Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma (original) (raw)
. Author manuscript; available in PMC: 2014 Nov 1.
Published in final edited form as: Nat Genet. 2014 Apr 6;46(5):457–461. doi: 10.1038/ng.2925
Diffuse intrinsic pontine glioma (DIPG) are highly infiltrative malignant glial neoplasms of the ventral pons, which due to their location within the brain, make them unsuitable for surgical resection and consequently have a universally dismal clinical outcome. The median survival is 9-12 months, with neither chemotherapeutic nor targeted agents showing any substantial survival benefit in clinical trials in children with these tumours1. We report the identification of recurrent activating mutations in the ACVR1 gene, which encodes a type I activin receptor serine/threonine kinase, in 21% of DIPG samples. Strikingly, these somatic mutations (R206H, R258G, G328E/V/W, G356D) have not been reported previously in cancer, but are identical to those found in the germline of patients with the congenital childhood developmental disorder fibrodysplasia ossificans progressiva (FOP)2, and have been shown to constitutively activate the BMP/TGF-β signalling pathway. These mutations represent novel targets for therapeutic intervention in this otherwise incurable disease.
Recent high-throughput sequencing approaches have revealed a striking prevalence of K27M mutations in the genes encoding the histone variants H3.3 (H3F3A) or H3.1 (HIST1H3B) in the childhood brain tumour DIPG3. This K-to-M substitution confers a trans-dominant ablation of global H3K27 trimethylation, which likely profoundly alters gene expression through de-repression of polycomb repressive complex 2 (PRC2) target genes4. Despite these advances in our understanding of the distinct biology of these tumours1, approaches for desperately-needed specific novel therapeutic interventions are not clear, and little has been reported of the additional mutations accompanying these changes.
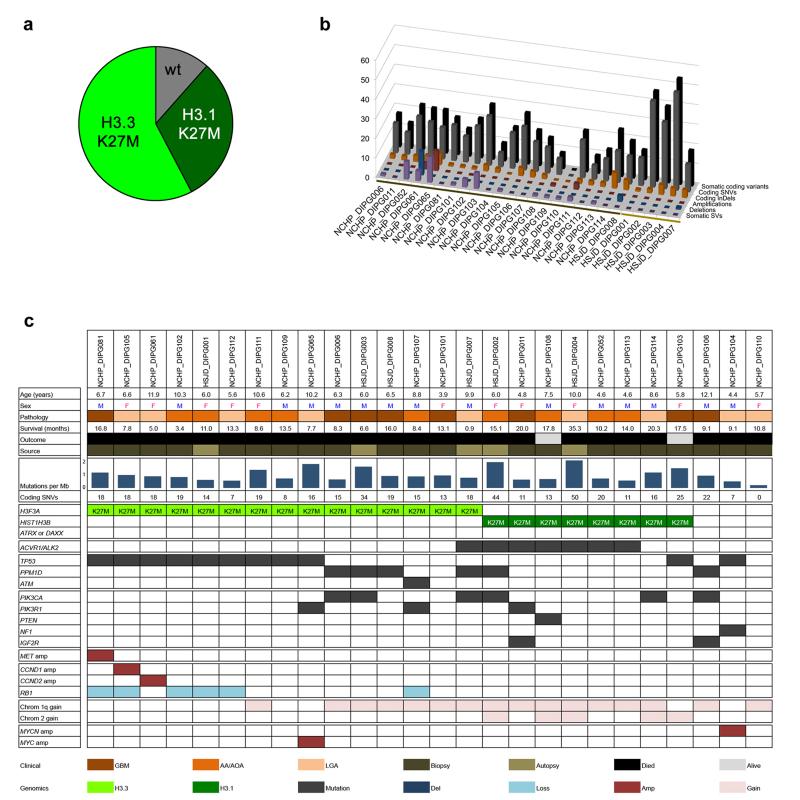
We carried out whole genome sequencing (WGS) on a unique series of 20 pre-treatment biopsy samples of DIPG, for which the patients underwent a safe stereotactic procedure5, and whole exome sequencing (WES) on a further biopsy case as well as five samples obtained at autopsy (Supplementary Table 1). Histone H3 K27M mutations were observed in 23/26 (88%) cases, comprising 15/26 (58%) H3F3A and 8/26 (31%) HIST1H3B (Figure 1a). These were not found in concert with mutations in the chaperones ATRX/DAXX as has been described for supratentorial paediatric glioblastoma (pGBM)6. There was also an absence of other known glioma-related molecular abnormalities such as IDH1/2, BRAF, FGFR1 mutations and gene fusions. The mutational spectrum of the untreated biopsy cases was not significantly different from the autopsies (Figure 1b), although the treatment-naïve samples had a low overall mutation rate, with a mean of 14.8 somatic single nucleotide variants (SNVs) per sample (range 0-25), significantly lower than observed in the radiation-treated autopsy cases (mean=32.0, range 14-50, p=0.004, t-test). There was a similarly significantly lower overall mutation rate in untreated samples taken at biopsy compared with autopsy cases (mean=0.76 vs 1.2 mutations per Mb, p=0.023, t-test).
