The genetic landscape of mutations in Burkitt lymphoma (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 6.
Published in final edited form as: Nat Genet. 2012 Nov 11;44(12):1321–1325. doi: 10.1038/ng.2468
Abstract
Burkitt lymphoma is characterized by deregulation of MYC, but the contribution of other genetic mutations to the disease is largely unknown. Here, we describe the first completely sequenced genome from a Burkitt lymphoma tumor and germline DNA from the same affected individual. We further sequenced the exomes of 59 Burkitt lymphoma tumors and compared them to sequenced exomes from 94 diffuse large B-cell lymphoma (DLBCL) tumors. We identified 70 genes that were recurrently mutated in Burkitt lymphomas, including ID3, GNA13, RET, PIK3R1 and the SWI/SNF genes ARID1A and SMARCA4. Our data implicate a number of genes in cancer for the first time, including CCT6B, SALL3, FTCD and PC. ID3 mutations occurred in 34% of Burkitt lymphomas and not in DLBCLs. We show experimentally that ID3 mutations promote cell cycle progression and proliferation. Our work thus elucidates commonly occurring gene-coding mutations in Burkitt lymphoma and implicates ID3 as a new tumor suppressor gene.
Burkitt lymphoma is characterized by deregulation of the MYC gene through its translocation to one of the immunoglobulin loci. The role of collaborating genetic mutations that contribute to Burkitt lymphoma remains unknown1,2. Whereas gene expression profiles of Burkitt lymphoma and the more common DLBCL have shown that these two diseases have vast molecular differences1,3, the genetic underpinnings of these differences are not known.
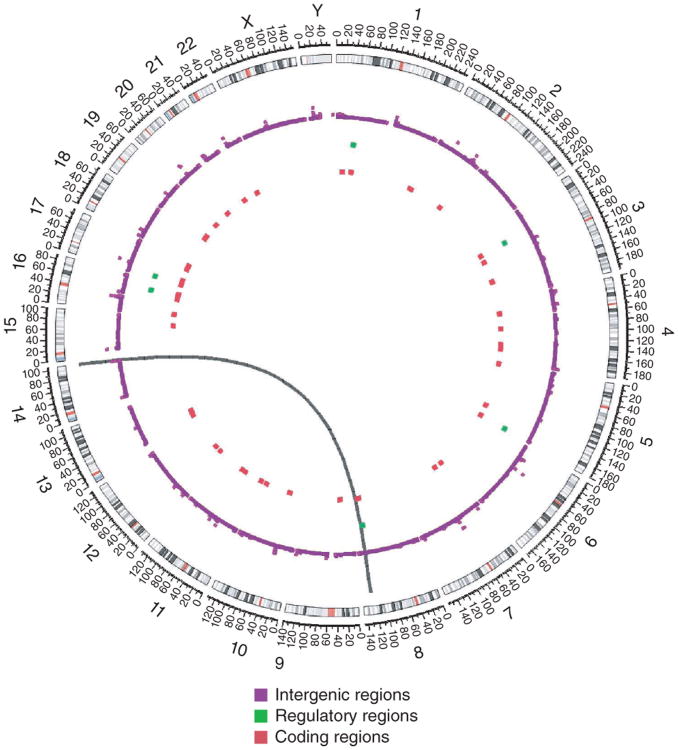
We identified a classic case of Burkitt lymphoma4 and performed whole-genome sequencing of tumor and germline DNA from the same affected individual using the Illumina platform. The distribution of somatic mutations observed in the Burkitt lymphoma genome is depicted in a Circos5 diagram (Fig. 1; summarized in Supplementary Table 1). The vast majority of somatic alterations were in intergenic regions. We observed 6 mutations in potential regulatory regions (loci within 2 kb of a transcriptional start site) and 42 in gene-coding regions. Through the analysis of paired-end reads6, we also identified the presence of the t(8;14) translocation (Supplementary Fig. 1) that is a defining feature of Burkitt lymphoma. Thus, in this single genome, nearly all the known hallmarks of Burkitt lymphoma were identified, including the translocation and mutation of the MYC gene.
Figure 1.
Results from whole-genome sequencing of a Burkitt lymphoma tumor and germline DNA. The Circos diagram5 summarizes the somatically acquired genetic variants in a Burkitt lymphoma genome. The outermost ring depicts the chromosome ideogram oriented clockwise, p terminus to q terminus. Centromeres are shown in red. The next three rings indicate somatically acquired mutations falling in intergenic regions, potential regulatory regions and the exome, respectively. The black arc connecting chromosomes 8 and 14 signifies a t(8;14) translocation detected in sequencing data.
We further characterized the diversity of mutations in Burkitt lymphoma by performing exome sequencing on 59 affected individuals, including 51 primary Burkitt lymphoma tumors, 14 with paired normal tissue, and 8 Burkitt lymphoma cell lines, using the Illumina platform and Agilent reagents. We verified adequate sequencing quality and coverage throughout the exome (Supplementary Fig. 2). We identified genetic variants and further classified these as synonymous, missense, nonsense and small insertions and/or deletions (indels).
