that free Rpn12 exists at low levels in WT yeast and builds up in lid mutants. Despite their very different mutations and presumed positions in the lid, mutation of every tested lid subunit caused an increase in free Rpn12. This suggests that stable binding of Rpn12 depends on the integrity of all other lid subunits, and that addition of Rpn12 to the assembling lid may be a late (or final) step in lid assembly.
A nearly complete lid accumulates in rpn12 mutants
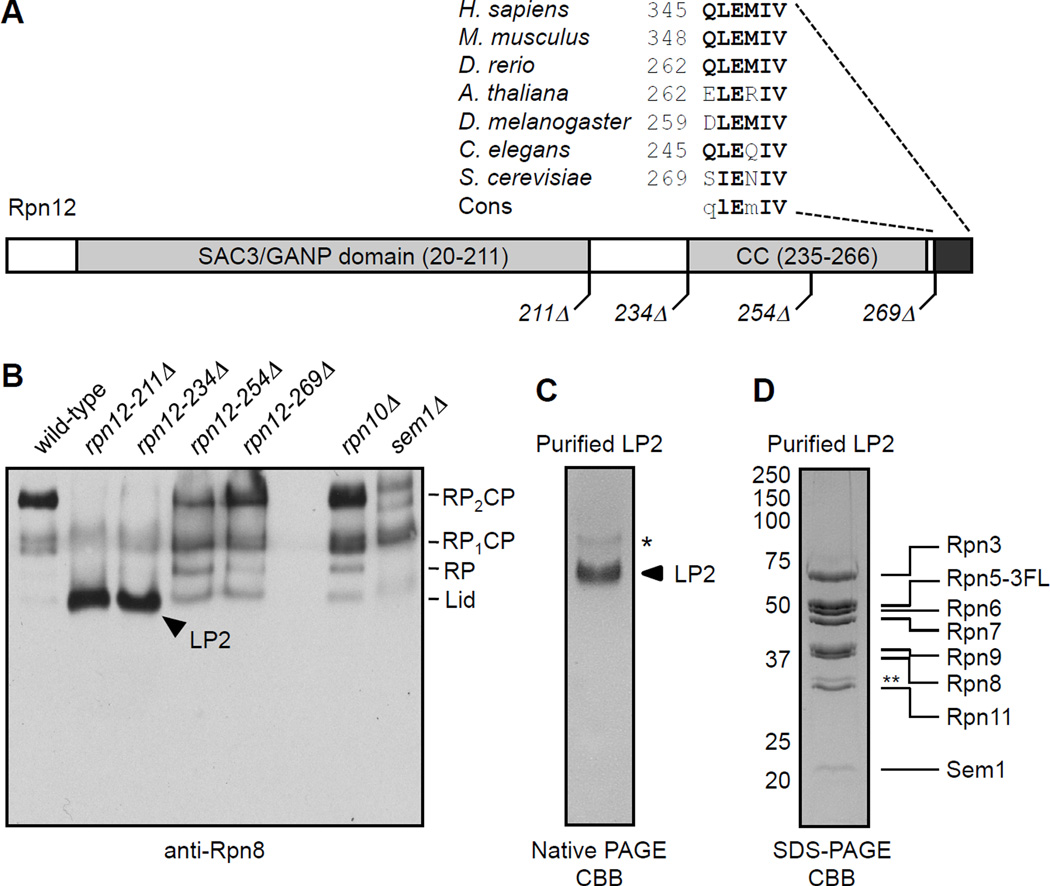
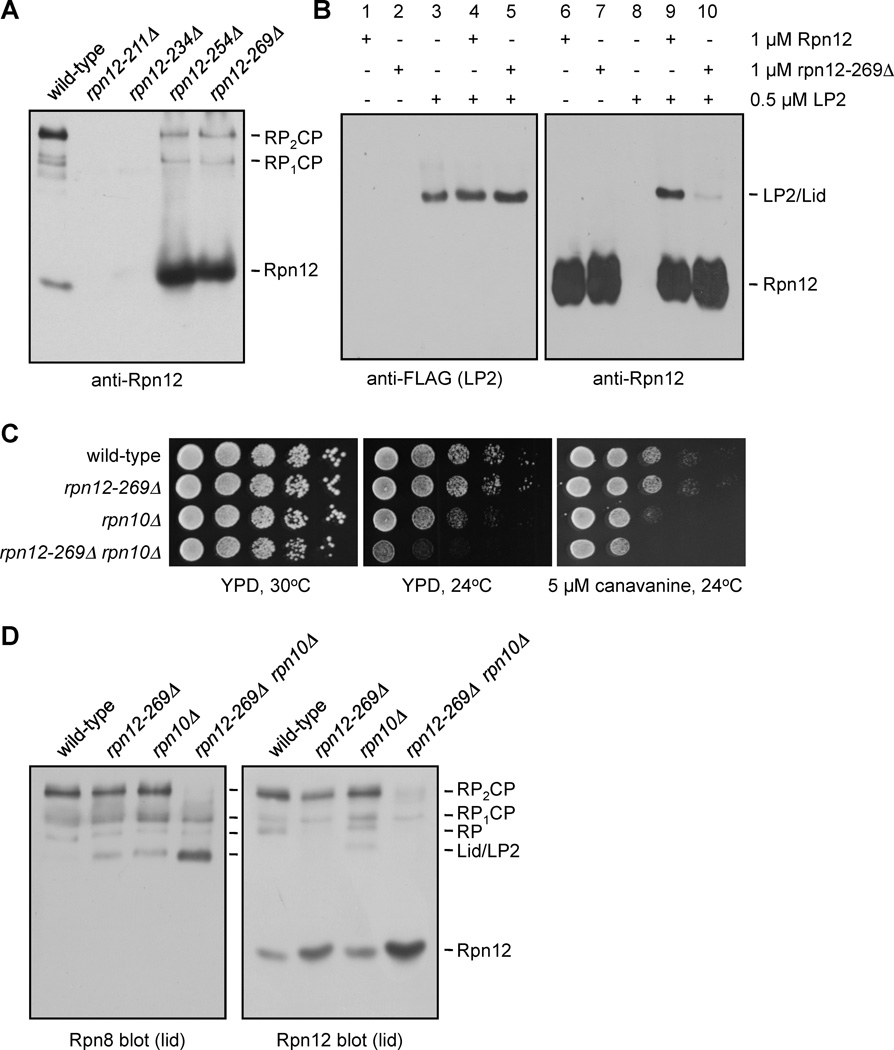
If Rpn12 incorporation completes lid assembly, then a lid subparticle containing all lid subunits except Rpn12 might accumulate in rpn12 mutants. To test this, we created a series of rpn12 alleles in the normal chromosomal locus that expressed C-terminally truncated forms of Rpn12 (Fig. 3A). Rpn12 has an N-terminal SAC3/GANP domain (amino acids 20–211) and a C-terminal domain containing a putative coiled coil (residues 235–266). Our sequence analysis also revealed that the very C-terminus of Rpn12 is highly conserved (Fig. 3A). Based on this analysis, we constructed four truncation mutants: rpn12-211Δ, rpn12-234Δ, rpn12-254Δ, and rpn12-269Δ. The rpn12-211Δ strain lacks all sequences after the SAC3/GANP domain and behaves similarly to the rpn12-1 mutation (Kominami and Toh-e, 1994). The rpn12-234Δ and -254Δ mutants are truncated immediately before or in the middle of the putative coiled coil, respectively, and the rpn12-269Δ mutant lacks only the five highly conserved C-terminal residues. These strains displayed distinct growth deficiencies, with the severity correlating with the extent of the deletion (Supp. Fig. S3A and B).
Figure 3. LP2 contains all lid subunits except Rpn12.
(A) Domain organization of Rpn12, with the locations of C-terminal truncations utilized in this study. The highly conserved C-terminal peptide of Rpn12 and its orthologs is highlighted.
(B) Immunoblot analysis of native PAGE-separated WCEs from rpn12 truncation mutants. LP2 (arrowhead) is an Rpn8-containing species that accumulates in the rpn12 truncation mutants but not a sem1Δ lid mutant or an rpn10Δ mutant.
(C) Native PAGE analysis of purified LP2 displays a single major species. *, a native PAGE-induced LP2 multimer not observed via analytical ultracentrifugation.
(D) SDS-PAGE analysis of purified LP2, indicating its eight protein components. Proteins listed were identified by LC-MS/MS analysis in a separate purification. *, an Rpn5 fragment not found in all purifications.