Figure 1. The genomic landscape of DIPG.
(a) Pie chart showing breakdown of histone H3 mutations in our series of 26 DIPG samples (H3F3A K27M – 15/26, 58%; HIST1H3B K27M – 8/26, 31%, wild-type 3/26, 11%). (b) The mutational spectrum of DIPG. Barchart showing total somatic coding variants (black), coding SNVs (grey) and InDels (orange), amplifications (red), deletions (blue) and SVs (purple) for each DIPG case. Number of events are plotted along the z axis. Biopsy cases are marked by the dark brown bar, autopsy cases by light brown. (c) Summary of major alterations found. Clinicopathological information of the 26 DIPG samples are provided along with mutation rate and number of somatic coding SNVs. Mutations, amplifications and deletions are noted for the histone H3 genes and ATRX/DAXX; ACVR1; ATM/TP53/PPM1D axis; members of the PI3K/MAPK signalling pathways; receptor tyrosine kinases; members of RB pathway, chromosome 1q and 2 single copy gains, and amplification of MYC/MYCN.
11/26 (42%) DIPGs harboured somatic TP53 mutations, with a further six cases (23%) shown to have SNVs in PPM1D, regulator of p38 mitogen-activated protein kinase (p38-MAPK)-p53 signalling in response to cellular stress, and an additional case with a somatic ATM mutation (Supplementary Figure 1), revealing non-overlapping targeting of a DNA damage response pathway in 18/26 (69%) DIPG (Supplementary Figure 2). We further identified non-overlapping recurrent alterations in the PI3-kinase pathway targeting PIK3CA, PIK3R1 and PTEN through SNVs and microdeletion (Supplementary Figure 3), in addition to amplification of MET (1/26, 4%) as previously described7,8, and truncating mutation of NF1 (1/26, 4%) (Figure 1c). We also identified novel recurrent somatic mutations in IGF2R (2/26, 8%), although these mutations are concurrent with others in the pathway, so their significance is unknown. In total, 12/26 (46%) DIPG cases harboured some form of alteration predicted to activate the RTK/PI3K/MAPK pathways (Supplementary Figure 4).
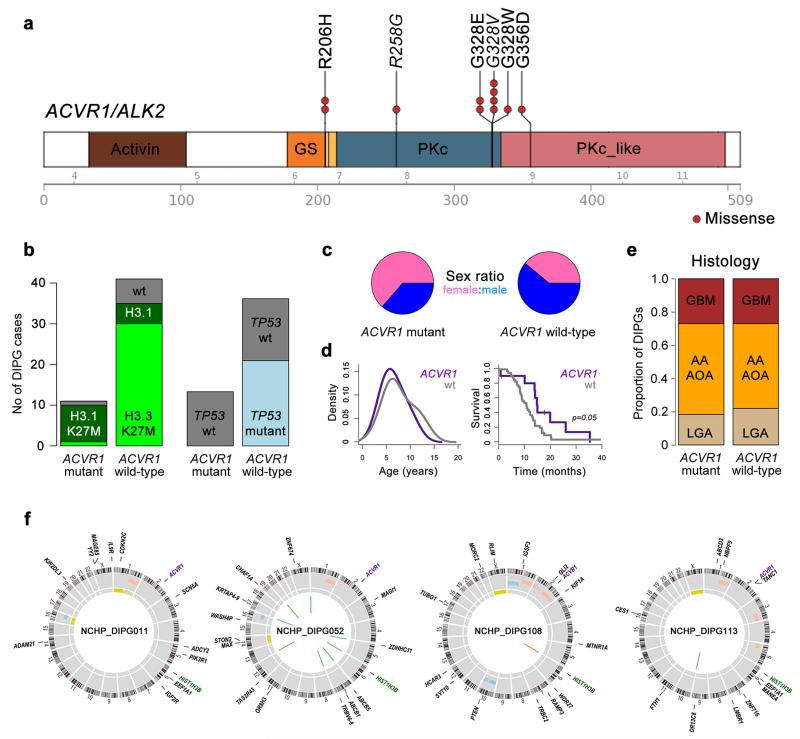
Heterozygous somatic coding mutations in the gene ACVR1, which encodes the activin A type I receptor ALK2, were observed in 7/26 (27%) cases (Figure 1c). These were restricted to the specific codons 328 (c.983G>T, p.G328V, two cases; c.983G>A, p.G328E, two cases), 258 (c.772C>T, p.R258G, one case), and 356 (c.1067G>A p.G356D), all within the serine/threonine kinase domain; and 206 (c.617G>A, p.R206H, one case), within the glycine-serine (GS)-rich domain. Screening an extended series of 26 DIPG biopsy samples by Sanger sequencing identified further recurrences of these mutations, and an additional variant at position 328 (c.982G>T, p.G328W) (Supplementary Figure 5). Overall, we identified 11/52 (21%) DIPG samples to harbour mutation in ACVR1 at four different codons (Figure 2a). These mutations appear highly specific to DIPG. SNVs in the ACVR1 coding region are present in the Catalogue of Somatic Mutations in Cancer (COSMIC9) database at an overall frequency of 20/5965 (0.3%), with no individual tumour type harbouring more than 2% frequency, and no mutations observed at any of the residues described in the present study, suggestive of a ‘passenger’ effect in other cancers.