We verified the accuracy of genetic variant identification from our deep sequencing data by performing Sanger sequencing on 108 missense and 16 frameshift and/or indel mutations (Supplementary Note). We found that the two methods agreed for over 80% of the variants assessed (Supplementary Table 2), confirming that our sequencing and bioinformatics methods generated accurate results.
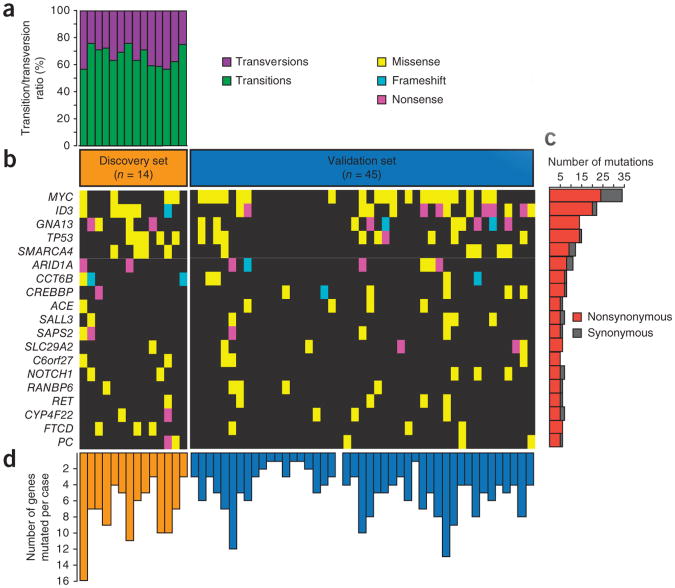
We designated the 14 Burkitt lymphoma samples with paired germline DNA our discovery set, and the remaining 45 Burkitt lymphoma samples were designated the validation set. We noted that transitions were the predominant form of somatically acquired genetic variation in the discovery set (P < 1 × 10−6, _χ_2 test; Fig. 2a and Supplementary Fig. 3)7. We identified 1,241 variants in 1,104 unique genes that were somatically mutated in at least 1 tumor-germline pair in the discovery set. We then identified additional genetic variants for these 1,104 genes in the validation set, which were similarly rare variants that were not present in databases of normal variation, including dbSNP135 (ref. 8), publicly available data from healthy individuals9–12 (Supplementary Note) and data from 19 additional exomes that we sequenced from control individuals without lymphoma.
Figure 2.
Exome sequencing in Burkitt lymphoma. (a) The ratio of somatically acquired transitions and transversions for samples with paired normal tissue are shown for all 14 discovery set samples. (b) The heatmap indicates the mutation patterns of the 19 most frequently implicated genes out of the 70 genes mutated in Burkitt lymphoma. Each column represents an affected individual, and each row represents a gene. (c) The bar graph shows the frequency of variants found per gene across all samples, divided into nonsynonymous and synonymous counts. (d) The bar graph shows the frequency of mutations in all 70 genes mutated in Burkitt lymphoma in each case: orange bars represent samples in the discovery set, and blue bars represent samples in the validation set.
For the 1,104 somatically mutated genes, we identified candidate mutated genes in the validation set of 45 Burkitt lymphomas. We annotated the 2,318 variants that were nonsynonymous and did not occur in normal controls. For a gene to be classified as being mutated in Burkitt lymphoma, it needed to have recurrent variants that were already in the Catalogue of Somatic Mutations in Cancer (COSMIC)13 or recurrent variants in close proximity to each other or affecting the same protein domain (Supplementary Fig. 4 and Supplementary Note).
We identified 70 recurrently mutated genes in Burkitt lymphoma (Supplementary Tables 3 and 4), including 16 genes that have been conclusively implicated in cancer13. The number and types of mutations in genes that were mutated in 10% or more of the Burkitt lymphomas (n = 19) are shown (Fig. 2b,c). We noted considerable heterogeneity in the number of mutated genes, which ranged up to 16 per lymphoma of these 70 genes (Fig. 2d). Gene expression data confirmed that all of these genes were measurably expressed in Burkitt lymphomas, DLBCLs or mature B cells (an example of expression is depicted in Supplementary Fig. 5).
The most frequently mutated genes in Burkitt lymphoma were MYC (40%) and ID3 (34%). Other frequently mutated genes included the known suppressor genes ARID1A, SMARCA4 and TP53, as well as the oncogene PIK3R1 and NOTCH1. In the recurrently mutated genes in Burkitt lymphoma, silencing events, such as nonsense and frameshift mutations, constituted a substantial proportion (∼30% or more) of the events in ID3, GNA13, ARID1A, CREBBP and CCT6B, suggesting that the genetic alterations may result in loss of function.
