Direct Binding of Occupied Urokinase Receptor (uPAR) to LDL Receptor-related Protein Is Required for Endocytosis of uPAR and Regulation of Cell Surface Urokinase Activity (original) (raw)
Abstract
Low-density lipoprotein receptor-related protein (LRP) mediates internalization of urokinase:plasminogen activator inhibitor complexes (uPA:PAI-1) and the urokinase receptor (uPAR). Here we investigated whether direct interaction between uPAR, a glycosyl-phosphatidylinositol–anchored protein, and LRP, a transmembrane receptor, is required for clearance of uPA:PAI-1, regeneration of unoccupied uPAR, activation of plasminogen, and the ability of HT1080 cells to invade extracellular matrix. We found that in the absence of uPA:PAI-1, uPAR is randomly distributed along the plasma membrane, whereas uPA:PAI-1 promotes formation of uPAR-LRP complexes and initiates redistribution of occupied uPAR to clathrin-coated pits. uPAR-LRP complexes are endocytosed via clathrin-coated vesicles and traffic together to early endosomes (EE) because they can be coimmunoprecipitated from immunoisolated EE, and internalization is blocked by depletion of intracellular K+. Direct binding of domain 3 (D3) of uPAR to LRP is required for clearance of uPA-PAI-1–occupied uPAR because internalization is blocked by incubation with recombinant D3. Moreover, uPA-dependent plasmin generation and the ability of HT1080 cells to migrate through Matrigel-coated invasion chambers are also inhibited in the presence of D3. These results demonstrate that GPI-anchored uPAR is endocytosed by piggybacking on LRP and that direct binding of occupied uPAR to LRP is essential for internalization of occupied uPAR, regeneration of unoccupied uPAR, plasmin generation, and invasion and migration through extracellular matrix.
INTRODUCTION
The urokinase plasminogen activator (uPA):plasmin system is involved in enhancing cell migration and invasion (Cohen et al., 1991; Reiter et al., 1993) during embryogenesis, wound healing, and metastasis (reviewed by Andreasen et al., 1994; Dano et al., 1985). Our current understanding of the uPA system is that uPA is activated upon binding to its cell surface receptor, uPAR (Cubellis et al., 1989;), a glycosyl-phosphatidylinositol (GPI)–anchored membrane protein (Nielsen et al., 1988; Ploug_et al._, 1991) with three homologous extracellular domains (D1, 2, and 3) (Behrendt et al., 1991). After activation, uPA catalyzes the conversion of plasminogen to plasmin (Ellis et al., 1989), which has a broad substrate specificity and is able to degrade many extracellular matrix proteins (Moser et al., 1993). uPA-dependent plasminogen activation is inhibited by specific uPA inhibitors with the best studied being plasminogen activator inhibitor type-1 (PAI-1) (Blasi_et al._, 1987; Saksela and Rifkin, 1988). Inactive uPA:PAI-1 complexes are rapidly internalized by the LDL receptor-related protein (LRP) (Herz et al., 1988, 1992) and degraded in lysosomes (Olson et al., 1992; Conese et al., 1995). Clearance of uPA:PAI-1 by LRP is accompanied by decreased uPAR at the cell surface, suggesting that uPAR is also internalized by LRP (Herz et al., 1992; Olson et al., 1992; Williams et al., 1994; Conese et al., 1995). It has also been demonstrated that when binding of uPA:PAI-1 to LRP is prevented with antibodies against LRP, cell surface plasminogen activity is reduced, presumably by delaying internalization of uPAR and preventing regeneration of unoccupied uPAR capable of binding uPA (Zhang et al., 1998).
Although this is the widely accepted model for regulation of the uPA system, the detailed mechanisms of how uPAR and LRP cooperate in clearance of uPA:PAI-1 as well as the trafficking itinerary of uPAR are not well understood. For example, it is not clear whether uPAR directly interacts with LRP or whether uPA-PAI-1 bridges uPAR and LRP. Moreover, the pathway by which uPAR is internalized has not yet been established.
LRP, like other members of the LDL receptor family, is internalized via clathrin-coated pits (Herz et al., 1988; Chen et al., 1990; Krieger and Herz, 1994), whereas uPAR, a GPI-anchored protein, has been shown to be distributed randomly along the plasma membrane (PM) under steady-state conditions (Conese et al., 1995; Maxfield and Mayor, 1997; Nykjaer et al., 1997).
To achieve a better understanding of the interplay between uPAR and LRP in regulation of cell surface plasminogen activator activity, we have investigated the interaction between uPAR and LRP and the trafficking of uPAR and LRP in HT1080 cells, a human fibrosarcoma cell line. We demonstrate here that uPA:PAI-1 initiates direct binding of domain 3 (D3) of uPAR to LRP on the PM and redistribution of uPAR to clathrin-coated pits and that this interaction is necessary for cointernalization of the two receptors into early endosomes (EE). We also show that binding of uPAR to LRP is essential for clearance of uPA:PAI, regeneration of unoccupied uPAR, and plasmin generation at the cell surface.
MATERIALS AND METHODS
Reagents
Human two-chain-uPA was purchased from American Diagnostica (Greenwich, CT) and human PAI-1 from Calbiochem (San Diego, CA). Holo-transferrin (Tf) and CHAPS were obtained from Sigma (St. Louis, MO), protein G-agarose from Amersham Pharmacia Biotech (Piscataway, NJ), and the thiol cleavable cross-linker 3,3′-dithiobis [sulfo succinimidyl-propionate] (DTSSP) from Pierce (Rockford, IL). Chromozym PL was purchased from Roche Molecular Biochemicals (Indianapolis, IN).
Antibodies
Mouse mAb against human uPAR was provided by Dr. Marc Shuman (University of California, San Francisco). A rabbit polyclonal antibody against recombinant soluble human uPAR1-274 (465) was generated as previously described (Orlando and Farquhar, 1993). Anti-LRP (456) was raised in rabbits against human LRP (Orlando and Farquhar, 1994), and anti-LRP (1073) was raised against a 14 amino acid peptide from the cytoplasmic tail of human LRP (Czekay et al., 1995). Mouse mAb against the ectodomain of human LRP (8G1) was obtained from Dr. Dudley Strickland (Holland Laboratory, American Red Cross, Rockville, MD). A hybridoma cell line expressing mAb 11H4 (recognizes the cytoplasmic tail of human LRP) was obtained from American Type Culture Collection. Polyclonal antisera against the cytoplasmic tail of human transferrin (TF) receptor and human uPA were obtained from Dr. Ian Trowbridge (Salk Institute) and Oncogene (Cambridge, MA), respectively. Affinity-purified chicken polyclonal antibodies against a peptide from the N terminus of human caveolin-1 (Stan et al., 1997) were provided by Dr. Radu V. Stan (University of California, San Diego). Affinity-purified mAb against AP-2 α-adaptin (Beck et al., 1992) was provided by Dr. Sandra Schmid (Scripps Research Institute). HRP-coupled donkey anti-rabbit or anti-mouse (H+L) IgG, depleted of cross-reactivity, was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cell Culture
Normal rat kidney (NRK) cells and human fibrosarcoma HT1080 cells were purchased from American Type Culture Collection and cultured according to their instructions. Cells were grown to confluency over 3–6 d.
Expression and Purification of Recombinant Soluble uPAR and Domains 2 and 3 of Human uPAR
uPAR cDNA (IMAGE Consortium, accession number T75241) encoding amino acids 1–274 (lacking the GPI-anchor site), D2 (encoding amino acids 92–174), and D3 cDNAs (encoding amino acids 192–274) were amplified by PCR (primers available on request) and subcloned into pET28b (_Eco_RI/_Hin_dIII; Novagen, Madison, WI).Escherichia coli strain BL21-DE3 expressing soluble uPAR1-274 and the D2 and D3 domains of uPAR were grown and induced as described (Orlando and Farquhar, 1994). Bacteria were pelleted by centrifugation (2500 × g, 10 min, 4°C), resuspended in TBS (20 mM Tris, pH 7.4, 150 mM NaCl) containing 100 μg/ml lysozyme, and lysed in TBS containing 3%_N_-lauroyl sarcosine. Lysates were cleared by centrifugation (15,000 × g, 20 min, 4°C), and supernatants were incubated with Ni2+-affinity resin (ProBond; Invitrogen, Carlsbad, CA). Bound uPAR1-274, D2, and D3 were eluted with TBS, 0.1% _N_-lauroyl sarcosine, 500 mM imidazole, and dialyzed against TBS, 0.1% _N_-lauroyl sarcosine.
Immunofluorescence
For immunofluorescence cells were grown to ∼75% confluence on glass coverslips, fixed for 20 min with 2% formaldehyde in 0.1 M phosphate buffer, pH 7.4, quenched in PBS containing 0.1% BSA and 10 mM glycine for 30 min as described (McCaffery and Farquhar, 1995), and permeabilized with 0.05% Triton X-100 in PBS for 3 min. Cells were then incubated with primary rabbit, mouse, or chicken antibodies for 1 h followed by incubation with appropriate Alexa 594- or Alexa 488-conjugated goat anti-rabbit, anti-mouse, or anti-chicken IgG (Molecular Probes, Eugene, OR). In some cases anti-uPAR antibodies were bound to the surface at 4°C in the presence or absence of D3 before fixation (Myohanen et al., 1993). Cells were examined by deconvolution microscopy with the Applied Precision DeltaVision imaging system coupled to an inverted Nikon SE 200 fluorescence microscope. For cross-sectional images of cells, stacks of at least 50 sections (150-nm step width) of raw image data were obtained to optimize reconstruction of the center plane image. Deconvolution was done on a SGI workstation (UNIX) using DeltaVision reconstruction software, and images were processed as TIFF files, pseudocolored, and superimposed using Photoshop 5.0 (Adobe Systems, Mountain View, CA).