Figure 2. Recurrent ACVR1 mutations in DIPG.
(a) Cartoon showing recurrent missense mutations in ACVR1, overlaid with functional protein domains and exon boundaries. In total, 11/52 (21%) of DIPG harboured somatic mutations at four residues, all of which have been previously described in the germline of patients with fibrodysplasia ossificans progressiva. The specific base changes which may be unique to DIPG are highlighted in italics. Activin: activin types I and II receptor domain; GS: TGF-β glycine-serine rich domain; PKc: protein kinase catalytic domain; PKc_like: protein kinase catalytic domain-like. (b) Bar graphs showing segregation of activating mutations in ACVR1 with HIST1H3B K27M mutations (p<0.0001, Fishers exact test) and wild-type TP53 (p=0.0103, Fishers exact test) in our extended series of 52 DIPG patients. (c) Sex distribution of patients with ACVR1 mutations, showing a strong predominance of females in mutant samples. (d) Age distribution (left) and overall survival (right) of DIPG patients with ACVR1 mutations (purple), compared with wild-type (grey). (e) Barplot representing histological breakdown of ACVR1 mutant and wild-type samples. GBM: glioblastoma multiforme; AA: anaplastic astrocytoma; AOA: anaplastic oligoastrocytoma; LGA: low grade astrocytoma. Brown=WHO grade 4, orange=grade 3, tan=grade 2. (f) Circos plots representing the whole genome sequences of the four cases of concurrent ACVR1 mutant / HIST1H3B K27M mutant / TP53 wild-type DIPGs. Outer ring contains chromosomal ideograms, annotated for somatic SNVs in coding genes. Inner ring plots copy number derived from coverage data, dark red=amplification, pink=gain, dark blue=deletion, light blue=loss. Innermost ring represents loss of heterozygosity (LOH, yellow). Inside the circle are drawn SVs, red=interchromosomal translocations, blue=intrachromosomal translocations, orange=deletion, purple=inversion.
ACVR1 mutations were found to co-segregate with the less common HIST1H3B K27M mutation in the canonical histone H3.1 variant (p<0.0001, Fishers exact test) (Figure 2b), as well as wild-type TP53 (p=0.0103, Fishers exact test). There was also an association between H3.1 mutation and chromosome 2 gain (on which ACVR1 is found at 2q24.1, p=0.0009, Fishers exact test). ACVR1 mutations appear to mark a distinct subset of DIPG patients (Supplementary Table 2). There was a marked predominance of females in the ACVR1 mutant tumour group (1.75:1 vs 0.64:1, p=0.05, Fishers exact test) (Figure 2c), as well as a relatively restricted age of onset (Figure 2d), compared to wild-type. Patients whose tumours harboured ACVR1 mutations also had a longer overall survival (median=14.9 months vs 10.9 months) p=0.05, log-rank test) (Figure 2d), although outcome remained very poor. There were no significant differences in histology between the groups (Figure 2e). WGS biopsy samples exemplifying this genotype with concurrent ACVR1 and HIST1H3B mutations harboured an additional 10-19 somatic SNVs, and 0-9 SVs respectively (Figure 2f).