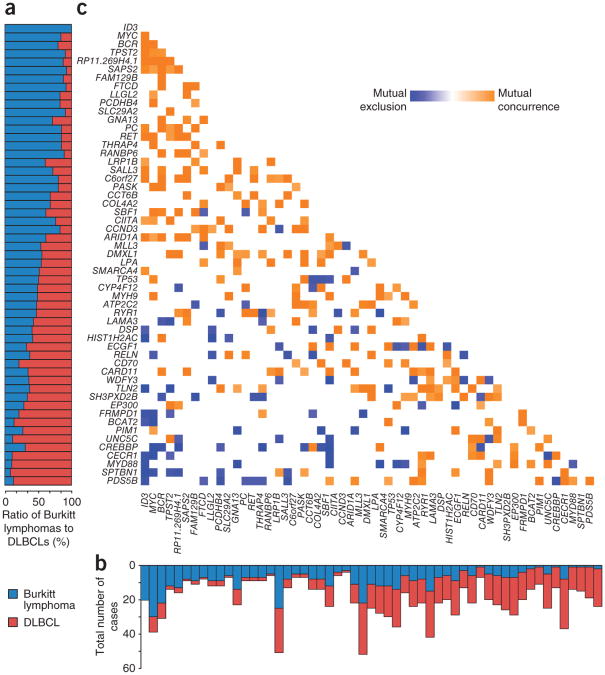
We further investigated the genetic differences between Burkitt lymphoma and DLBCL. Through similar analyses, we identified 351 recurrently mutated genes in DLBCL (Supplementary Note and J.Z. et al., unpublished data), a number of which overlapped with those identified in previously published studies of DLBCL14–16. We identified all genes that were recurrently mutated in either Burkitt lymphoma or DLBCL at a frequency of at least 10% in our study or one of the published studies of DLBCL. We plotted the relative and absolute frequencies of the gene alterations in Burkitt lymphoma and DLBCL (Fig. 3a,b). We found a number of genes, including ID3, MYC, TPST2 and RET, that were predominantly mutated in Burkitt lymphoma (P < 0.05, Fisher's exact test). In contrast, PIM1, CECR1 and MYD88 (ref. 17) were predominantly mutated in DLBCL. A number of genes had overlapping patterns of mutation in the two diseases, including MLL3, TP53 and LAMA3.
Figure 3.
Patterns of exonic mutations in Burkitt lymphoma compared to DLBCL. (a) The bar graph shows the proportion of Burkitt lymphoma and DLBCL samples containing a mutation in each gene. (b) The bar graph shows the number of cases that contain a mutation in the given gene in Burkitt lymphoma and DLBCL. (c) The heatmap shows the association between 55 genes found to be recurrently mutated in Burkitt lymphoma and/or DLBCL. Blue denotes negative association between genes, and orange denotes positive association.
We further examined the association between the occurrence of individual gene alterations in Burkitt lymphoma and DLBCL (Fig. 3c). Notably, we found that mutations in the SWI/SNF family members SMARCA4 and ARID1A occurred in a mutually exclusive fashion, suggesting that mutation in one of these genes by itself may be sufficient to deregulate the SWI/SNF chromatin-remodeling complex. The different mutational patterns of Burkitt lymphoma and DLBCL were also related in part to the lineage-derived subsets of DLBCL18. MYD88 and C_D79A_ were predominantly mutated in the activated B cell–like (ABC) DLBCLs compared to Burkitt lymphoma. GNA13, EZH2 and BCL2 showed overlapping mutational patterns in Burkitt lymphomas and DLBCLs derived from germinal center B cells.
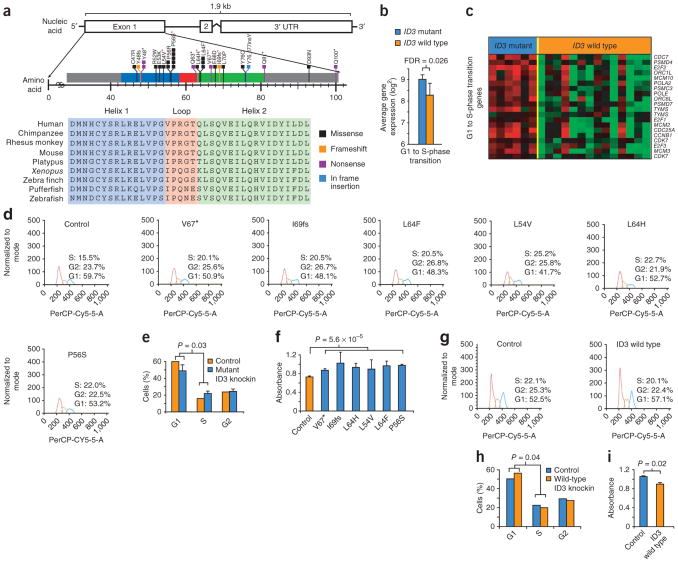
ID3 mutations affected nearly a third of the Burkitt lymphomas and were not present in any DLBCLs, including those containing MYC translocations (Supplementary Note). Nearly all of the alterations in ID3 affected the highly conserved helix-loop-helix (HLH) domain (Fig. 4a). Of these alterations, nearly 30% represented nonsense and frameshift mutations, suggesting that the mutations have a silencing effect on the gene.