Immunoblotting and Immunoprecipitation
For immunoblotting, cell lysates were prepared in 10 mM CHAPS in buffer A (20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM CaCl2) and processed for SDS-PAGE (Laemmli, 1970), immunoblotting, and detection by enhanced chemiluminescence (Pierce) as described (Czekay et al., 1997). Biotinylated proteins were detected using HRP-avidin (ABC-Biotinylation kit; Vector Laboratories, Burlingame, CA).
For immunoprecipitation, cell lysates were incubated with mAbs against either uPAR (3.9 μg) or LRP (2.6 μg), mixed with protein G-agarose beads (20 μl/1 ml of cell lysate) for 16 h at 4°C, and processed for SDS-PAGE, followed by autoradiography or immunoblotting and densitometry.
Radioiodination
Holo Tf, α2M, uPAR1-274, and D3 were radioiodinated with Na-125I (PerkinElmer Life Science Products, Boston, MA) using Iodo-Beads (Pierce). Specific activities were 4994, 1861, 3550, and 9821 cpm/ng, respectively. Cell surface proteins were radioiodinated using the lactoperoxidase method (Czekay et al., 1995).
Purification, Activation, and Degradation of α2M
α2-Macroglobulin (α2M) was purified from pooled citrated human plasma (San Diego Blood Bank, San Diego, CA) as described (Kurecki et al., 1979) except that elution buffer was modified to 0.01 M Na-acetate and 0.15 M NaCl, pH 5. Purified α2M was activated for receptor binding with 0.4 M methylamine in 0.1 M Tris-HCl, pH 8, for 2 h at room temperature and unbound methylamine removed by passage over a PD-10 column (Amersham Pharmacia Biotech).
Cells were incubated at 37°C with 125I-labeled α2M (2 nM) for 8 h in the presence or absence of unlabeled α2M (40 nM) in DMEM, 1 mg/ml BSA, 10 mM HEPES, pH 7.4. Medium was removed after 0, 2, 4, and 8 h, microfuged for 2 min, and supernatants were processed for trichloroacetic acid (TCA) precipitation and gamma counting (Czekay et al., 1997). Degradation was calculated as TCA-soluble cpm divided by specific activity of the radioiodinated ligand (Czekay et al., 1997) normalized to total cellular protein (BCA Protein Assay; Pierce).
Cell Surface Biotinylation
Cell monolayers were acid-washed in isoosmolar buffer B (50 mM glycine-HCl, pH 3.0, 100 mM NaCl) for 3 min at 4°C to release endogenous bound uPA from surface uPAR (Cubellis et al., 1989). After washing (twice in buffer B and twice in ice-cold PBS), surface proteins were biotinylated with 400 μM Biotin-XX (Molecular Probes) on ice for 35 min, and the reaction was quenched with 20 mM glycine in PBS, pH 7.4, for 15 min.
Cell Surface Binding and Uptake of Radioiodinated Tf and α2M
Cells were incubated with 125I-labeled holo-Tf (200 ng/ml) at 4°C (to radiolabel the cell surface pool of Tf receptor) and 18°C (to allow uptake of125I-labeled Tf into EE) (Czekay et al., 1997) after which the cells were acid-washed to release125I-labeled Tf bound to Tf receptors at the cell surface (Czekay et al., 1997).
125I-labeled α2M (2 nM) was bound to cells for 4 h at 4°C in the presence or absence of varying amounts of unlabeled α2M (0.06–40 nM), rinsed, and cell-associated radioactivity was quantified by gamma counting and normalized to total cellular protein (BCA Protein Assay; Pierce). Binding affinities (apparent _K_D) for125I-labeled α2M were determined as the concentration of unlabeled α2M which resulted in 50% inhibition of binding. Specificity was determined as the difference between total binding (without competition) and nonspecific binding (noncompetable). The amount of bound ligand was calculated as cpm divided by the specific activity of 125I-labeled α2M.
Cross-linking Experiments
125I-labeled, recombinant uPAR1-274 (25 nM) was bound to HT1080 cells in serum-free DMEM (containing 2% BSA, 20 mM HEPES, pH 7.4) for 2 h at 4°C in the presence or absence of unlabeled recombinant uPAR1-274 (2.5 μM) or the D2 (1.25 μM) or D3 (1.25 μM) domains of uPAR. Cells were washed and bound proteins were cross-linked using the thiol cleavable cross-linker DTSSP (1 mM) (Pierce) for 20 min at 4°C. The reaction was quenched with TBS for 10 min at 4°C, cell lysates were prepared in 10 mM CHAPS, and immunoprecipitation was carried out with anti-LRP mAb (11H4).
Binding of 125I-uPAR1-274 or125I-D3 to Purified LRP
Placental tissue (Scripps Clinic, La Jolla, CA) was finely minced, homogenized in buffer A (containing 10 μg/ml aprotinin, 0.5 mM PMSF) by using a TissueMizer (Tekmar, Cincinnati, OH) for 3 min, and centrifuged (15,300 × g, 30 min) at 4°C. Placental proteins were extracted with 10 mM CHAPS in buffer A for 2 h, and LRP was affinity purified on mAb 11H4 (5 mg) prebound to protein G-Sepharose beads (Oncogene). The bound material contained only LRP when analyzed by SDS-PAGE and silver staining.
Affinity-purified immobilized LRP was incubated with either125I-uPAR1-274 (100 nM) or125I-D3 (50 nM) in the presence or absence of unlabeled uPAR1-274 (4 μM), D2 (1.5 μM), or D3 (1.5 μM), and radioiodinated proteins were visualized by autoradiography followed by densitometry or PhosphorImager analysis (Czekay et al., 1995).
Internalization of uPAR after Binding uPA:PAI-1 and Cell Fractionation
uPA:PAI complexes were formed by incubation of PAI-1 (5 μM) with two-chain uPA (100 nM) for 1 h at room temperature as described (Cubellis et al., 1989). Cells were acid-washed, biotinylated (to specifically label the surface pool of uPAR), and incubated with uPA:PAI-1 (50 nM) for 1 h at 4°C or sequentially at 4 and 18°C, 1 h each, and cell fractionation was carried out as described (Czekay et al., 1997). In brief, postnuclear supernatants (1 ml) were top loaded onto isoosmolar 20%-Percoll (Amersham Pharmacia Biotech, Alameda, CA), and gradients were established by centrifugation (20,000 × g, 52 min) at 4°C. Fractions (1 ml) were collected from the top of the gradient, and the distribution of biotinylated uPAR was determined by immunoprecipitation with anti-uPAR mAb followed by blotting with HRP-coupled avidin and detection by chemiluminescence. In some cases, binding and internalization were done in the presence of either 200 μg/ml protein A-purified anti-LRP IgG (456), which blocks ligand binding to LRP (Orlando, unpublished observations), or 2.5 μM D3 to block uPAR binding to LRP. All solutions contained Ca2+, which is required for ligand binding to LRP (Moestrup et al., 1990). For K+ depletion, cells were incubated in 5 ml of hypotonic medium (1:1, DMEM/water) before uPA:PAI-1 binding followed by incubation for 30 min in isotonic K+-free buffer (Larkin et al., 1983) at room temperature.
Binding of 125I-uPA:PAI-1 to uPAR1-274
Recombinant purified human uPAR1-274 (2 nM) was incubated in PBS for 1 h at 4°C with125I-uPA:PAI-1 (10 nM) complexes in the presence or absence of D3 (100 nM), and samples were mixed with anti-uPAR (465) and protein G-Sepharose beads (Oncogene) for 16 h at 4°C. Bound proteins were processed for SDS-PAGE, autoradiography, and densitometry.
Immunoisolation of Early Endosomes
Postnuclear supernatants were fractionated on Percoll gradients as described above and gradient fractions 8–10 (containing EE) were pooled. mAb 11H4, specific for the C terminus of human LRP, was prebound to protein G-agarose beads in buffer B and subsequently incubated with pooled fractions 8–10 for 2 h at room temperature. After washing, immunoisolates were incubated for 1 h on ice in buffer B containing 10 mM CHAPS, beads were separated from supernatant by centrifugation, and Percoll in the supernatant was pelleted (Czekay et al., 1997). Proteins released into the supernatant and those remaining bound to the agarose beads were processed for immunoblotting.
uPA Activity at the Cell Surface
Cells were acid-washed as described above, incubated with uPA:PAI-1 (10 nM) at 4°C for 1 h, and, after removing unbound ligand, at 37°C for 1 h in the presence or absence of D3 (5 μg/ml). Subsequently, active uPA (2 nM) was bound to HT1080 cells at 4°C for 1 h in the presence of anti-LRP (456), 200 μg/ml. After washing, cells were incubated in 50 mM Tris-HCl, pH 7.4, 0.1 M NaCl with 0.2 μM human plasminogen (Calbiochem) and 0.5 mM Chromozym PL (Tosyl-Gly-ProLys-4-nitranilide-acetate; Roche Molecular Biochemicals), a chromogenic substrate for plasmin, according to the manufacturer's instructions. Hydrolysis of the substrate was measured at 405 nm, and absorbance was used to calculate the generation of plasmin (U/ml).