By native-PAGE immunoblotting with anti-Rpn8 (lid) antibodies, most Rpn8 was detected in doubly capped (RP2CP) 26S proteasomes (Fig. 3B). In the rpn12-254Δ and -269Δ mutants, increased levels of two particles, approximately the size of free RP and lid, were observed. For the rpn12-211Δ and -234Δ mutants, we observed a highly abundant particle that migrated close to the position of free lid; we named this particle LP2 (Fig. 3B, arrowhead). LP2 accumulation was accompanied by a near complete loss of detectable 26S proteasomes. By comparison, neither rpn10Δ nor sem1Δ mutants, which have lid-base attachment and lid assembly defects, respectively, showed a strong increase in LP2 levels; hence, LP2 is not accumulating due to a general defect in lid or RP assembly. To determine the composition of LP2, we introduced an RPN5-3xFLAG allele into the rpn12-234Δ strain (Supp. Fig. S3C). LP2 was purified by sequential FLAG affinity and anion exchange chromatography. Native PAGE analysis of purified LP2 displayed a single major species (Fig. 3C). Purified LP2 was completely unreactive with antibodies to the CP, base, or Rpn12 (not shown and below). LC-MS/MS peptide sequencing of purified LP2 identified Rpn3, Rpn5–9, Rpn11, and Sem1 (Supp. Table S4). These proteins were visible in roughly equal amounts by SDS-PAGE (Fig. 3D). These data suggest that LP2 is a structurally stable particle that contains all lid subunits other than Rpn12, consistent with a model in which Rpn12 completes lid assembly.
Rpn12 and LP2 are competent for assembly into 26S proteasomes
If lid assembly culminates with binding of Rpn12 to LP2, then purified LP2 and Rpn12 should associate with one another in vitro. We incubated purified recombinant His6-Rpn12 and purified LP2 alone or together, and determined their interaction by native PAGE analysis (Fig. 4A). Rpn12 by itself migrated relatively quickly (lane 1), similar to LP1 from yeast (Fig. 2A). As expected, purified LP2 did not react with anti-Rpn12 antibodies (lane 2). When mixed, a portion of Rpn12 displayed a retarded migration identical to that of LP2, indicating that Rpn12 binds LP2 in vitro (Fig.4A, lanes 3 and 7). The total fraction of LP2-bound Rpn12 was rather low; this likely reflects interference of binding by the His6 tag and linker (not shown) and potentially the absence of other factors that facilitate LP2-Rpn12 interaction in vivo. The amount of Rpn12 that bound LP2 was not affected by ATP, presumably because no ATPases are present in this purified system (Fig. 4A, lane 3 vs. lane 4).