Figure 4.
Recurrent ID3 mutations in Burkitt lymphomas. (a) Deep sequencing reads identify recurrent mutations affecting the HLH domain of ID3 in Burkitt lymphomas. Each colored line represents an individual somatic mutation or a rare genetic variant. The conservation of the HLH domain across species is also shown. Red asterisks identify the alterations that were functionally validated. (b) The bar graph shows significantly higher expression of genes involved in the G1 to S-phase transition in _ID3_-mutant Burkitt lymphoma samples compared to those with wild-type ID3 (FDR = 0.026), as determined in gene set enrichment analysis. Error bars, s.d. (c) Heatmap of genes corresponding to the gene set enrichment msigdb-derived list of those involved in the G1 to S-phase transition. Red denotes high relative expression, and green represents low relative expression across samples. (d) Cell cycle analysis of Jijoye cells expressing mutant ID3 proteins compared to control cells overexpressing GFP, where the red histogram denotes cells in G1 phase, orange denotes S phase, and green denotes G2 phase. The x axis shows the relative fluorescence in the PerCP-Cy5.5 channel. (e) The bar graph summarizes cell cycle analysis from an average of all cells expressing ID3 mutants compared to cells overexpressing GFP (P = 0.03, χ2 test). Error bars, s.d. (f) MTT cell proliferation assay performed on Jijoye cells expressing mutant ID3 or control GFP. Absorbance was read 24 h after plating, with higher absorbance indicating more cells and signifying faster proliferation. Error bars, s.d. (g) Cell cycle analysis of BL41 cells expressing wild-type ID3 compared to those expressing GFP control. (h) The bar graph summarizes cell cycle analysis for BL41 cells expressing wild-type ID3 relative to cells expressing GFP control. (i) MTT cell proliferation assay performed on BL41 cells expressing wild-type ID3 or GFP. Error bars, s.d.
To better understand the biological role of ID3 mutations in Burkitt lymphoma, we began by examining gene expression in 21 Burkitt lymphomas and 87 DLBCLs. We found that Burkitt lymphomas were characterized by twofold higher expression of ID3 compared to DLBCLs (P = 0.002; Supplementary Fig. 6). Both alleles seemed to be expressed at similar levels in cases with mutations (Supplementary Fig. 7). Gene set enrichment analysis19 identified genes associated with the G1 to S-phase transition as being significantly upregulated in lymphomas with ID3 mutations (false discovery rate (FDR) < 0.05; Supplementary Fig. 8). The expression of cell cycle pathway genes corresponding to the G1 to S-phase transition, including E2F1, CDK7 and MCM10, was significantly higher in _ID3-_mutant Burkitt lymphoma samples relative to those with wild-type ID3 (Fig. 4b, c). Samples with ID3 mutation also showed higher expression of known MYC target genes (Supplementary Fig. 9). These findings provided a working hypothesis that ID3 mutations promote the G1 to S-phase transition, which we then tested experimentally.
We designed constructs expressing six different mutant forms of the ID3 gene, encoding the Val67*, Ile69fs, Leu64Phe, Leu54Val, Leu64His and Pro56Ser variants. We expressed these mutant constructs using a lentiviral vector in Jijoye, a Burkitt lymphoma cell line with wild-type ID3, and confirmed their expression using protein blot analysis and fluorescence microscopy (Supplementary Fig. 10). Cells expressing each of the six mutant constructs had a greater proportion of cells in S phase and a reduced proportion of cells in G1 phase (Fig. 4d), differences that, when averaged together and plotted, were significant compared to control cells encoding wild-type ID3 (P = 0.03, paired t test; Fig. 4e). Cell-cycle analysis over 24 h showed higher cell proliferation in all cell lines expressing mutant ID3 (P = 5.6 × 10−5, Student's t test; Fig. 4f). These results suggest that mutations in ID3 result in increased G1 to S-phase cell cycle progression in Burkitt lymphoma.
Conversely, when we expressed wild-type ID3 in the BL41 cell line encoding mutant ID3 (with the p.Val67* alteration), we found that the proportion of cells in S phase was lower in cells expressing wild-type ID3 compared to control cells overexpressing only GFP (Fig. 4g,h). Similarly, we observed significantly lower cell proliferation in cells expressing wild-type ID3 at 24 h in culture (P = 0.02, Student's t test; Fig. 4i).
Thus, ID3 mutants increased cell cycle progression and cellular proliferation in Burkitt lymphoma cells, whereas expression of wild-type ID3 in mutant cells gave the opposite results. These experiments support a role for ID3 as a new tumor suppressor gene in Burkitt lymphoma.