Remarkably, these somatic mutations in ACVR1 are at identical residues to those described in the germline of patients with autosomal dominant congenital childhood developmental disorder fibrodysplasia ossificans progressiva (FOP, OMIM:135100)2. This debilitating disease is characterised by heterotopic ossification of soft connective tissue resulting in severe skeletal abnormalities10. Patients with classical clinical features of FOP carry heterozygous R206H mutations in the glycine and serine residue (GS) activation domain11, whilst atypical patients with a less severe phenotype have been shown to harbour either R258S12, G328E/R/W13, G356D14, or other heterozygous mutations in the GS and kinase domains2,15. This latter series of mutations may be exposed at the interface with the GS domain and abrogate interactions with the negative regulator FKBP1212,13,15. These mutations have been shown to constitutively activate the bone morphogenic protein (BMP)-dependent transforming growth factor (TGF)-β pathway in the absence of ligand binding, as evidenced by increased phosphorylation of Smad1/5/8 in vitro14,16.
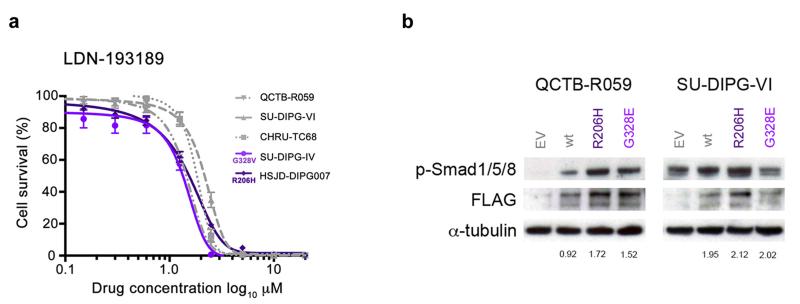
To investigate the specific role of ACVR1 mutations in the context of DIPG, we assembled a panel of four DIPG patient-derived primary cultures (and one thalamic paediatric GBM culture harbouring an H3F3A K27M mutation), representing two ACVR1 mutations (R206H and G328V) and three wild-type lines (Supplementary Table 3). RNAseq data demonstrated in these models that the mutant allele was expressed in approximately half the reads, also evidenced by Sanger sequencing of cDNA from patient sample NCHP_DIPG011 (Supplementary Figure 6). Treatment with the selective ALK2 inhibitor LDN-19318917 resulted in marked inhibition of cell viability in all cells, with GI50 values ranging from 0.86 – 2.1 μM, approximately 10-fold lower than the less potent parent compound dorsomorphin, with a trend towards increased sensitivity in the mutant cultures (p=0.10, F-test) (Figure 3a). Transfection of ACVR1 wild-type thalamic GBM and DIPG cells (both H3F3A K27M) with FLAG-tagged mutations conferred an increased signalling through phospho-Smad 1/5/8, particularly for R206H, and to a lesser extent for G328E (Figure 3b). ACVR1 mutation may only be one mechanism by which this pathway is activated in DIPG, however, as high basal levels of phospho-Smad 1/5/8 were also observed for the H3F3A K27M mutant, ACVR1 wild-type cells used in this study (Supplementary Figure 7). This may explain the lack of a more robust genotype-dependent response to the inhibitor, and also expand upon the population of patients which may benefit from targeting the receptor.
Figure 3. ACVR1 mutations are weakly activating and responsive to targeted inhibition.
(a) In vitro cytotoxicity of the ALK2 inhibitor LDN-193189. Primary cultures were treated with inhibitor for 72 hours and cell viability measured by CellTiter Glo. The cells used were HSJD-DIPG007 (DIPG, ACVR1 R206H, H3F3A K27M), SUDIPG-IV (DIPG, ACVR1 G328V, HIST1H3B K27M), CHRU-TC68 (DIPG, ACVR1 wt, H3F3A K27M), SU-DIPG-VI (DIPG, ACVR1 wt, H3F3A K27M), QCTB-R059 (thalamic paediatric GBM, ACVR1 wt, H3F3A K27M). (b) ACVR1 mutations confer increased signalling through phospho-Smad 1/5/8. QCTB-R059 and SU-DIPG-VI cells were transfected with FLAG-tagged ACVR1 R206H and G328E mutations, and assessed for phospho-Smad 1/5/8 by Western blot. EV: empty vector; wt: wild-type ACVR1. α-tubulin is included as a loading control. Figures are given for phospho-Smad 1/5/8 levels quantitated relative to FLAG expression.