To test whether free Rpn12 and purified LP2 could be incorporated into 26S proteasomes, we exploited the observations that 1) rpn11-1 yeast contain high amounts of free Rpn12 but cannot form LP2 due to the truncated Rpn11 protein; 2) rpn12-234Δ cells have the complementary defect, bearing only minimally functional Rpn12; and 3) neither strain has detectable 26S proteasomes by native PAGE (Figs. 1A and 3B). LP2 migrated as a single major species by anti-FLAG immunoblotting (Rpn5-3xFLAG) (Fig. 4B lane 1). When LP2 was added to rpn11-1 WCEs, both LP2 and Rpn12 rapidly shifted into two slowly-migrating species characteristic of 26S proteasomes (lanes 5 and 10). These complexes were reactive with CP and base subunits and they rapidly hydrolyzed the proteasome substrate suc-LLVY-AMC (not shown), indicating they were fully formed, active 26S proteasomes. No FLAG-reactive subcomplexes were observed that migrated faster than LP2, suggesting that LP2 incorporated into 26S proteasomes intact. While purified lid rapidly assembled into 26S proteasomes in rpn12-234Δ WCEs (not shown), LP2 did not (lane 4). This indicates that LP2 depends upon Rpn12 incorporation for subsequent assembly steps.
These in vitro data demonstrate first, that Rpn12 and LP2 directly associate, thereby completing lid assembly, and second, that they are competent intermediates for assembly into active proteasomes. Importantly, neither can assemble efficiently into proteasomes in the absence of the other. The in vivo and in vitro results are summarized in the provisional assembly pathway depicted in Fig. 4C.
Function of the conserved C-terminal Rpn12 tail in RP assembly
To understand why LP2 depends on Rpn12 for further RP assembly steps, we reexamined our rpn12 truncation mutants by native gel immunoblotting for Rpn12 (Fig. 5A). Surprisingly, both the rpn12-254Δ and rpn12-269Δ strains suffered a substantial loss of Rpn12 from fully-formed proteasomes and a large increase in free Rpn12 (Fig. 5A). The purified rpn12-269Δ protein also bound much more poorly than WT Rpn12 to LP2 in native gel binding assays (Fig. 5B). Therefore, the C-terminal pentapeptide is necessary for stable incorporation of Rpn12 into the assembling lid both in vivo and in vitro.
Figure 5. The conserved Rpn12 C-terminal tail is required for efficient proteasome assembly.
(A) Impaired in vivo incorporation of C-terminally truncated Rpn12 into the proteasome. Proteins from the indicated strains were resolved by native PAGE and visualized by anti-Rpn12 immunoblotting. Rpn12 is not detected in the rpn12-211Δ and rpn12-234Δ mutants due to epitope loss.
(B) Recombinant Rpn12 missing its five C-terminal residues binds purified LP2 poorly.
(C) Enhanced phenotypic defects in an rpn12-269Δ rpn10Δ double mutant. Serial dilutions of strains were spotted onto the indicated media and incubated for 3 d.
(D) Immunoblot analysis of native PAGE-separated proteins from the indicated strains.
The proteasomal ubiquitin receptor Rpn10 interacts directly with Rpn12 in S. pombe (Riedinger et al., 2010). In S. cerevisiae, deletion of RPN10 destabilizes interaction between the lid and base (Glickman et al., 1998), and the rpn12-1 mutation is lethal when combined with rpn10Δ (Fujimuro et al., 1998), suggesting a partially redundant function for these proteins. RPN10 deletion was also lethal with rpn12-211Δ, rpn12-234Δ, and rpn12-254Δ (not shown), but the rpn10Δ rpn12-269Δ double mutant was viable (Fig. 5C). The double mutant displayed enhanced growth defects relative to the single mutants at 24°C and on plates containing the amino acid analog canavanine, implying an overlapping function for Rpn10 and the Rpn12 C-terminal tail, perhaps in lid-base stabilization. In contrast, rpn12-269Δ did not interact genetically with the sem1Δ lid mutant (Supp. Fig. S4), suggesting that the synthetic phenotype with rpn10Δ might reflect a function for the Rpn12 tail beyond a generalized lid assembly defect.
The synthetic growth defects of the rpn10Δ rpn12-269Δ mutant were paralleled by defects in proteasome assembly. As expected, the rpn10Δ mutants displayed a modest increase in free lid (Fig. 5D). An Rpn8-reactive species migrating identically to LP2 was present at high levels in the double mutant. This species was not reactive with antibodies to Rpn12, base or CP (Fig. 5D and not shown) and thus is likely LP2. The rpn10Δ rpn12-269Δ mutant also had increased free Rpn12 compared to either single mutant (Fig. 5D), suggesting that, while Rpn10 is not necessary for incorporation of WT Rpn12 into proteasomes, it helps to stabilize proteasome-associated Rpn12 when its pentapeptide tail is missing. Together, these results suggest an overlapping function of the Rpn12 tail with Rpn10 in lid-base attachment.