The role of MYC as a human oncogene was first discovered in Burkitt lymphoma20, and its importance has since been shown in a number of different malignancies, including carcinomas of the lung21, breast22, cervix22, ovary23 and colon24. Little is known about the role of other genetic alterations that collaborate with MYC deregulation in Burkitt lymphoma.
Inhibitor of DNA binding (ID) proteins have been shown to be regulators of normal cellular development25. These proteins lack a DNA-binding domain and inhibit transcription through the formation of nonfunctional heterodimers with other basic helix-loop-helix (bHLH) proteins. Our data implicate ID proteins, for the first time to our knowledge, in Burkitt lymphoma and cancer, with ID3 mutations affecting over a third of Burkitt lymphomas. Predominantly silencing mutations in ID3 were associated with increased cell cycle progression and the expression of proliferation-associated genes. The ability of wild-type ID3 to decrease cell proliferation in Burkitt lymphoma suggests the possibility of using ID3 mimetics as a potential therapeutic approach in Burkitt lymphoma and other bHLH-driven cancers. The role of ID3 also highlights the importance of context in shaping the effect of genetic alterations in cancer. Affecting a single gene, mutations in ID3 seem unlikely to have a clear oncogenic role in most cancers. It is only in the setting of deregulation of MYC (and perhaps other oncogenic bHLH proteins) that inactivating ID3 mutations might have a role by significantly amplifying the actions of these oncogenes. Similar context-dependent roles may be carried out by a number of other oncogenes and tumor suppressor genes.
Our study newly implicates a number of other genes in Burkitt lymphoma. Mutations in SWI/SNF family members ARID1A and SMARCA4 occurred in a mutually exclusive fashion in Burkitt lymphoma, affecting nearly 25% of the tumors. Lineage also seems to have a key role in determining the mutations acquired in Burkitt lymphomas. GNA13, which encodes a guanine nucleotide–binding G protein, was mutated through predominantly silencing events in nearly 15% of the lymphomas and has been shown to be specifically mutated in germinal center B cell–derived DLBCLs (ref. 26 and J.Z. et al., unpublished data). Thus, alterations in GNA13 seem to be a germinal center B cell–specific oncogenic event in lymphomas, similar to those described for EZH2 (ref. 26), which was also mutated in 7% of Burkitt lymphomas. We also observed recurrent mutations in the RET, BRAF, NOTCH1 and PI3KR1 genes and their associated pathways. These findings suggest new therapeutic possibilities in Burkitt lymphoma that can be tested in clinical trials in conjunction with approaches that assay for these mutations. Our data also implicate a number of genes for the first time in cancer, including CCT6B, SALL3, FTCD and PC. These genes likely have roles in other cancers that remain to be explored.
Exome sequencing has emerged as a powerful approach for the delineation of gene-coding mutations in malignancies. However, this approach does not capture every important aspect of tumor biology. Not every gene will have adequate coverage in every instance. Exome sequencing also does not assay for structural genetic alterations, mutations in regulatory regions and epigenetic alterations that could also make critical contributions to observed tumor phenotypes. Nevertheless, exome sequencing provides a cost-effective means to identify broad patterns of mutation in diseases at a resolution that was unthinkable just a few years earlier.
Our work thus provides an important starting point for understanding the genetic landscape of mutations in Burkitt lymphomas.
URLs
Picard, http://picard.sourceforge.net/; GATK base quality score recalibration, http://www.broadinstitute.org/gatk; mpileup settings, http://samtools.sourceforge.net/mpileup.shtml; Mutation Assessor, http://mutationassessor.org/; Circos, http://circos.ca/software/download/; Novoalign V2.06.09, http://novocraft.com/; VCFtools, http:// vcftools.sourceforge.net/.
Online Methods
Sample collection and processing
Burkitt lymphoma tumors (n = 51) and normal tissues (n = 14) were obtained from the institutions that constitute the Hematologic Malignancies Research Consortium (HMRC)27. Information for the individuals from whom these samples were obtained is shown in Supplementary Table 5. All tumors contained over 90% malignant cells. Samples were anonymized, shipped to Duke University and processed in accordance with a protocol approved by the institutional review board at Duke University. Informed consent was obtained from all patients, except for those cases determined by the Duke Institutional Review Board to be exempt. In addition, eight well-characterized Burkitt lymphoma cell lines were included in the study. Genomic DNA was extracted from the tissues and cell lines using a previously described column-based method27.