There are no reports to our knowledge of coincident FOP and DIPG, although the clinical features of both typical and atypical cases of FOP can commonly include neurological symptoms and have been reported in children to include cerebellar and brain stem abnormalities15,18, including demyelinated lesions in the pons both of patients and mouse models19. It will nonetheless be a challenge to identify the mechanism by which the temporal and spatial context of BMP/TGF-β pathway activation confer such differing clinical phenotypes. In experimental models of FOP, ACVR1 mutations are associated with defects in stem cell maintenance, reprogramming and differentiation, offering links with cancer-related cellular processes. First generation ALK2 inhibitors such as dorsomorphin20 and LDN-19318917 have been shown to downregulate intracellular BMP/TGF-β signalling and reduce heterotypic ossification, opening the tantalising possibility of CNS-penetrant compounds showing a similar potential in a childhood brain tumour otherwise devoid of efficacious treatment options.
ONLINE METHODS
Tumour cohort
DIPG samples and matched peripheral blood were available from 21 patients who underwent a stereotactic biopsy at the Neurosurgery Department of Necker Sick Children’s Hospital in Paris, France, 20 of whom were subjected to whole genome sequencing. All patients were clinically diagnosed as diffuse intrinsic pontine glioma based on clinical presentation and radiography as part of a multidisciplinary assessment. These patients had diffuse intrinsic tumour centred to the pons and occupying at least 50% of the volume of this structure, and an associated short clinical history of less than 3 months. DNA from an additional 26 biopsy samples were available as a validation cohort. A further five DIPG cases with matched peripheral blood were obtained at autopsy at the Hospital Sant Joan de Déu, Barcelona, Spain, and were sequenced after exome capture using Agilent SureSelect. All patient material was collected after informed consent and subject to local research ethics committee approval. There were 23 girls and 29 boys (1:1.26 ratio). The median age of the patients was 6.6 years and the median overall survival was 11.6 months. A summary of the tumour cohort and clinicopathological information is provided in Supplementary Table 2.
Whole genome / exome sequencing
Exome capture was carried out on the four autopsy cases using the 50Mb Agilent SureSelect platform (Agilent, Santa Clara, CA, USA), and paired-end-sequenced on an Illumina HiSeq2000 (Illumina, San Diego, CA, USA) with a 100bp read length. Library preparation for the biopsy samples was carried out by the Illumina FastTrack service, and the entire genomes paired-end-sequenced on an Illumina HiSeq2000. The median coverage for the tumour genomes was 37-67× (matched normal genomes 34-41×). Reads were mapped to the hg19 build of the human genome using bwa (bio-bwa.sourceforge.net), and PCR duplicates removed with PicardTools 1.5 (picard.sourceforge.net).
Genome analysis
Somatic single nucleotide variants were called using the Illumina Genome Network (IGN) Cancer Normal pipeline version 1.0.2 and the Genome Analysis Tool Kit v2.4-9 (www.broadinstitute.org/gatk/). Structural variants were called using IGN and SV detect (svdetect.sourceforge.net). Variants were annotated using the Ensembl Variant Effect Predictor v71 (www.ensembl.org/info/docs/variation) incorporating SIFT (sift.jcvi.org) and PolyPhen (genetics.bwh.harvard.edu/pph2) predictions, COSMIC v64 (www.sanger.ac.uk/genetics/CGP/cosmic/) and dbSNP build 137 (www.ncbi.nlm.nih.gov/sites/SNP) annotations. Copy number was obtained by calculating log2 ratios of tumour/normal coverage binned into exons of known genes, smoothed using circular binary segmentation (www.bioconductor.org) and processed using in-house scripts. Loss of heterozygosity (LOH) was calculated using APOLLOH (compbio.bccrc.ca/software/apolloh/). Cartoons showing locations of recurrent mutations were produced by the St Jude Washington University Protein Paint tool (http://www.explorepcgp.org). Statistical analysis was carried out using R3.0.0 (www.r-project.org). Continuous variables were analysed using Student’s t-test.
Count data was compared using a Fisher’s exact test.