Transwell Invasion Assays
Cells were detached from tissue culture plates with PBS, pH 7.4, containing 10 mM EDTA, and resuspended in DMEM (10% fetal calf serum). Cells (5 × 105/500 μl) were added to the upper compartment of Matrigel-coated invasion chambers, 8-μm pore size (Becton Dickinson, Bedford, MA) in the presence or absence of soluble uPAR1-274 (2.5 μM), D3 (1.25 μM), or D3 and active uPA (8 nM). Culture medium alone (750 μl) was added to the lower part of the invasion chamber. After incubation at 37°C for 36 h, cells were removed from the upper side of the filter membrane by using cotton swabs. Filters were fixed, stained, and mounted according to the manufacturer's instructions. Cells that invaded through the Matrigel and migrated to the under side of the filter were counted. The values obtained were calculated by averaging the total number of cells from three filters per condition.
RESULTS
LRP Expressed on the Surface of HT1080 Cells Is Functionally Active
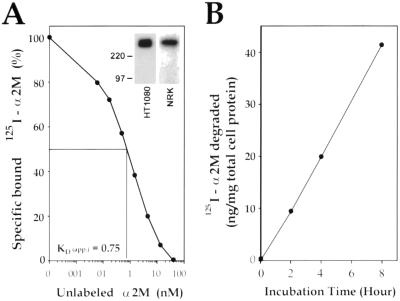
To study the interactions between uPAR and LRP we used HT1080 cells, a human fibrosarcoma cell line, known to express functional uPAR on the PM (Xue et al., 1997). Because LRP expression has not been previously studied in this cell line, we first verified that it is expressed and functionally active at the cell surface. LRP was clearly detected after cell surface radioiodination of HT1080 cells followed by immunoprecipitation with anti-LRP mAb (11H4), and it comigrated with LRP from NRK cells (Herz et al., 1990) (Figure 1A, inset). We then tested whether HT1080 cells are capable of binding and internalizing α2M, a well-established specific ligand for LRP.125I-labeled α2M bound to HT1080 cells with an apparent _K_D of 0.75 nM (Figure 1A), and 42 ng of 125I-labeled α2M/mg of total cell protein was degraded in a linear manner over 8 h (Figure 1B). Thus, the ability of HT1080 cells to bind and degrade ligand is comparable to noncancer cells (Moestrup and Gliemann, 1989).
Figure 1.
Expression of LRP by HT1080 cells. (A) LRP on HT1080 cells shows a high affinity for α2M (apparent_K_D = 0.75 nM). HT1080 cells were incubated with 125I- α2M (2 nM), an LRP-specific ligand, at 4°C in the presence or absence of varying amounts of unlabeled α2M (0.06–40.0 nM). Cell-associated radioactivity was quantitated by gamma counting, normalized to total cell protein, and the affinity was calculated as described in MATERIALS AND METHODS. (Inset) LRP immunoprecipitated from HT1080 cells comigrates with LRP from NRK cells. HT1080 cells and NRK cells (used as control) and cell lysates were radioiodinated and processed for immunoprecipitation with anti-LRP mAb (11H4). (B) HT1080 cells take up and degrade 125I-α2M linearly up to 8 h. Cells were incubated at 37°C with125I-α2M (2 nM) for 8 h. TCA-soluble radioactivity released into the incubation medium was used to quantify the amount of ligand degraded as described in MATERIALS AND METHODS.
LRP Is Located in Clathrin-coated Pits on the Plasma Membrane of HT1080 Cells
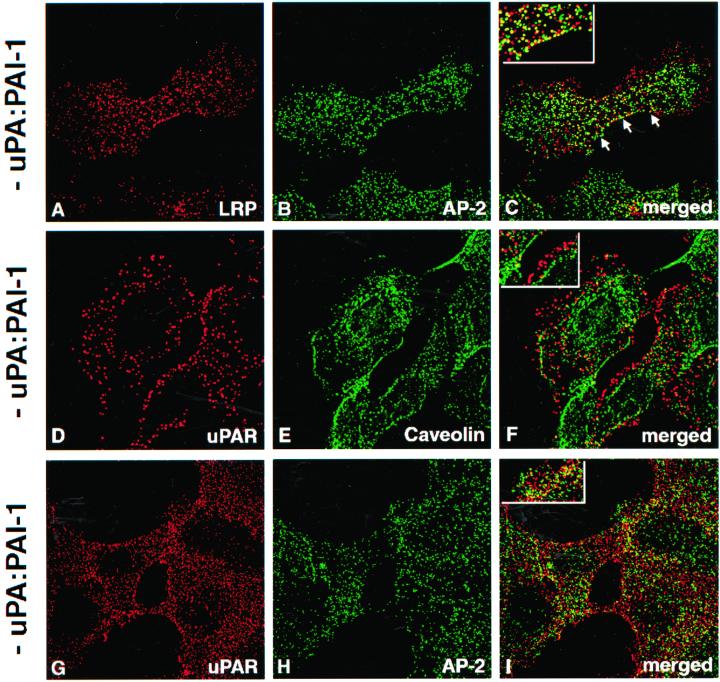
Next, we investigated the distribution of LRP on the PM of HT1080 cells under steady-state conditions by immunofluorescence. Because it has been assumed that LRP is localized in clathrin-coated pits (Herz_et al._, 1988; Krieger and Herz, 1994), we carried out double labeling for LRP and AP-2, a well-established marker for clathrin-coated pits and vesicles associated with the PM (Brodsky, 1997). Staining for LRP (Figure 2A) was concentrated along the PM where it showed a striking overlap with AP-2 (Figure 2B), confirming its location in clathrin-coated pits.
Figure 2.
Distribution of LRP and uPAR on the surface of HT-1080 cells at steady state. (A–C) Double labeling showing significant overlap of LRP (A) and AP-2 (B) by immunofluorescence. (D–F) Little or no overlap is seen between uPAR (D) and caveolin (E). (G–I) Similarly, there is little or no overlap between uPAR (G) and AP-2 (H). Cells were incubated with polyclonal anti-uPAR (3936) or anti-LRP mAb (456) at 4°C, fixed with 2% formaldehyde, permeabilized with Triton X-100 and subsequently incubated with anti-AP-2 (mAb) or anti-caveolin (chicken), followed by detection with appropriate secondary antibodies.
Distribution of uPAR on the Plasma Membrane of HT1080 Cells
To determine the distribution of uPAR we carried out double labeling for uPAR and either AP-2 or caveolin-1, a marker for caveolae (Rothberg et al., 1992). Staining for uPAR (Figure 2, D and G), caveolin-1 (Figure 2E), and AP-2 (Figure 2H) was seen on the PM, but there was little or no overlap between uPAR and either caveolin or AP-2. We conclude that under steady-state conditions LRP is associated with clathrin-coated pits, whereas uPAR is not associated with either clathrin-coated pits or caveolin-enriched microdomains.
Redistribution of uPAR on the PM in the Presence of uPA:PAI-1 Complexes
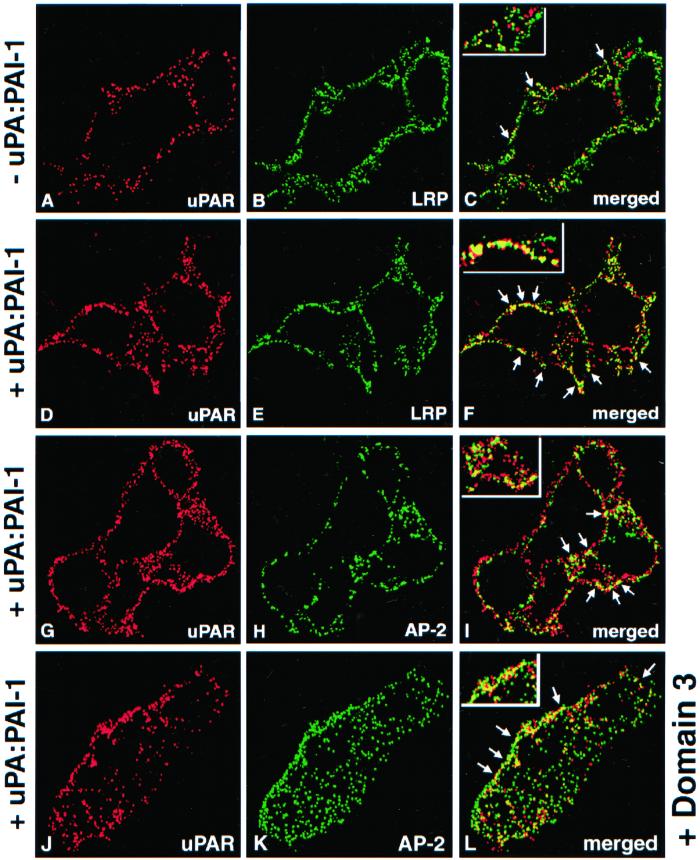
Binding of exogenous uPA:PAI-1 complexes to uPAR has been shown to initiate internalization of both uPA:PAI-1 and uPAR (Olson_et al._, 1992). To investigate the fate of uPAR after uPA:PAI-1 binding we carried out immunofluorescence on HT1080 cells that had been acid-washed to release endogenous bound ligand followed by incubation in the presence or absence of uPA:PAI-1 complexes at 4°C, and double labeling for uPAR and LRP. We found that in the absence of uPA:PAI-1, there was little overlap in staining for uPAR and LRP (Figure 3, A–C). However, in the presence of uPA:PAI-1, the overlap in staining for uPAR with both LRP (Figure 3, D–F) and AP-2 (Figure 3, G–I) was strikingly increased. These data indicate that binding of uPA:PAI-1 to uPAR induces redistribution of occupied uPAR on the PM to clathrin-coated pits, suggesting that uPA:PAI-1 may bridge uPAR and LRP.