The Rpn12 pentapeptide tail bridges lid and base
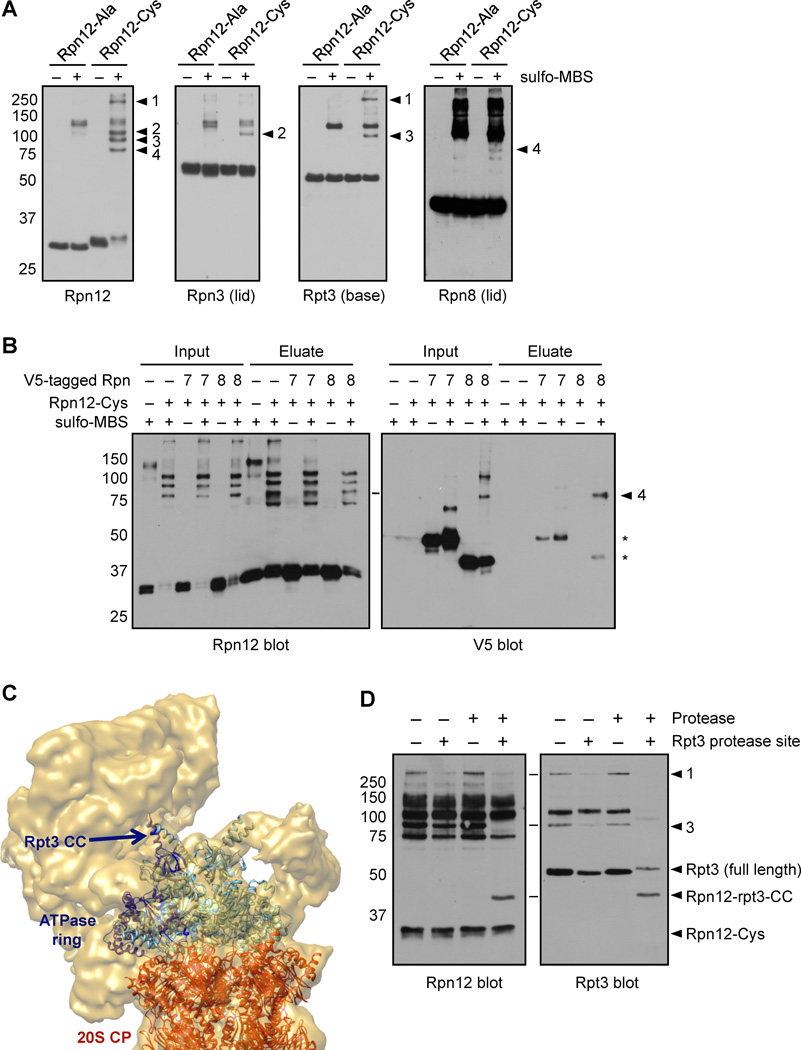
If the Rpn12 tail contributes to lid-base attachment by interacting with the base, it would be expected to be near the interface of these RP subcomplexes. We tested this using a novel site-directed protein crosslinking approach. We engineered strains with RPN12 alleles driven by the normal promoter but encoding either a C-terminal cysteine-rich extension (GCCC-6His, “rpn12-Cys”) or a control extension (GAA-6His, “rpn12-Ala”). The cysteines at the Rpn12 tail can be targeted by sulfhydryl-specific crosslinkers. Purified proteasomes from these strains contained all subunits in equal amounts (Supp. Fig. S5A). We treated the proteasomes with the sulfhydryl- and amine-reactive heterobifunctional crosslinker sulfo-MBS (7.3 Å linker length). Both proteasomes and crosslinker were at low concentration to limit interproteasomal crosslinks. Sulfo-MBS treatment followed by anti-Rpn12 immunoblotting (Fig. 6A, left panel) revealed several slow-migrating species in both rpn12-Ala and rpn12-Cys proteasomes that were not seen without sulfo-MBS: a faint band at ~110 kDa and several closely-migrating bands at ~125 kDa. Most importantly, four additional bands unique to rpn12-Cys were detected (Fig. 6A, numbered arrowheads). Based on their sizes, bands 2–4 were likely Rpn12-containing heterodimers, whereas band 1 was probably a higher order multimer. The same four bands were observed in experiments using proteasomes with a single cysteine in the extension (Supp. Fig. S5B), indicating that the crosslinks could occur with the cysteine closest to the tail.
Figure 6. The Rpn12 C-terminal tail is positioned at the lid-base interface.
(A) Purified proteasomes containing Rpn12 subunits with either an alanine- or cysteine-containing C-terminal tag were incubated with the sulfo-MBS crosslinker, and SDS-PAGE-resolved proteins were analyzed by immunoblotting. Numbered arrowheads indicate four cysteine-dependent crosslinked species.
(B) Purified proteasomes containing His10-rpn12-Ala or His10-rpn12-Cys and the indicated V5-tagged subunits were incubated with sulfo-MBS, denatured in 8 M urea, and Rpn12 (and subunits crosslinked to it) were purified via the His10 tag. *, small amounts of contaminating uncrosslinked lid subunits.
(C) The Rpt3 N-terminal domain points toward the proteasomal lid. The position of the Rpt3 coiled coil (Rpt3-CC) modeled within the cryo-EM structure of the Drosophila 26S proteasome (Bohn et al., 2010) is shown. Image rendered with Chimera software.
(D) Proteasomes containing rpn12-Cys and either WT Rpt3 or Rpt3 with a protease cleavage site immediately following the coiled coil domain were crosslinked as in (A) before protease treatment, followed by immunoblotting with Rpn12 or Rpt3 antibodies. Rpn12-Rpt3-CC, Rpn12 crosslinked to the cleaved N-terminal domain of Rpt3 (predicted MW ≈ 44 kDa). The Rpt3 antibody used (Enzo #PW8250) is directed toward the N-terminus of Rpt3, and did not recognize the C-terminal fragment of Rpt3 generated upon protease treatment.