Whole-genome sequencing
Up to 3 μg of genomic DNA from a tumor and paired normal tissue was sheared to a target size of 500 bp using Covaris adaptive-focused acoustics with duty cycle of 5%, intensity of 3, frequency of 200 cycles/burst, duration of 80 s and water bath temperature of 4 °C. For each sample, sheared DNA was column purified, resuspended in 10 mM Tris-HCl, pH 8.5, and quantified on a Bioanalyzer (Agilent) using the DNA 1000 chip. DNA ends were repaired using T4 DNA ligase, Klenow enzyme and T4 polynucleotide kinase (PNK) (NEB), with samples incubated at 20 °C for 30 min. Poly-A tails were added to the 3′ ends using Klenow exo‒ fragment (NEB), with samples incubated at 37 °C for 30 min. A ratio of 2 ill of Illumina paired-end adapters (J.Z. et al., unpublished data) per 1 µg of DNA was added to the fragments using Quick Ligase (NEB), with samples incubated at 20 °C for 15 min. The library was PCR amplified for 6–8 cycles using Illumina paired-end PCR primers and 2× Phusion High-Fidelity Master Mix (NEB). The resulting library was purified and assayed on a Bioanalyzer to determine size and concentration. Final libraries were diluted to 5 pM for Illumina clustering, and paired-end sequencing was carried out over 9 d.
Whole-genome sequence alignment
Using the Illumina platform, we generated 178 Gb and 99 Gb of sequence from tumor and normal samples, respectively. We estimated the average per-base sequencing coverage at 48-fold for the tumor and 26-fold for the paired germline DNA.
Reads in fastq format28 were initially processed with GATK29 version 1.0.3954 to remove Illumina adaptor sequences (analysis type -T ClipReads, -XF illumina.adapters.fa) and Phred-scaled base qualities of ≤10 (-QT 10). After GATK processing, reads were mapped using the Burrows-Wheeler Aligner (BWA)30 version 0.5.8c with a -q15 setting for read trimming, which removes the 3′ portion of reads from an alignment if it is below the quality threshold specified. The alignments (sai files) were used to generate SAM (Sequence Alignment/Map) paired-end read files using bwa sample and were sorted by the leftmost coordinate with SAMtools version 0.1.12-4. All alignments were saved as BAM files31 (the binary equivalent of the SAM format) and merged using Picard (see URLs). PCR and optical duplicates and multiple reads likely to have been read from a single cluster on the flow-cell image were marked with Picard. Base quality recalibration was performed using GATK to generate a more accurate base quality score that took into account the reported quality score in the original fastq file, position within the read and sequence context, for example, AC and TG dinucleotides (see URLs).
To improve the accuracy and quality of the calls, localized indel realignments were performed using GATK29, which infers the consensus indel call from multiple reads mapping to suspected indel genomic regions, rather than considering each read independently. Regions that needed to be realigned were identified using the GATK Realigner Target Creator. SNVs and indel variants were called using SAMtools31. First, reads with mapping quality of at least 1 were extracted with samtools view, and information was collated for each base pair by genomic location using pileup. SNPs and short indels were filtered from the pileup file using the samtools.pl v.0.3.3 helper script varFilter. Read alignments were visualized with the Integrative Genomics Viewer (IGV)32 version 1.5.05. The complete list of variants is documented in Supplementary Table 1.
To compare the whole genome of the Burkitt lymphoma and its matching normal sample, SAMtools mpileup with settings -C50 and -m3 -F0.0002 was concurrently run for the pair of pileup files and converted to a single VCF file. The mpileup settings (see URLs) were set to -C50 to limit the contribution of reads with many mismatches and -m3 -F0.0002 to maximize the sensitivity of indel discovery by requiring three supporting reads at a minimum of 0.02% abundance, rather than using the default abundance cutoff of 0.2%.
Individual SNVs and indels were annotated with gene names, and their effects on function were predicted by the Sequence Variant Analyzer33, which uses genomic coordinates and variants as input and yields functional annotation, including where within genes the alteration lies (for example, in the 5′ UTR, 3′ UTR or coding sequences) and the predicted result of the alteration (for example. amino-acid change, frameshift and stop codon gain). To distinguish new variants discovered in our study from those previously found, all variants were intersected with variant databases, including dbSNP135, HapMap 3 allele frequencies and 1000 Genome Project pilot 1 allele frequencies, and Consensus Coding Sequence (CCDS) Gene IDs were determined using BEDTools and intersectBed and formatted using awk and custom Python scripts. Predictions of the phenotypic severity of variants not previously annotated were determined using Mutation Assessor34 (see URLs), Polyphen-2 (ref. 35) and SIFT 36. The final variant calls are summarized in Supplementary Table 4.
Circos
Circos5 (see URLs) was used to depict the whole-genome Burkitt lymphoma copy-number alterations and somatically acquired mutations separated by region (intergenic, regulatory and exonic). Somatically acquired mutations in intergenic, regulatory and exonic regions were counted in 250-kb bins and are depicted in Figure 1. Bins in which no mutations were detected were not plotted.
Exon capture and sequencing
Up to 3 μg of genomic DNA was sheared to a target size of 250 bp by Covaris adaptive-focused acoustics with duty cycle of 10%, intensity of 5, frequency of 200 cycles/burst, duration of 135 s and water bath temperature of 4 °C. As in whole-genome library preparation, purified sheared DNA was end repaired, A tailed, ligated to Illumina paired-end adapters (J.Z. et al., unpublished data) and amplified as described in the Agilent SureSelect Target Enrichment System for Illumina Paired-End Sequencing Library. The ligated paired-end library was PCR amplified using Illumina paired-end PCR primers and 2× Phusion High-Fidelity Master Mix (NEB). The library was column purified and assayed on a Bioanalyzer to determine size and concentration.