Figure 3.
Distribution of uPAR in the presence of uPA:PAI-1 complexes and D3. (A–C) In HT1080 cells incubated in the absence of uPA:PAI:1 complexes there is little overlap in staining for LRP (A) and uPAR (B). (D–E) In the presence of uPA:PAI-1 overlap in the staining for uPAR (D) and LRP (E) is dramatically increased. There is also increased overlap in staining for uPAR (G) and AP-2 (H), indicating redistribution of uPAR to clathrin-coated pits. (J and K) Redistribution of uPAR to clathrin-coated pits in the presence of uPA:PAI-1 is not affected by the presence of D3. HT1080 cells were acid-washed, incubated at 4°C in the absence or presence of uPA:PAI-1 complexes and D3 (J–L), and processed for immunofluorescence as described in Figure 2 legend.
uPAR Coprecipitates with LRP after uPA:PAI-1 Binding to the Cell Surface
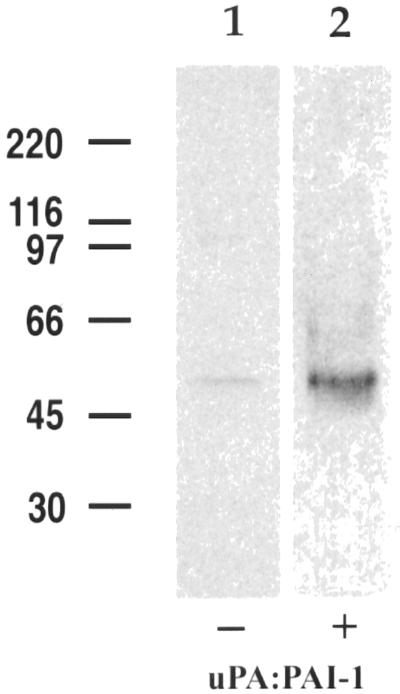
To find out whether uPAR and LRP form a molecular complex after binding uPA and PAI-1, we carried out immunoprecipitation with anti-LRP mAb (11H4) on lysates of HT1080 cells incubated at 4°C in the presence or absence of uPA:PAI-1 followed by immunoblotting for uPAR. We found considerable uPAR in precipitates obtained with anti-LRP from cells incubated in the presence of uPA:PAI-1 (Figure 4, lane 2), whereas only traces of uPAR could be detected in precipitates from controls incubated in the absence of uPA:PAI-1 (Figure 4, lane 1). These data indicate that LRP and uPAR form immunoprecipitable complexes at the surface of HT1080 cells in the presence of uPA:PAI-1.
Figure 4.
LRP and uPAR form detergent-stable complexes at the surface of HT1080 cells in the presence of uPA:PAI-1. In the absence of uPA:PAI-1 (lane 1), only traces of uPAR are detected in immunoprecipitates obtained with anti-LRP, whereas in the presence of uPA:PAI-1 immunoprecipitates contain considerable uPAR (lane 2). HT1080 cells were surface radioiodinated, incubated in the presence or absence of uPA:PAI-1, and processed for immunoprecipitation with anti-LRP mAb (11H4). Precipitated proteins were subsequently immunoblotted with anti-uPAR (465).
Coimmunoprecipitation of uPAR1-274 with Endogenous LRP
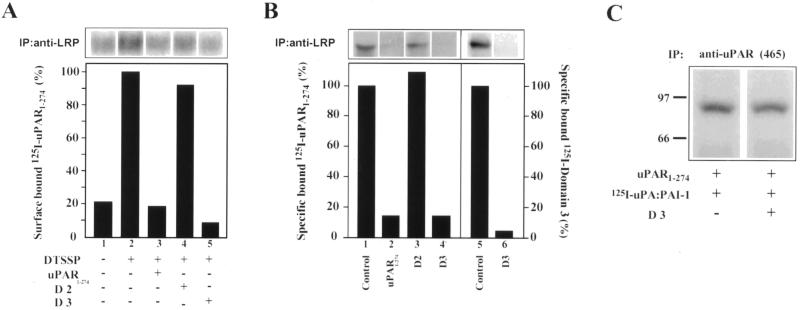
Next, we tested whether uPAR binds to endogenous LRP on the PM by carrying out cross-linking experiments in the presence of recombinant, soluble (lacking a GPI-anchor) uPAR. When125I-uPAR1-274 was bound to the cell surface of acid-washed cells at 4°C followed by cross-linking and immunoprecipitation with anti-LRP mAb (11H4), considerable 125I-uPAR1-274 coprecipitated with LRP (Figure 5A, lane 2). The amount of 125I-uPAR that coprecipitated was greatly reduced (>85%) in the presence of unlabeled uPAR1-274 (Figure 5A, lane 3). When similar experiments were carried out in the presence of recombinant D3 or D2, D3 greatly reduced (>90%) uPAR1-274 binding (Figure 5A, lane 5), whereas D2 had no significant effect (Figure 5A, lane 4). These findings suggest that uPAR binds to LRP through D3.
Figure 5.
Direct binding of soluble uPAR to LRP via D3 of uPAR. (A) Considerable 125I-uPAR1-274 can be detected in anti-LRP immunoprecipitates (lane 2) after cross-linking. The amount of cross-linked 125I-uPAR1-274 is reduced (80–90%) in the presence of unlabeled uPAR1-274 (lane 3) and D3 (lane 5). D2 (lane 4) has no significant effect on uPAR1-274 binding to LRP. Acid-washed cells were incubated with 25 nM recombinant 125I-uPAR1-274 at 4°C in the presence (+) or absence (−) of 2.5 μM unlabeled uPAR1-274, or 1.25 μM recombinant D2, or D3 followed by cross-linking with DTSSP (lanes 2–5) and immunoprecipitation with anti-LRP mAb (11H4). Immunoprecipitated proteins were processed for autoradiography and densitometry. (B) Binding of125I-uPAR1-274 to affinity-purified LRP (lane 1) is reduced by >85% in the presence of unlabeled uPAR1-274 (lane 2) or D3 (lane 4). D2 (lane 3) has no effect on 125I-uPAR1-274 binding to LRP. In addition, 125I-D3 binds to affinity-purified LRP (lane 5) and the binding is reduced by >95% in the presence of unlabeled D3 (lane 6). Purified LRP was incubated with125I-uPAR1-274 (100 nM) or 125I-D3 (50 nM) at 4°C in the absence or presence of unlabeled uPAR1-274 (4 μM), D2 (1.5 μM), or D3 (1.5 μM). Immunoprecipitated proteins were processed for autoradiography and densitometry. (C) Binding of 125I-uPA:PAI-1 to recombinant uPAR1-274 (lane 1) is not affected by D3 (lane 2). uPAR1-274 (2 nM) was incubated with125I-uPA:PAI-1 (10 nM) in PBS in the presence or absence of D3 (100 nM) and processed for immunoprecipitation with anti-uPAR (465). Samples were processed for autoradiography and densitometry.
uPAR1-274 Binds Directly to Affinity-purified LRP through Domain 3
Next, we incubated affinity-purified, immobilized LRP with125I-uPAR1-274 in the presence or absence of unlabeled uPAR1-274, D2, or D3. We found that125I-uPAR1-274 bound directly to LRP (Figure 5B, lane 1), and the amount bound was greatly reduced (>85%) in the presence of unlabeled uPAR1-274 (Figure 5B, lane 2) or D3 (Figure 5B, lane 4). D2 had no effect on the binding (Figure 5B, lane 3). Furthermore, we found that 125I-D3 also bound to immobilized LRP (Figure 5B, lane 5), and binding was reduced (>90%) in the presence of unlabeled D3. Collectively, these results demonstrate that 1) uPAR binds directly to LRP on the surface of HT1080 cells, and 2) the binding site is located in D3.