To identify the proteins crosslinked to the Rpn12 tail, we immunoblotted the same crosslinking reaction products with antibodies to RP base and lid subunits. We tested all RP subunits either by subunit-specific immunoblotting, or in the case of Rpn13, deletion of the chromosomal locus, which did not affect the crosslinking pattern (not shown). Two RP subunits were clearly part of Rpn12-crosslinked products: the Rpn3 lid subunit and the Rpt3 base ATPase. Both Rpn3 and Rpt3 interact with Rpn12 in yeast two-hybrid assays (Chen et al., 2008; Fu et al., 2001). Interaction between Rpn12 and Rpn3 was further supported by our findings that purification of overexpressed Rpn12 from yeast copurified a small amount of Rpn3 but no other subunits (Supp. Fig. S5C). A weak cysteine- and crosslinker-dependent species (arrowhead 4) was observed in Rpn8 blots (Fig. 6A). This species was also observed in V5 immunoblots from a strain expressing Rpn8-V5 from its chromosomal locus, and copurified with His-tagged Rpn12 under denaturing conditions (Fig. 6B), confirming it as an Rpn12-Rpn8 crosslink. Together, these results place the Rpn12 tail near lid subunits in both module 1 and LP3 and indicate it also lies near the lid-base interface, consistent with a role in lid attachment.
Guided by protein sequence information and crystal structures of the archaeal ATPase homolog PAN, the six eukaryotic ATPases have been docked into the cryo-EM density of the 26S proteasome (Bohn et al., 2010), and their model agrees with our experimentally determined ATPase arrangement (Tomko et al., 2010). In this pseudo-atomic model, the N-terminal domain of Rpt3, which contains a coiled coil domain, points toward the center of the most distal mass of the proteasome, presumed to be the lid (Fig. 6C). To localize the site of the Rpn12-Rpt3 crosslink, we engineered a protease site into Rpt3 that upon cleavage separates the N-terminal domain from the rest of the protein. Protease treatment of crosslinked proteasomes resulted in loss of the Rpn12-Rpt3 crosslink (arrowhead 3) in both Rpn12 and Rpt3 blots, and a new species at ≈ 44 kDa appeared (Fig. 6D), as expected for a crosslink between Rpn12 and the N-terminal domain of Rpt3. This provides the first insight into the location of Rpn12 within the RP, and places the Rpn12 tail close to the N-terminus of Rpt3 and thus the lid-base interface.
The Rpn12 C-terminal tail contributes to lid attachment
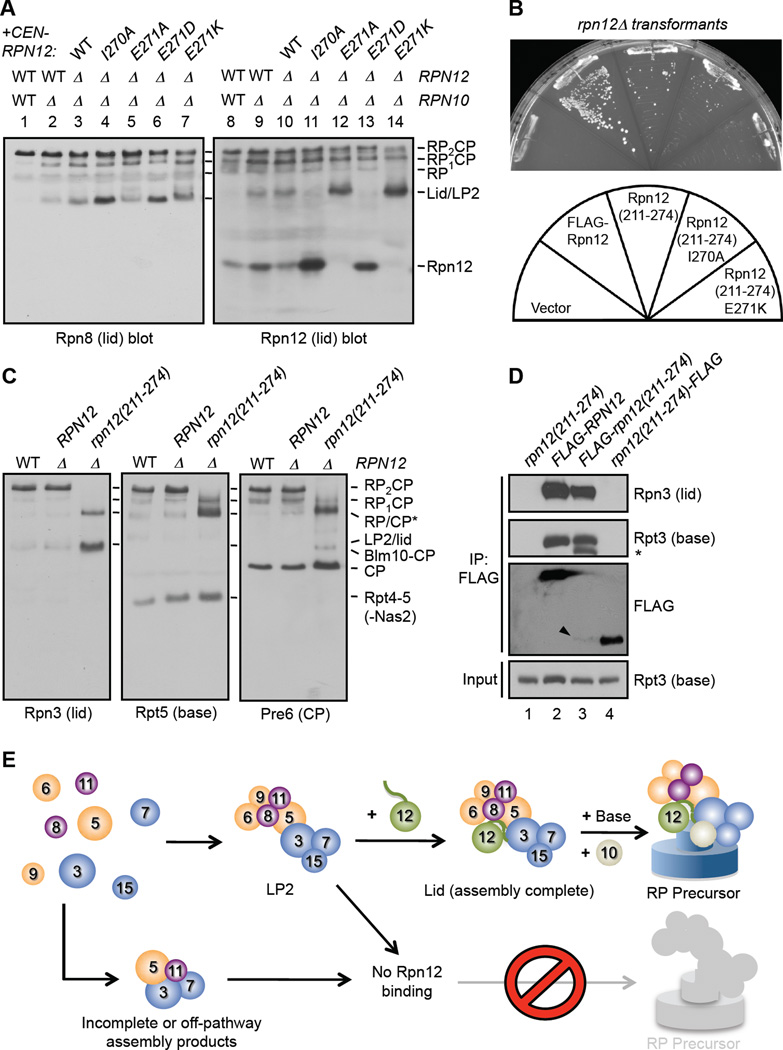
We attempted to verify the putative lid assembly and lid-base attachment functions of the Rpn12 tail by mutagenesis. We generated rpn12 alleles bearing point mutations in the five C-terminal residues and introduced them by plasmid shuffle into an rpn10Δ rpn12Δ strain, which depends upon the Rpn12 tail for efficient proteasome assembly (Fig. 5D). The mutant proteins were expressed similarly to WT Rpn12 (Supp. Fig. S6A). Although highly conserved, mutation of the two C-terminal amino acids to alanine either alone or in combination revealed no apparent assembly defects (not shown). In contrast, the rpn10Δ rpn12-I270A mutant accumulated high levels of free Rpn12 and an Rpn8-containing species devoid of Rpn12, likely LP2 (Fig. 7A). This suggests that Rpn12-I270 contributes primarily to LP2-Rpn12 association. Strikingly, the E271A and E271K mutants had no detectable free Rpn12 and accumulated increased amounts of an Rpn8- and mutant Rpn12-containing species, likely the lid. The charge-conserving E271D mutant showed only a small increase in the Rpn8-reactive species and a mild accumulation of free Rpn12, suggesting the Glu271 residue may normally interact with a positively charged site, possibly in the base.