Paired-end libraries (65 ng each) were pooled and prepared according to the protocol provided by Agilent SureSelect Target Enrichment. They were hybridized against Agilent SureSelect Human All Exome 50MB biotinylated baits in a thermocycler at 65 °C for 24 h. The DNA that hybridized to baits was purified on SPRI magnetic beads (Agencourt) according to the Agilent SureSelect protocol. The captured product was PCR amplified with Herculase II Fusion DNA Polymerase (Stratagene) for 12 cycles, and the molarity and size distribution were measured by Bioanalyzer using the DNA High Sensitivity chip. Captured libraries were diluted to 5 pM for Illumina clustering, and paired-end sequencing was carried out over 9 d.
Exome sequence alignment
Alignment steps were performed as described for whole-genome sequence processing. Because of the shorter insert target size of this sequence relative to that generated in whole-genome sequencing, some 100-bp paired-end reads were read through the insert and into the opposite adaptor. The process was modified as follows: after the first alignment step with BWA, any discordantly mapped or unmapped read pair was extracted with SAMtools and realigned using Novoalign V2.06.09 (see URLs), a Needleman-Wunsch algorithm–based aligner37, with a SoftClip setting that permits clipping of the read to the best local alignment to help align reads from shorter library inserts. Remaining unmapped reads were clipped to 35 bp to remove adaptor-matched sequence at the 3′ end of the read and realigned with BWA. All alignments were merged using Picard.
Merging of data from different samples was performed using GATK, and variants that fell within CCDS exons38 were then extracted using BEDTools IntersectBed39. Overlaps between samples were computed using VCFtools (see URLs) and AWK scripts.
SAMtools pileup files were generated for all 59 Burkitt lymphomas and 14 matching normal tissues, 94 DLBCLs and 34 matching normal tissues, and 275 control exomes from individuals without cancer. The 275 control exomes consisted of 19 prepared in house, as well as 256 from publicly available data sets (Supplementary Note), all of which were processed from raw fastq sequencing reads using methods identical to those used for the sequenced exomes.
Sequence variants were annotated by gene, and their effects on function were predicted using the Sequence Variant Analyzer33. These data were collapsed by unique genomic positions for intersection with known variants, as described for whole-genome analysis. SAMtools mpileup with settings -C50 and -m3 -F0.0002 was concurrently run on data from all lymphoma cases and used to generate a single VCF file.
Sanger sequencing validation
DNA regions of interest were amplified using primers targeting exonic regions containing the variant (Supplementary Table 6), as described previously40. At least 50 ng of DNA was PCR amplified using 2× HotStar Master Mix (Qiagen, 203443) and 300 nM of each primer. A touchdown PCR method was carried out, with reactions incubated at 98 °C for 10 min and 94 °C for 2 min, followed by 10 cycles at 94 °C for 10 s, 50 °C for 1 min 10 sec and 72 °C for 45 sec and 20 cycles at 94 °C for 15 s, 50 °C for 30 s, 74 °C for 45 sec, incremented by 5 °C per cycle, and 72 °C for 2 min 30 sec. The amplified fragments were verified by agarose gel and purified with Agencourt Ampure XP beads according to the manufacturer's instructions (Beckman Coulter Genomics, A63881).
Gene expression microarray analysis
Gene expression profiling on 21 Burkitt lymphomas was performed using standard Affymetrix protocols as described previously1. Briefly, 1 μg of total RNA was reverse transcribed, using oligo(dT) primer to synthesize cDNA. T7 primer was used for in vitro transcription, resulting in labeled cRNA, which was fragmented and hybridized to Affymetrix Whole-Genome Gene 1.0 ST microarrays. Microarrays were washed and scanned, and the data were normalized as described previously27.
Expression of ID3 in cell lines
Wild-type and mutant ID3 constructs were cloned into the pLEGFP-N1 vector (BD Biosciences) to generate GFP-fused proteins. Briefly, RT-PCR was performed using high-capacity RNA-to-cDNA Master Mix (Applied Biosystems) on mRNA from normal lymph node and samples from affected individuals with mutations corresponding to p.Val67*, p.Ile69fs, p.Pro56Ser, p.Leu64His and p.Leu64Phe alterations for cDNA synthesis. ID3 mutants were PCR amplified using the _ID3_F and _ID3_R primers (Supplementary Table 6) with Phusion High-Fidelity PCR Master Mix. PCR products were purified and digested with HindIII and BamHI and cloned into pLEGFP-N1 digested with the same enzymes. The cloned ID3 mutants were confirmed by Sanger sequencing.