uPAR with Bound uPA:PAI-1 and LRP Are Internalized and Delivered to Early Endosomes
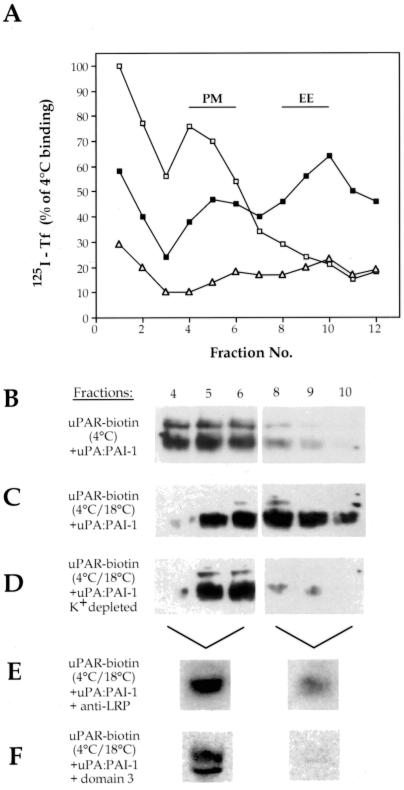
To investigate the trafficking of uPAR:LRP complexes in the presence of uPA:PAI-1, we carried out subcellular fractionation by using 125I-Tf as a marker to identify PM and EE fractions. When HT1080 cells were incubated with125I-Tf for 1 h at 4°C and homogenates were fractionated on isoosmotic Percoll gradients, the majority of the125I-Tf sedimented in gradient fractions 4–6 (Figure 6A, open squares), indicating the location of PM. After incubation of cells at 18°C for 1 h to allow uptake into EE, the majority of the 125I-Tf sedimented in fractions 8–10 (Figure 6A, closed squares), defining fractions containing EE.
Figure 6.
Internalization of uPAR requires binding to LRP. (A) In cells incubated with 125I-Tf at 4°C (□), Tf peaks in fractions 4–6, indicating the location of PM. After incubation at 18°C (▪), 125I-Tf peaks in fractions 8–10 where it defines the location of EE. In cells subjected to K+ depletion (▵), uptake of 125I-Tf is blocked, because no 125I-Tf cosedimented with EE. (B) In cells incubated at 4°C with uPA:PAI-1, biotinylated uPAR sediments in PM fractions 4–6. (C) After 60-min incubation at 18°C, the distribution of a substantial fraction of the biotinylated uPAR shifts to EE fractions (8–10). (D) After K+ depletion and incubation at 18°C the majority of the biotinylated uPAR is found in PM fractions (4–6), and little or no uPAR is detected in EE fractions (8–10), indicating that internalization of uPAR is blocked. After incubation for 60 min at 18°C in the presence of 200 μg/ml anti-LRP (456) (E), the majority of the biotinylated uPAR remains associated with PM fractions. (F) In the presence of 1.25 μM D3, virtually all detectable, biotinylated uPAR (∼95%) is found in PM fractions after incubation at 18°C. HT1080 cells were acid-washed, surface biotinylated, and incubated with either 125I-Tf (200 ng/ml) or uPA:PAI-1 complexes (50 nM) at 4°C (60 min) or for 4°C (60 min) followed by 18°C (60 min). In some cases D3 (1.25 μM) or anti-LRP antibodies (456; 200 μM/ml) were added or intracellular K+ was depleted as described in MATERIALS AND METHODS. Postnuclear supernatants were fractionated on Percoll gradients, and fractions were processed for gamma counting of 125I-Tf or for immunoprecipitation of uPAR and by detection of the biotinylated, cell surface pool of uPAR by blotting with HRP-coupled avidin. In E and F, PM fractions (4–6) and EE fractions (8–10), were pooled before analysis.
To follow the fate of uPAR after uPA:PAI-1 binding, HT1080 cells were surfaced biotinylated, incubated with uPA:PAI-1, and fractionated on Percoll gradients after incubation at 4°C or binding at 4°C followed by incubation at 18°C for 1 h. After uPA-PAI-1 binding at 4°C, the majority of the biotinylated uPAR cosedimented with125I-Tf in PM fractions (4–6) (Figure 6B). However, when cells were subsequently incubated at 18°C after binding the peak of the biotinylated uPAR shifted to EE fractions (8–10) (Figure 6C). When similar experiments were carried out on cells incubated in the presence of anti-LRP (456) (Figure 6E) at 18°C, uPAR remained largely associated with PM-containing fractions, confirming previous reports that anti-LRP blocks internalization of uPAR (Zhang_et al._, 1998). From these results we conclude that uPAR is internalized into an EE compartment after uPA:PAI-1 binding, and internalization occurs via LRP.
uPAR Is Internalized via Clathrin-coated Vesicles
LRP is known to be internalized via clathrin-coated vesicles (Chen_et al._, 1990), but the route of internalization of uPAR has not yet been established. To obtain information on this point we used a procedure (i.e., hypotonic shock and depletion of intracellular K+) that selectively blocks clathrin-mediated endocytosis (Larkin et al., 1983) but has no effect on internalization via caveolae (Roettger et al., 1995). When cells were K+ depleted and incubated at 18°C as described above, uptake of both 125I-Tf (Figure6A, open triangles) and uPAR (Figure 6D) was completely abolished. These results together with our immunocytochemical data demonstrate that internalization of occupied uPAR is clathrin mediated.
D3 Blocks Internalization of uPAR but Does Not Affect Binding of uPAR:PAI-1
Next, we tested the effects of D3 on internalization of uPAR. We found that internalization of uPAR was blocked in the presence of D3, because uPAR remained largely associated with PM-containing fractions (Figure 6F).
Because previous studies have suggested that regions within D3 are important for high-affinity binding of uPA to uPAR (Ploug, 1998), it was necessary to determine whether D3 blocks binding of uPA:PAI-1 to uPAR. Thus, we carried out experiments in which we incubated125I-uPA:PAI-1 complexes (10 nM) with recombinant purified human uPAR1-274 (2 nM) in the presence or absence of D3 (100 nM). We found that D3 had no effect on the binding of uPA:PAI-1 to uPAR1-274 (Figure 5C).
We also found that D3 does not block redistribution of uPAR induced by uPA:PAI-1 complexes (Figure 3, J–L). We conclude that D3 blocks internalization of uPAR but has no affect on the binding of uPA:PAI-1 to uPAR or on the redistribution of uPAR with bound uPA:PAI-1 to clathrin-coated pits.
Presence of uPAR:LRP Complexes in Early Endosomes
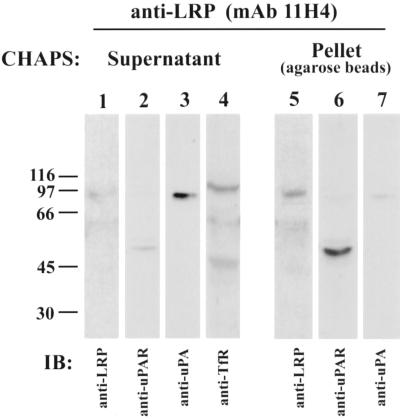
Next, we investigated whether uPAR and LRP remain associated during internalization into EE. We prepared EE fractions (8–10) from surface biotinylated cells incubated at 18°C in the presence of uPA:PAI-1 and used them as starting material for immunoisolation of EE on mAb 11H4 (specific for the cytoplasmic tail of LRP), which had been prebound to protein G-agarose. Vesicles that had been immunoadsorbed to the beads were then detergent solubilized (to release non-LRP–associated proteins), and the beads with bound LRP were pelleted and analyzed by immunoblotting for LRP-associated proteins. As shown in Figure7, most (>90%) of the uPA:PAI-1 (lane 3) and Tf receptor (lane 4) were released into the supernatant after detergent treatment, whereas most (>90%) of the LRP (lane 5) and uPAR (lane 6) remained bound to the beads. These results show that uPAR and LRP are internalized together and remain associated in EE, whereas most of the uPA:PAI-1 is no longer bound to uPAR in EE.
Figure 7.
Presence of LRP and uPAR complexes in EE. The majority of the uPAR (lane 6) coprecipitates with LRP (lane 5) and is found in the pellet, which contains only trace amounts of uPA: PAI-1 (lane 7). The majority of the uPA:PAI-1 complexes (lane 3) and Tf receptor (lane 4) do not coprecipitate with LRP and are found in the supernatant, which contains only traces of LRP (lane 1) and uPAR (lane 2). Thus, uPAR and LRP form a complex that can be coprecipitated from EE. uPA:PAI-1 was bound to biotinylated HT1080 cells at 4°C, and the cells were fractionated as in Figure 6. EE fractions were pooled and LRP-containing vesicles were immunoisolated on mAb 11H4 (against the cytoplasmic tail of LRP) bound to protein G-agarose beads. Beads were treated with 10 mM CHAPS, and the proteins released into the supernatant and those remaining bound to the beads were analyzed by immunoblotting with anti-LRP (1073), anti-uPAR (465), anti-uPA, and anti-Tf receptor.