Figure 7. The Rpn12 C-terminal tail is involved both in LP2 binding and lid-base joining.
(A) Mutation of the Rpn12 C-terminal tail selectively disrupts either Rpn12 binding to LP2 or lid association with base. In lanes 5 and 7, increased free lid is not obvious by Rpn8 blotting; it is possible these structurally perturbed lids aren’t recognized efficiently by the Rpn8 antibody.
(B) A C-terminal fragment of Rpn12 lacking the SAC3/GANP domain – rpn12(211-274) – is sufficient for viability. Mutant rpn12Δ cells bearing plasmids expressing the indicated alleles from a GPD promoter were struck on 5-fluoroorotic acid to evict the resident WT RPN12 plasmid and grown at 24°C for 4 d.
(C) Mutant rpn12(211-274) cells can form a species that migrates near RP1CP and is positive for lid, base, and CP subunits. The indicated strains were analyzed by native gel immunoblotting.
(D) The rpn12(211-274) fragment is incorporated into the RP. FLAG-rpn12(211-274), but not rpn12(211-274)-FLAG, efficiently precipitates lid and base subunits. Arrowhead marks FLAG-rpn12(211-274), which is functional but present at very low levels, even in immunoprecipitates. *, cleavage product of Rpt3.
(E) Hierarchical model for Rpn12-dependent lid assembly and lid-base joining. Rpn12 binds tightly only when the full LP2 lid precursor has formed properly, and lid subunits only associate efficiently with the base if Rpn12 has incorporated into the lid. The Rpn12 C-terminal tail (wavy green line) interacts with components of both lid subcomplexes (Rpn3 of LP2 and Rpn8 of module 1) and facilitates lid-base joining by interaction with the Rpt3 ATPase.
The above data suggest that the C-terminal pentapeptide tail is necessary for stable lid-base interaction (in the absence of Rpn10). We also asked whether the Rpn12 segment C-terminal to the SAC3/GANP domain was sufficient for RP assembly and function. If the major point of contact between Rpn12 and the base is in the C-terminus and this region also contributes to lid association of Rpn12, then the C-terminal domain of Rpn12 may also be sufficient for viability. We created rpn12Δ strains with high-copy plasmids encoding full-length Rpn12 or a fragment lacking the SAC3/GANP domain, rpn12(211-274). Expression of these alleles was driven from a strong promoter because the rpn12(211-274) fragment was expressed at extremely low levels (see below). Yeast expressing the rpn12(211-274) construct were viable at 24°C, but grew slowly (Fig. 7B). Introduction of the I270A or E271K mutations further compromised growth, consistent with the observed biochemical defects in lid assembly and attachment.
Although the rpn12(211-274) mutant had substantially reduced levels of 26S proteasomes, we observed a low level of RP1CP and two closely-migrating species that appears to represent a mixture of RP and (uncharacterized) CP-containing complexes (RP/CP*) (Fig. 7C). This RP-like species contained all lid and base subunits tested as well as the RP chaperone Nas6, but not Rpn10 (not shown), indicating that the N-terminal domain of Rpn12 may be required for stable binding of Rpn10 to proteasomes. While we could not detect the rpn12(211-274) protein in these complexes due to its very low expression, FLAG-rpn12(211-274) co-precipitated base and lid subunits from WCEs nearly as well as when full-length FLAG-Rpn12, despite the very low relative abundance of the fragment (Fig. 7D, lanes 2 and 3). In contrast, a C-terminally FLAG-tagged fragment could not efficiently precipitate RP subunits, further supporting the importance of the Rpn12 C-terminus in RP assembly, especially in the absence of the N-terminal SAC3/GANP domain. Thus, the C-terminal domain of Rpn12 is sufficient for yeast viability and formation of proteasomes.
Together, these data demonstrate that the C-terminal tail of Rpn12 functions in RP assembly by promoting both stable Rpn12 incorporation into the lid and lid-base association.
Discussion
Our data suggest a hierarchical mechanism of lid assembly and base attachment that is mediated by Rpn12 (Fig. 7E). Identification of the three lid intermediates LP1/Rpn12, LP2, and LP3 supports an assembly model in which LP3 associates with lid module 1 to form LP2, which can only then bind Rpn12 stably and thereby complete lid assembly (Fig. 4C). LP2 and Rpn12 depend on each other for incorporation into 26S proteasomes, consistent with the idea that integration of Rpn12 into the lid is coupled to lid-base attachment. Rpn12 thus provides a checkpoint function in RP assembly. This role of Rpn12 is mediated, at least in part, via its highly conserved C-terminal tail, which localizes near the lid-base interface and is well positioned to transmit lid assembly status to the base.
Hierarchical lid assembly model
A recent analysis of purified lid suggests that the complex can be split into two structural modules (Sharon et al., 2006). Here we have demonstrated the formation in vivo of an Rpn3-Rpn7-Sem1 complex (LP3), which is similar to module 2 but lacks Rpn12. Our in vivo and in vitro data indicate that Rpn12, rather than completing module 2 to allow subsequent joining to module 1, only joins after module 1 has bound LP3 to form LP2 (Fig. 4C). This may be because Rpn12 binds with modest affinity to sites in both module 1 and LP3 or because LP3 binding to module 1 triggers conformational changes that enhance Rpn12 binding (or both). Consistent with a multisite-binding model, the C-terminal tail of Rpn12 appears to contact subunits in both module 1 (Rpn8) and LP3 (Rpn3). This does not rule out conformational changes that enhance or are driven by Rpn12 binding, and preliminary analyses suggest that Rpn12-dependent structural changes might indeed be occurring (Tomko et al., unpublished). By either model, Rpn12 incorporation is a key restriction point in lid assembly.