Viral particle production was conducted following the vendor's recommendations (Clontech's Retroviral Gene Transfer and Expression User Manual) using ID3 constructs, pVSV-G plasmid and the GP2-293 packaging cell line. Briefly, the packaging cell line was transfected with ID3 constructs and pVSV-G using Lipofectamine 2000 (Invitrogen). After 48 h, the cell culture supernatant was collected and filtered through a 0.45-micron filter. Harvested virus was used to transduce targeted cell lines. Geneticin (Invitrogen/Gibco) was added to a final concentration of 50 μg/ml 24 h after viral transduction to select transductants. Expression of the (wild-type or mutant) ID3-GFP fusion proteins was confirmed by protein blot analysis and fluorescence microscopy (Supplementary Fig. 10).
Cell cycle analysis using flow cytometry
Between 1 and 2 million cells were washed 3 times in PBS containing 2% FBS. Cells were resuspended in 0.5 ml of PBS, added dropwise to 3 ml of ice-cold 100% ethanol while vigorously vortexing and fixed overnight at −20 °C. Cells were spun down at 250_g_ for 5 min and washed twice in PBS. Cell pellets were resuspended in 1 ml of pro-pidium iodide and allowed to stain at 4 °C for 3 h. Cells were analyzed for the presence of PerCP-cy5-5-A. FlowJo software (Tree Star) was used to generate cell cycle data and figures.
MTT cell proliferation assay
Cells were plated at equal density in a 96-well plate in 100 μl of medium. After 24 h, 10 μl of MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) reagent was added to the cells, which were then incubated at 37 °C for 3 h. After 3 h, 75 μl of detergent was added, and samples were incubated for 3 h at room temperature away from light. Plates were read at 510 nm.
Supplementary Material
S2
S3
S4
S5
S6
S7
supplement
Acknowledgments
The authors thank S. Sunay and the Georgia Cancer Coalition for support in sample collection. A.B.M. was supported by the Hertz Foundation. This work was supported through grants R21CA1561686 and R01CA136895 from the National Cancer Institute (S.S.D.). S.S.D. was also supported by the American Cancer Society. We gratefully acknowledge the generous support of C. Stiefel and D. Stiefel.
Footnotes
Accession codes. All primary sequencing data will be made publicly available through dbGAP (phs000562.v1.p1), and gene expression data are available at the Gene Expression Omnibus (GEO; GSE22898).
Author Contributions: C.L., R.M., K.L.R., C.H.D., W.W.L.C., G.S., P.L.L., D.A.R., A.S.L., L.B.-M., K.P.M., C.R.F., K.N.N., A.M.E., A.C., L.I.G., M.B.C., J.I.G., E.D.H., J.Z., G.L., A.G., M.M., S.L. and S.S.D. performed research and edited the manuscript. C.L., J.Z., A.B.M., D.J., Z.S., V.G., A.B., D.B.D. and S.S.D. analyzed data. C.L. and S.S.D. wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Note: Supplementary information is available in the online version of the paper.
References
- 1.Dave SS, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 2.Schiffman JD, et al. Genome wide copy number analysis of paediatric Burkitt lymphoma using formalin-fixed tissues reveals a subset with gain of chromosome 13q and corresponding miRNA over expression. Br J Haematol. 2011;155:477–486. doi: 10.1111/j.1365-2141.2011.08883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hummel M, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 4.Swerdlow SH, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th. IARC Press; Lyon, France: 2008. [Google Scholar]
- 5.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 11.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 13.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright G, et al. A gene expression–based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 21.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplifcation and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 22.Münzel P, Marx D, Kochel H, Schauer A, Bock KW. Genomic alterations of the c-myc protooncogene in relation to the overexpression of c-erbB2 and Ki-67 in human breast and cervix carcinomas. J Cancer Res Clin Oncol. 1991;117:603–607. doi: 10.1007/BF01613296. [DOI] [PubMed] [Google Scholar]
- 23.Wang ZR, Liu W, Smith ST, Parrish RS, Young SR. c-myc and chromosome 8 centromere studies of ovarian cancer by interphase FISH. Exp Mol Pathol. 1999;66:140–148. doi: 10.1006/exmp.1999.2259. [DOI] [PubMed] [Google Scholar]
- 24.Augenlicht LH, et al. Low-level c-myc amplifcation in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 1997;57:1769–1775. [PubMed] [Google Scholar]
- 25.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 26.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jima DD, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifes hundreds of novel microRNAs. Blood. 2010;116:e118–e127. doi: 10.1182/blood-2010-05-285403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ fle format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parmigiani G, et al. Design and analysis issues in genome-wide somatic mutation studies of cancer. Genomics. 2009;93:17–21. doi: 10.1016/j.ygeno.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge D, et al. SVA: software for annotating and visualizing sequenced human genomes. Bioinformatics. 2011;27:1998–2000. doi: 10.1093/bioinformatics/btr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Henikoff S, Ng PC. the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 37.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 38.Pruitt KD, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S2
S3
S4
S5
S6
S7
supplement