D3 Blocks Cell Surface uPA Activity
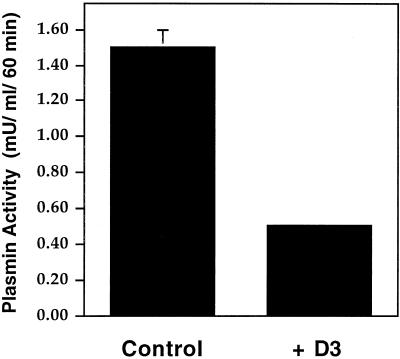
Previously, it was shown that blocking uPA:PAI-1 binding to LRP with RAP impairs regeneration of unoccupied uPAR and reduces uPA activity at the cell surface, presumably by blocking endocytosis of uPAR (Zhang et al., 1998). Thus, we reasoned that blocking uPAR binding to LRP should also lead to decreased cell surface uPA activity and subsequent generation of plasmin in HT1080 cells. To determine whether this is the case we quantified uPA-dependent plasmin activation in the presence and absence of D3 using a chromagenic assay. After incubation with uPA:PAI-1 in the presence of recombinant D3 at 4°C (1 h) or 37°C (1 h), there was a ∼60% reduction in surface-bound uPA activity (0.5 ± 0.02 mU/ml plasmin activity) compared with controls (1.5 ± 0.1 mU/ml) over 60 min (Figure8). These results suggest that preventing the binding of uPAR to LRP with recombinant D3 results in decreased uPA activity at the cell surface, presumably by preventing clearance of occupied uPAR and regeneration of unoccupied uPAR capable of binding activated uPA at the cell surface.
Figure 8.
Inhibition of uPAR/LRP interaction reduces plasminogen activation. Plasmin activity at the cell surface of HT1080 cells is reduced from 1.5 mU (control) to 0.5 mU/ml/60 min when cells are incubated in the presence of D3. Acid-washed cells were incubated sequentially for 1 h at 4°C and 1.5 h at 37°C with uPA:PAI-1 (10 nM) in the presence or absence of D3 (5 μg/ml) and subsequently incubated at 4°C with proteolytically active uPA (2 nM) for 1 h in the presence of anti-LRP antibodies (456; 200 μg/ml). Plasminogen (0.2 μM) was bound to the cell surface at 4°C, and uPA-dependent plasmin generation was calculated as absorbance change at 405 nm over 60 min using Chromozym PL as a plasmin substrate.
D3 Inhibits Migration of HT1080 Cells through Matrigel-coated Transwell Chambers
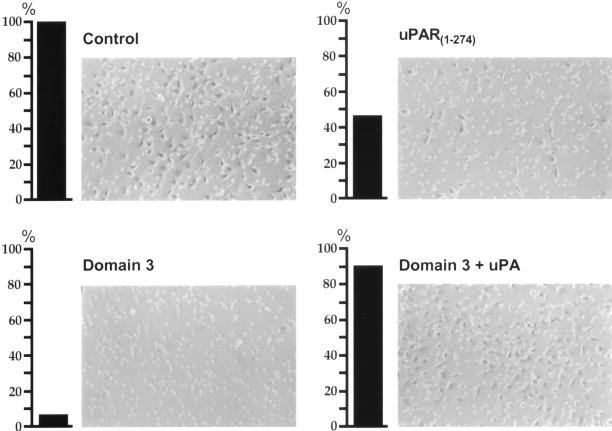
Because blocking LRP:uPAR interaction reduces cell surface uPA activity it might also be expected to slow or prevent the ability of cells to migrate through the extracellular matrix. To determine whether this is the case, we carried out a well-established cell invasion assay (Kramer et al., 1986; Albini et al., 1987; Chen_et al._, 1997) in which we analyzed the ability of HT1080 cells to migrate through Matrigel-coated Transwell invasion chambers in the presence or absence of soluble uPAR1-274 or D3. In the absence of uPAR1-274 or D3 (Figure9) 115 (± 12) cells (100%) migrated through the Matrigel layer. However, in the presence of recombinant uPAR1-274 or D3 the number of cells that migrated through the filter was reduced to 54 (± 3) cells (47%) and 9 (± 3) cells (8%), respectively. Addition of proteolytically active uPA to cells incubated in the presence of D3 restored the number of HT1080 cells able to migrate through the Matrigel-coated filters to control levels (105 [± 7] cells; 91%), indicating that the effect can be attributed in large part to uPA. No significant affect on the number migrating through the Matrigel layer was detectable when D2 was added to the assay. These data indicate that inhibiting uPAR's interaction with LRP and preventing endocytosis of uPAR significantly reduces the ability of HT1080 cells to invade and to migrate through extracellular matrix.
Figure 9.
Blocking uPAR/LRP interaction inhibits migration of HT1080 cells through Matrigel-coated filters. Cells (5 × 105) were seeded onto the top of Matrigel-coated filter membranes and incubated at 37°C in the presence or absence of uPAR1-274 (2.5 μM), D3 (1.25 μM), or D3 in combination with active uPA (8 nM). Cells that migrated through the filters and appeared at the under side of the filters were counted after 36 h of incubation. In the presence of uPAR1-274 and D3 the number of invading cells is decreased to 46.5 and 7.5% of control, respectively. Coincubation of cells with D3 and active uPA restores the ability of the cells to migrate to near control levels.
DISCUSSION
It has been established that LRP mediates the clearance of uPA:PAI-1 from the extracellular environment and that this process is accompanied by internalization of uPAR (Conese et al., 1995), but the clearance mechanism and the intracellular trafficking itinerary of uPAR have remained unknown. In this article we investigated the nature of the interaction between uPAR and LRP during clearance of uPA:PAI-1 and their endocytic trafficking. We have shown that in HT1080 cells, uPAR directly interacts with LRP in the presence of uPA:PAI-1 and that LRP and occupied uPAR form stable complexes that are cointernalized into EE via clathrin-coated vesicles. We have further demonstrated that blocking the interaction between the two receptors results in decreased clearance of uPA:PAI-1 and decreased plasmin generation at the cell surface and significantly reduces the ability of the cells to migrate through extracellular matrix in a standard cell invasion assay.
Our immunofluorescence results indicate that at steady-state LRP and uPAR are localized in different microdomains of the PM. LRP, like other members of the LDL receptor gene family, is concentrated in clathrin-coated pits, whereas uPAR is not associated with either clathrin-coated vesicles or caveolae because it did not colocalize with either caveolin or with AP-2, markers for caveolae and clathrin-coated pits, respectively (Brodsky, 1997). The latter finding is in keeping with previous reports indicating that at steady-state uPAR as well as other GPI-anchored proteins are rather broadly distributed at the cell surface (Conese et al., 1995; Maxfield and Mayor, 1997;Nykjaer et al., 1997). However, we found that when cells are incubated in the presence of uPA:PAI, which binds to uPAR, uPAR forms a complex with LRP and moves into clathrin-coated pits. That binding is direct and occurs at the D3 domain of uPAR was suggested by cross-linking experiments and the fact that addition of recombinant D3 completely blocked both the binding and internalization of occupied uPAR. This was confirmed by demonstrating that recombinant uPAR binds to affinity-purified LRP. The demonstration of direct interaction between uPAR and LRP is a novel finding, because it has been assumed or implied that uPA-PAI-1 bridges the two receptors and bridging is sufficient for endocytosis of the complexes (Andreasen et al., 1994). There has been no previous work showing a requirement for direct interaction between uPAR and LRP.
It has been shown previously (Behrendt et al., 1991) that uPAR is composed of three homologous domains (designated D1, D2, and D3 from the N terminus) of which D1 contains the ligand-binding domain. Currently, little is known about the physiological functions of D2 and D3. However, it has been hypothesized that they might increase the specificity of uPAR for its ligand and facilitate uPAR interaction with adhesion receptors at the sites of focal contacts (Dear and Medcalf, 1998). Our findings demonstrate D3 contains a binding site for LRP and provide novel insights into the function of D3.
We also established that when occupied by uPA:PAI-1 the GPI-anchored protein uPAR is internalized into EE via clathrin-coated vesicles, because internalization does not occur when clathrin-mediated endocytosis is blocked by K+ depletion. Furthermore, uPAR remains associated with LRP during internalization to EE even after dissociation of uPA:PAI-1 as demonstrated by our immunoprecipitation analysis on immunoisolated EE. LRP is known to internalize ligands through a clathrin-dependent pathway mediated by the NPXY motifs in its cytoplasmic tail, whereas GPI-anchored proteins such as uPAR lack this motif and are usually assumed to be internalized via caveolae (Lisanti et al., 1993; Anderson, 1998). To our knowledge, this is the first demonstration of a GPI-anchored protein piggybacking with another receptor to be internalized via clathrin-coated vesicles. Indirect evidence suggests that the same might apply to uPAR and certain integrins (Memmo and McKeown-Longo, 1998; Simon et al., 2000) and to prions, which have been postulated to interact with unknown transmembrane receptors (Shyng et al., 1994).