In contrast to the CP and base, no lid-specific assembly chaperones have yet been identified. General molecular chaperones such as Hsp90 have been suggested to mediate lid assembly (Imai et al., 2003), although we did not detect non-proteasomal proteins in our purified lid subcomplexes. Lid assembly may proceed by stochastic self-assembly of subunits coupled with conformational changes or generation of high avidity interactions that drive its ordered assembly. Similar hierarchical assembly mechanisms would be predicted for the related CSN (Sharon et al., 2009) and may be a general feature of the PCI/MPN protein complexes. In fact, the I270 and E271 residues that contribute to lid assembly and attachment are conserved in CSN8, the Rpn12 paralog in the CSN (Supp. Fig. S6B). These residues may contribute to CSN assembly and perhaps also to proposed CSN interactions with the proteasome (Huang et al., 2005) or the hexameric ATPase p97/VCP (Cayli et al., 2009), by analogy to lid-base attachment within the RP.
Rpn12 incorporation is required for efficient lid-base attachment
Biogenesis of the RP from its 19 subunits represents a substantial challenge. Recent EM structures suggest a dense mesh of protein-protein interactions within the RP (Bohn et al., 2010; da Fonseca and Morris, 2008), implying that the lid and base may be closely apposed at multiple positions. The apparent requirement for Rpn12 incorporation prior to lid-base joining suggests that premature attachment of lid intermediates may be problematic for subsequent assembly steps. In this regard, restraining lid attachment until completion of its assembly may act as a failsafe to minimize formation of aberrant or inactive RP complexes. Alternatively, Rpn12 incorporation into incomplete RP complexes that contain both base and lid subunits may be sterically hindered, requiring disassembly of these metastable complexes. Identification of the positions of lid and base subunits within the proteasome should allow evaluation of these possibilities.
Coordination of the ubiquitin-deconjugating, ATPase, and proteolytic activities of the proteasome is essential for properly regulated degradation of substrates. The deubiquitylating activity of the Rpn11 lid subunit is activated allosterically through association with the base and is thought to be active only in the full RP or 26S proteasome (Yao and Cohen, 2002; Verma et al., 2002). Premature docking of Rpn11-containing lid assembly intermediates to the base might aberrantly activate Rpn11, leading to deubiquitylation of proteasome substrates without unfolding or degradation. Immunolabeling of Rpn11 within the 26S proteasome EM structure has placed it very close to the base ATPase ring (Bohn et al., 2010). However, our results suggest that Rpn11 does not associate with the base prior to Rpn12 incorporation. Therefore, monitoring the completion of lid assembly prior to base attachment via this Rpn12-dependent checkpoint may limit Rpn11 activity until the full RP can form properly.
The Rpn12 C-terminal tail drives LP2 binding and lid-base association
Our results indicate that the Rpn12 C-terminal tail has two distinct functions in RP assembly. First, the tail was required for efficient binding of Rpn12 to LP2 in vitro and for incorporation of Rpn12 into proteasomes in vivo. Second, it was essential for stable and efficient lid-base association, although this function was only revealed when cells lacked Rpn10, another factor with a role in lid-base stabilization. Based on our site-specific crosslinking analysis, the Rpn12 tail probably interacts with Rpn3 and Rpn8 to promote completion of lid assembly. Crosslinking of the tail to the base subunit Rpt3 indicates that the tail lies near the interface of the lid and the base, at least in mature proteasomes. As noted earlier, a mammalian RP subparticle that contained the Rpn2, Rpt3, and Rpt6 base subunits associated with a fully formed lid has been reported (Thompson et al., 2009). An interaction between the Rpn12 tail and Rpt3 may mediate formation of this lid-base precursor complex and might even facilitate base assembly under some conditions.
Parallels to other assembly pathways
During proteasome biogenesis, C-terminal peptide tails of several subunits have emerged as important regulators of the assembly process. Besides the Rpn12 tail, the C-terminal residues of several of the RP ATPases are thought to bind to surface pockets in a preformed CP platform and appear to be necessary for efficient RP assembly in vivo, at least in yeast (Park et al., 2009). Such a mechanism could account for the proposed templating function of the CP in base assembly (Kusmierczyk et al., 2008). Another example is seen in the final assembly of the CP from two half-CP intermediates. A unique C-terminal extension of the β7 subunit binds a surface groove of the opposite half-mer, driving proper alignment and dimerization (Li et al., 2007; Marques et al., 2007).
The CP β7 subunit tail also provides another example of the hierarchical nature of proteasome assembly. Just as Rpn12 is likely the last subunit to join the assembling lid, thereby promoting attachment of the completed lid to the base, so β7 is the last subunit to join the CP half-mer precursor, allowing the completed α7β7 double-ring complexes to dimerize (Li et al., 2007). Notably, in both examples, partially redundant mechanisms work to join assembly subcomplexes. The Rpn12 tail only becomes important for lid-base joining when Rpn10 is deleted. Similarly, the β7 tail is only essential for CP dimerization when the β5 propeptide, an intramolecular chaperone, is deleted (Li et al., 2007). These data suggest that proteasome assembly does not depend on a single rigidly ordered pathway even though certain routes appear to be preferred.
Hierarchical self-assembly mechanisms are not unique to the proteasome. A well-known example is bacterial 30S ribosome assembly from purified components (Stern et al., 1989; Traub and Nomura, 1968). The 16S ribosomal RNA associates with 20 different proteins. Certain proteins bind more readily to the unfolded RNA, helping to drive further RNA conformational changes and promote binding of other proteins (Stern et al., 1989). Here too there is not a single unique pathway of protein-binding steps but a dynamic “assembly landscape” that is altered by sequential protein-binding events (Talkington et al., 2005). It will be interesting to see how proteasome assembly can bypass potential assembly bottlenecks such as Rpn12-stimulated lid-base joining, and whether alternative pathways lead to higher frequencies of misassembled complexes.
Experimental Procedures
In vitro assembly assays
Assembly assays using only purified components were performed by mixing the indicated proteins at 30°C for 30 min before analysis. Yeast extract-based assembly assays were performed in 50 µL final volumes in 26S Buffer (50 mM Tris-HCl, pH 7.4, 10% glycerol, 5 mM MgCl2) containing 1 µg of LP2, 10 mM ATP, and 120 µg of the indicated WCEs prepared in 26S Buffer. Reactions were mixed, and incubated for 30 min at 30°C. Reactions were immediately cooled to 0°C on ice, and 15µL reaction aliquots were separated by native PAGE for immunoblotting. Reactions containing only purified components were spiked with 1 mg/mL BSA as carrier protein.
Chemical crosslinking of purified proteasomes and denaturing purifications
Proteasomes were purified from RPN5-6xGly-3xFLAG rpn12Δ strains carrying RPN12 plasmids pRT388, pRT389, pRT390, pRT464, or pRT465; FLAG affinity chromatography in Buffer HA was performed as described above. Proteasomes (400 nM) were treated for 10 min at 30°C with 0.1 mM sulfo-MBS (_m_-maleimidobenzoyl-_N_-hydroxysulfosuccinimide ester; Thermo Scientific). Crosslinking was terminated with 80 mM glycine, pH 7.2. Treated proteasomes were separated on 8% SDS-PAGE gels, transferred to PVDF (Millipore), and probed with proteasome subunit-specific antibodies. For denaturing purifications, proteasomes were crosslinked as above, denatured in Buffer HA+UBT (Buffer HA supplemented with 8 M urea, 1 mg/mL BSA, and 0.5% Tween-20), and bound to TALON resin (Clontech) for 30 minutes at 24°C. After two washes in Buffer HA+UBT and one wash in Buffer HA containing 1% SDS, samples were eluted in Buffer HA containing 1% SDS and 500 mM imidazole and analyzed by immunoblotting.
Protease cleavage assays
RPN12 plasmid pRT390 and RPT3 plasmids pRT421 or pRT478 were introduced into MHY7121 by plasmid shuffle. Proteasomes were then purified and crosslinked as above. Reactions were then supplemented with 0.1% SDS, boiled for five minutes, and treated for 20 hours at 4°C with buffer or ≈ 20 µg of purified PreScission protease.
Supplementary Material
01
Acknowledgements
We thank Eric M. Rubenstein, Eri Sakata, and Christopher M. Hickey for helpful comments; Eri Sakata for rendering Fig. 6C; and Dan Finley, Akio Toh-e, Teresa Rinaldi, and Eric Bailly for reagents. This work was supported by NIH grant GM083050 to MH. RJT Jr. was supported in part by an American Cancer Society New England Division – Mass Biotech Council Cancer Research Challenge – Millennium: The Takeda Oncology Company Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, Forster F, Baumeister W, Nickell S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayli S, Klug J, Chapiro J, Frohlich S, Krasteva G, Orel L, Meinhardt A. COP9 signalosome interacts ATP-dependently with p97/valosin-containing protein (VCP) and controls the ubiquitination status of proteins bound to p97/VCP. J Biol Chem. 2009;284:34944–34953. doi: 10.1074/jbc.M109.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang C, Chen S, Liang J, Lin W, Ke G, Zhang H, Wang B, Huang J, Han Z, et al. Subunit-subunit interactions in the human 26S proteasome. Proteomics. 2008;8:508–520. doi: 10.1002/pmic.200700588. [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M, Tanaka K, Yokosawa H, Toh-e A. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1998;423:149–154. doi: 10.1016/s0014-5793(98)00084-2. [DOI] [PubMed] [Google Scholar]
- Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochemical and biophysical research communications. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Huang X, Hetfeld BK, Seifert U, Kahne T, Kloetzel PM, Naumann M, Bech-Otschir D, Dubiel W. Consequences of COP9 signalosome and 26S proteasome interaction. The FEBS journal. 2005;272:3909–3917. doi: 10.1111/j.1742-4658.2005.04807.x. [DOI] [PubMed] [Google Scholar]
- Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, Toh-e A. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Saeki Y, Yokosawa H, Toh-e A. Rpn7 Is required for the structural integrity of the 26 S proteasome of Saccharomyces cerevisiae. J Biol Chem. 2004;279:27168–27176. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]
- Isono E, Saito N, Kamata N, Saeki Y, Toh EA. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Kominami K, Toh-e A. Characterization of the function of the NIN1 gene product of Saccharomyces cerevisiae. Experimental cell research. 1994;211:203–211. doi: 10.1006/excr.1994.1079. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J Biol Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedinger C, Boehringer J, Trempe JF, Lowe ED, Brown NR, Gehring K, Noble ME, Gordon C, Endicott JA. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem. 2010;285:33992–34003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon M, Mao H, Boeri Erba E, Stephens E, Zheng N, Robinson CV. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J Biol Chem. 2009;284:24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochemistry and Biophysics. 2011;60:13–20. doi: 10.1007/s12013-011-9178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01