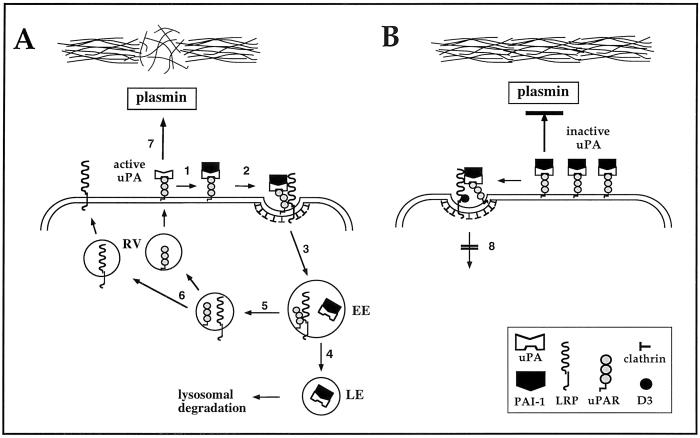
Taken together, our results provide evidence for the model of uPA:PAI-1 clearance and uPAR internalization shown in Figure10. According to this model, formation of uPA:PAI-1 complexes on uPAR bridges occupied uPAR and LRP at the cell surface, promotes relocation of occupied uPAR into clathrin-coated pits, and initiates binding of LRP to the D3 domain of uPAR. This results in generation of a quaternary complex, uPA:PAI-1/uPAR:LRP, which is then internalized into EE via clathrin-coated vesicles. uPA:PAI-1 is released from the receptors in EE and traffics through late endosomes to lysosomes for degradation (Cubellis et al., 1990). Unoccupied uPAR (Nykjaer et al., 1997) and LRP recycle to the cell surface. Unoccupied uPAR is available for binding and activation of pro-uPA, which catalyzes the generation of plasmin, promoting degradation of extracellular matrix and cell invasion. When binding of occupied uPAR to LRP is prevented (i.e., in the presence of recombinant D3), uPA:PAI-1 is still capable of bridging the two receptors and uPAR redistributes into clathrin-coated pits. However, internalization of occupied uPAR is prevented, leading to accumulation of occupied uPAR in clathrin-coated pits on the PM. Accumulation of redistributed uPAR occupied by inactive uPA at the cell surface decreases plasmin generation and degradation and invasion of extracellular matrix.
Figure 10.
Model depicting proposed events in the interaction and trafficking of uPAR and LRP. (A) Active uPA binds to uPAR on the PM. 1) PAI-1 binds to and inhibits uPA. 2) uPA:PAI-1 bridges occupied uPAR and LRP, which leads to relocation of occupied uPAR into clathrin-coated pits and promotes a stable interaction between D3 of uPAR and LRP. 3) These quaternary complexes are internalized via clathrin-coated pits and delivered to EE. 4) uPA:PAI-1 dissociates from uPAR and LRP in EE and traffics through late endosomes (LE) to lysosomes for degradation. 5) Unoccupied uPAR and LRP also dissociate and 6) return to the cell surface via recycling vesicles (RV). This makes available unoccupied uPAR capable of binding pro-uPA. 7) Presence of active uPA at the cell surface catalyzes the generation of plasmin, which can degrade extracellular matrix. (B) When binding of occupied uPAR to LRP is prevented, uPA:PAI-1 bridges uPAR and LRP and occupied uPAR moves into clathrin-coated pits. However, clearance of occupied uPAR is blocked (8) and occupied inactive uPAR accumulates on the cell surface. As a consequence, generation of plasmin and degradation and invasion of extracellular matrix are decreased.
Because activation of secreted pro-uPA requires its binding to cell surface uPAR (Ellis et al., 1989; Stephens et al., 1989) and unoccupied uPAR constitutes the sole binding and activation site for pro-uPA at the cell surface (Vassalli et al., 1985), it follows that the number of unoccupied uPAR at the cell surface is a limiting factor in uPA activation and activity.
The net effect of the interaction of between occupied uPAR and LRP is to remove uPAR with inactive protease (uPA) activity from the cell surface and replace it with unoccupied uPAR capable of binding pro-uPA and initiating a new cycle of plasminogen activation. The importance of LRP in regeneration of cell surface protease activity and cell migration has previously been demonstrated by the finding that RAP and anti-LRP antibody impair the regeneration of unoccupied uPAR and decrease uPA activity of human trophoblast cells (Zhang et al., 1998). Moreover, it was reported that LRP-deficient cultured fibroblasts (Weaver et al., 1997) and cells in which LRP expression has been down-regulated by tumor-promoting phorbol esters (Picone et al., 1989) have increased amounts of uPAR on their surfaces compared with normal or untreated cells (Picone et al., 1989; Weaver et al., 1997). Our data validate the unique role of LRP in regulation of extracellular plasminogen activation and further demonstrate that uPAR binding to LRP is a mandatory step in the internalization of these receptors and in the regeneration of unoccupied uPAR.
ACKNOWLEDGMENTS
We thank Ileana Popa for preparation of activated α2M. This work was supported by a Department of Defense Breast Cancer Research Program Grant DAMD-96-1-6317 to M.G.F., Fellowship DAMD17-96-1-6318 to R.-P.C., and a Peter Lippincola Fellowship to T.K.
Abbreviations used:
α2M
α2-macroglobulin
D2
domain 2 of uPAR
D3
domain 3 of uPAR
EE
early endosomes
LDL
low density lipoprotein
LRP
LDL receptor-related protein
PAI-1
plasminogen activator inhibitor type-1
PM
plasma membrane
Tf
transferrin
uPA
urokinase-type plasminogen activator
uPAR
uPA receptor
REFERENCES
- Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- Anderson RGW. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- Andreasen PA, Sottrup-Jensen L, Kjøller L, Nykjaer A, Moestrup SK, Petersen CM, Gliemann J. Receptor-mediated endocytosis of plasminogen activators and activator/inhibitor complexes. FEBS Lett. 1994;338:239–245. doi: 10.1016/0014-5793(94)80276-9. [DOI] [PubMed] [Google Scholar]
- Beck KA, Chang M, Brodsky FM, Keen JH. Clathrin assembly protein AP-2 induces aggregation of membrane vesicles: a possible role for AP-2 in endosome formation. J Cell Biol. 1992;119:787–796. doi: 10.1083/jcb.119.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt N, Ploug M, Patthy L, Houen G, Blasi F, Dano K. The ligand-binding domain of the cell surface receptor for urokinase-type plasminogen activator. J Biol Chem. 1991;266:7842–7847. [PubMed] [Google Scholar]
- Blasi F, Vassalli JD, Danø K. Urokinase-type plasminogen activator: proenzyme, receptor, and inhibitors. J Cell Biol. 1987;104:801–804. doi: 10.1083/jcb.104.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM. New fashions in vesicle coats. Trends Cell Biol. 1997;7:175–179. doi: 10.1016/S0962-8924(97)01038-6. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Chen H, Paradies NE, Fedor-Chaiken M, Brackenbury R. E-cadherin mediates adhesion and suppresses cell motility via distinct mechanisms. J Cell Sci. 1997;110:345–356. doi: 10.1242/jcs.110.3.345. [DOI] [PubMed] [Google Scholar]
- Cohen RL, Xi XP, Crowley CW, Lucas BK, Levinson AD, Shuman MA. Effects of urokinase receptor occupancy on plasmin generation and proteolysis of basement membrane by human tumor cells. Blood. 1991;78:479–487. [PubMed] [Google Scholar]
- Conese M, Nykjaer A, Petersen CM, Cremona O, Pardi R, Andreasen PA, Gliemann J, Christensen EI, Blasi F. Alpha-2 macroglobulin receptor/LDL receptor-related protein (LRP)-dependent internalization of the urokinase receptor. J Cell Biol. 1995;131:1609–1622. doi: 10.1083/jcb.131.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubellis MV, Andreasen P, Ragno P, Mayer M, Dano K, Blasi F. Accessibility of receptor-bound urokinase to type-1 plasminogen activator inhibitor. Proc Natl Acad Sci USA. 1989;86:4828–4832. doi: 10.1073/pnas.86.13.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubellis MV, Wun TC, Blasi F. Receptor-mediated internalization and degradation of urokinase is caused by its specific inhibitor PAI-1. EMBO J. 1990;9:1079–1085. doi: 10.1002/j.1460-2075.1990.tb08213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czekay RP, Orlando RA, Woodward L, Adamson ED, Farquhar MG. The expression of megalin and LRP diverges in differentiating F9 embryonal carcinoma cells. J Cell Sci. 1995;108:1433–1441. doi: 10.1242/jcs.108.4.1433. [DOI] [PubMed] [Google Scholar]
- Czekay RP, Orlando RA, Woodward L, Lundstrom M, Farquhar MG. Endocytic trafficking of megalin/RAP complexes: dissociation of the complexes in late endosomes. Mol Biol Cell. 1997;8:517–532. doi: 10.1091/mbc.8.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dano K, Andreasen PA, Grondahl-Hansen J, Kristensen P, Nielsen LS, Skriver L. Plasminogen activators, tissue degradation, and cancer. Adv Cancer Res. 1985;44:140–266. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- Dear AE, Medcalf RL. The urokinase-type-plasminogen-activator receptor (CD87) is a pleiotropic molecule. Eur J Biochem. 1998;252:185–193. doi: 10.1046/j.1432-1327.1998.2520185.x. [DOI] [PubMed] [Google Scholar]
- Ellis V, Scully MF, Kakkar VV. Plasminogen activation initiated by single-chain urokinase-type plasminogen activator. Potentiation by U937 monocytes. J Biol Chem. 1989;264:2185–2188. [PubMed] [Google Scholar]
- Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kD liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Kowal RC, Goldstein JL, Brown MS. Proteolytic processing of the 600 kD low density lipoprotein receptor-related protein (LRP) occurs in a trans-Golgi compartment. EMBO J. 1990;9:1769–1776. doi: 10.1002/j.1460-2075.1990.tb08301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RH, Bensch KG, Wong J. Invasion of reconstituted basement membrane matrix by metastatic human tumor cells. Cancer Res. 1986;46:1980–1989. [PubMed] [Google Scholar]
- Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP) Annu Rev Biochem. 1994;63:601–637. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- Kurecki T, Kress LF, Laskowski M., Sr Purification of human plasma alpha 2 macroglobulin and alpha 1 proteinase inhibitor using zinc chelate chromatography. Anal Biochem. 1979;99:415–420. doi: 10.1016/s0003-2697(79)80026-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larkin JM, Brown MS, Goldstein JL, Anderson RG. Depletion of intracellular potassium arrests coated pit formation and receptor-mediated endocytosis in fibroblasts. Cell. 1983;33:273–285. doi: 10.1016/0092-8674(83)90356-2. [DOI] [PubMed] [Google Scholar]
- Lisanti MP, Tang ZL, Sargiacomo M. Caveolin forms a hetero-oligomeric protein complex that interacts with an apical GPI-linked protein: implications for the biogenesis of caveolae. J Cell Biol. 1993;123:595–604. doi: 10.1083/jcb.123.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, Mayor S. Cell surface dynamics of GPI-anchored proteins. Adv Exp Med Biol. 1997;419:355–364. doi: 10.1007/978-1-4419-8632-0_47. [DOI] [PubMed] [Google Scholar]
- McCaffery JM, Farquhar MG. Localization of GTPases by indirect immunofluorescence and immunoelectron microscopy. Methods Enzymol. 1995;257:259–279. doi: 10.1016/s0076-6879(95)57031-4. [DOI] [PubMed] [Google Scholar]
- Memmo LM, McKeown-Longo P. The αvβ5 integrin functions as an endocytic receptor for vitronectin. J Cell Sci. 1998;111:425–433. doi: 10.1242/jcs.111.4.425. [DOI] [PubMed] [Google Scholar]
- Moestrup SK, Gliemann J. Purification of the rat hepatic α2-macroglobulin receptor as an approximately 440-kDa single chain protein. J Biol Chem. 1989;264:15574–15577. [PubMed] [Google Scholar]
- Moestrup SK, Kaltoft K, Petersen CM, Pedersen S, Gliemann J, Christensen EI. Immunocytochemical identification of the human alpha 2-macroglobulin receptor in monocytes and fibroblasts: monoclonal antibodies define the receptor as a monocyte differentiation antigen. Exp Cell Res. 1990;190:195–203. doi: 10.1016/0014-4827(90)90185-d. [DOI] [PubMed] [Google Scholar]
- Moser TL, Enghild JJ, Pizzo SV, Stack MS. The extracellular matrix proteins laminin and fibronectin contain binding domains for human plasminogen and tissue plasminogen activator. J Biol Chem. 1993;268:18917–18923. [PubMed] [Google Scholar]
- Myohanen HT, Stephens RW, Hedman K, Tapiovaara H, Ronne E, Hoyer-Hansen G, Dano K, Vaheri A. Distribution and lateral mobility of the urokinase-receptor complex at the cell surface. J Histochem Cytochem. 1993;41:1291–301. doi: 10.1177/41.9.8394852. [DOI] [PubMed] [Google Scholar]
- Nielsen LS, Kellerman GM, Behrendt N, Danø PRK, Blasi F. A 55,000-60,000 Mr receptor protein for urokinase-type plasminogen activator. J Biol Chem. 1988;236:2358–2363. [PubMed] [Google Scholar]
- Nykjaer A, Conese M, Christensen EI, Olson D, Cremona O, Gliemann J, Blasi F. Recycling of the urokinase receptor upon internalization of the uPA:serpin complexes. EMBO J. 1997;16:2610–2620. doi: 10.1093/emboj/16.10.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson D, Pöllänen J, Høyer-Hansen G, Rønne E, Sakaguchi K, Wun T-C, Appella E, Danø K, Blasi F. Internalization of the urokinase-plasminogen activator inhibitor type-1 complex is mediated by the urokinase receptor. J Biol Chem. 1992;267:9129–9133. [PubMed] [Google Scholar]
- Orlando RA, Farquhar MG. Identification of a cell line that expresses a cell surface and a soluble form of the gp330/receptor-associated protein (RAP) Heymann nephritis antigenic complex. Proc Natl Acad Sci USA. 1993;90:4082–4086. doi: 10.1073/pnas.90.9.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando RA, Farquhar MG. Functional domains of the receptor-associated protein (RAP) Proc Natl Acad Sci USA. 1994;91:3161–3165. doi: 10.1073/pnas.91.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone R, Kajtaniak EL, Nielsen LS, Behrendt N, Mastronicola MR, Cubellis MV, Stoppelli MP, Pedersen S, Danø K, Blasi F. Regulation of urokinase receptors in monocytelike U937 cells by phorbol ester phorbol myristate acetate. J Cell Biol. 1989;108:693–702. doi: 10.1083/jcb.108.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploug M, Rønne E, Behrendt N, Jensen AL, Blasi F, Danø K. Cellular receptor for urokinase plasminogen activator. Carboxyl-terminal processing and membrane anchoring by glycosyl-phosphatidylinositol. J Biol Chem. 1991;266:1926–1933. [PubMed] [Google Scholar]
- Ploug M. Identification of specific sites involved in ligand binding by photoaffinity labeling of the receptor for the urokinase-type plasminogen activator. Residues located at equivalent positions in uPAR domains I and III participate in the assembly of a composite ligand-binding site. Biochemistry. 1998;37:16494–16505. doi: 10.1021/bi981203r. [DOI] [PubMed] [Google Scholar]
- Reiter LS, Kruithof EK, Cajot JF, Sordat B. The role of the urokinase receptor in extracellular matrix degradation by HT29 human colon carcinoma cells. Int J Cancer. 1993;53:444–450. doi: 10.1002/ijc.2910530316. [DOI] [PubMed] [Google Scholar]
- Roettger BF, Rentsch RU, Pinon D, Holicky E, Hadac E, Larkin JM, Miller LJ. Dual pathways of internalization of the cholecystokinin receptor. J Cell Biol. 1995;128:1029–1041. doi: 10.1083/jcb.128.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Saksela O, Rifkin DB. Cell-associated plasminogen activation: Regulation and physiological functions. Ann Rev Cell Biol. 1988;4:93–126. doi: 10.1146/annurev.cb.04.110188.000521. [DOI] [PubMed] [Google Scholar]
- Shyng SL, Heuser JE, Harris DA. A glycolipid-anchored prion protein is endocytosed via clathrin-coated pits. J Cell Biol. 1994;125:1239–1250. doi: 10.1083/jcb.125.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DI, Wei Y, Zhang L, Rao NK, Xu H, Chen Z, Liu Q, Rosenberg S, Chapman HA. Identification of a urokinase receptor-integrin interaction site. Promiscuous regulator of integrin function. J Biol Chem. 2000;275:10228–10234. doi: 10.1074/jbc.275.14.10228. [DOI] [PubMed] [Google Scholar]
- Stan RV, Roberts WG, Predescu D, Ihida K, Saucan L, Ghitescu L, Palade GE. Immunoisolation and partial characterization of endothelial plasmalemmal vesicles (caveolae) Mol Biol Cell. 1997;8:595–605. doi: 10.1091/mbc.8.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RW, Pollanen J, Tapiovaara H, Leung KC, Sim PS, Salonen EM, Ronne E, Behrendt N, Dano K, Vaheri A. Activation of pro-urokinase and plasminogen on human sarcoma cells: a proteolytic system with surface-bound reactants. J Cell Biol. 1989;108:1987–1995. doi: 10.1083/jcb.108.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli JD, Baccino D, Belin D. A cellular binding site for the Mr 55,000 form of the human plasminogen activator, urokinase. J Cell Biol. 1985;100:86–92. doi: 10.1083/jcb.100.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Hussaini IM, Mazar A, Henkin J, Gonias SL. Embryonic fibroblasts that are genetically deficient in low density lipoprotein receptor-related protein demonstrate increased activity of the urokinase receptor system and accelerated migration on vitronectin. J Biol Chem. 1997;272:14372–14379. doi: 10.1074/jbc.272.22.14372. [DOI] [PubMed] [Google Scholar]
- Williams SE, Kounnas MZ, Argraves KM, Argraves WS, Strickland DK. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein and the receptor-associated protein. An overview. Ann NY Acad Sci. 1994;737:1–13. doi: 10.1111/j.1749-6632.1994.tb44297.x. [DOI] [PubMed] [Google Scholar]
- Xue W, Mizukami I, Todd R, Petty HR. Urokinase-type plasminogen activator receptors associate with beta1 and beta3 integrins of fibrosarcoma cells: dependence on extracellular matrix components. Cancer Res. 1997;57:1682–1689. [PubMed] [Google Scholar]
- Zhang JC, Sakthivel R, Kniss D, Graham CH, Strickland DK, McCrae KR. The low density lipoprotein receptor-related protein/alpha2-macroglobulin receptor regulates cell surface plasminogen activator activity on human trophoblast cells. J Biol Chem. 1998;273:32273–32280. doi: 10.1074/jbc.273.48.32273. [DOI] [PubMed] [Google Scholar]