Cell culture and drug sensitivity
Primary cultures were derived from DIPG patient samples taken at either biopsy or autopsy at multiple centres, representing both ACVR1 mutant and wild-type, and both H3F3A and HIST1H3B K27M, in addition to cells from a paediatric glioblastoma specimen arising in the thalamus with an H3F3A K27M mutation. A summary of the Cells were grown under adherent stem cell conditions using laminin (Sigma, Poole, UK)-coated flasks in neurobasal medium (Invitrogen, Paisley, UK) supplemented with B-27 (Invitrogen) and growth factors EGF, b-FGF, PDGF-AA and PDGF-BB (all Shenandoah Biotech, Warwick, PA, USA). The ALK2 inhibitors LDN-193189 (Sigma) and dorsomorphin (Abcam, Cambridge, UK) were tested for effects on cell viability in the cells using a highly sensitive luminescent assay measuring cellular ATP levels (CellTiter-Glo™; Promega, Madison, WI, USA). Drug was added in various concentrations and the cells assayed in triplicate after 72 hours. Statistical analysis was carried out using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA).
Allelic expression of ACVR1
SU-DIPG-IV cells were subjected to full transcriptome sequencing as part of the DIPG Preclinical Consortium. Counts of reads aligned to the ACVR1 coding region in NCBI_36 were analysed for ratio of mutant sequence to wild-type, and visualised in Genome Browse (Golden Helix, Bozeman, MT, USA). NCHP_DIPG011 primary tumour RNA was reverse-transcribed, PCR-amplified, and Sanger sequenced to determine if both mutant and wild-type alleles were expressed (Supplementary Table 4).
Overexpression of mutant ACVR1
ACVR1 mutations R206H and G328E were cloned into pcDNA3.1 by site-directed mutagenesis as previously described16 and transfected into primary cells QCTBR059 and SU-DIPG-VI using lipofectamine (Invitrogen), with protein collected after 24 hours using standard procedures. Western blots were carried out for anti-FLAG HRP (#A8592, Sigma; 1:1000 dilution) and phosphorylated Smad1/5/8 (#9511, Cell Signalling; 1:1000) under standard conditions. Relative levels of phosphorylated Smad1/5/8 were measured by Image J software (National Institute of Mental Health, Bethesda, MD, USA).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA) and R 3.0.1 (www.r-project.org). Comparison between number of coding SNVs and mutation rate in biopsy and autopsy cases was performed by t-test. For analysis of categorical association between patients with ACVR1 mutations and mutations in HIST1H3B or TP53, sex and histology, Fishers exact test was used. Differences in survival were analysed by the Kaplan-Meir method and significance determined by the log-rank test. All tests were two-sided and a p value of less than 0.05 was considered significant. A sum-of-squares F test was used to assess differences in dose-response curves for ACVR1 mutant cells versus wild-type.
Supplementary Material
Supplementary Figures 1-7 and Supplementary Tables 2-4
Supplementary Table 1
ACKNOWLEDGEMENTS
This study was funded by the Cancer Research UK Genomics Initiative (A14078), and makes use of data generated by the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. We are grateful to the DIPG Preclinical Consortium funded by The Cure Starts Now and The Lyla Nsouli Foundation for RNAseq data. This work is supported by The Stavros Niarchos Foundation, Abbie’s Army, The Lyla Nsouli Foundation, the Royal Marsden Hospital Childrens Department Fund, and Fondo Alicia Pueyo. MM gratefully acknowledges funding by National Institutes of Neurological Disease and Stroke (NINDS grant K08NS070926), Alex’s Lemonade Stand Foundation, McKenna Claire Foundation and the Dylan Jewett Memorial Fund. CP acknowledges funding from the Agence National pour la Recherche. NT, CP and JG acknowledge funding from the charity l’Etoile de Martin, NE-W acknowledges support from Enfants et Santé. AMC acknowledges funding from the Fundación Cientifica de la aecc. WJI acknowledges funding from Children’s Health Foundation Queensland and the Brainchild Foundation. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation, the Canadian Institutes for Health Research, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust [092809/Z/10/Z]. KRT, AM, MV, DC, DH and CJ acknowledge NHS funding to the National Institute of Health Research Biomedical Research Centres.
Footnotes
ACCESSION NUMBERS: Raw data has been submitted to the European Genome-phenome Archive (www.ebi.ac.uk/ega/), accession numbers EGAS00001000524 (whole genome sequencing) and EGAS00001000572 (exomes).
REFERENCES
- 1.Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol. 2012;9:400–13. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 2.Katagiri T. Recent topics in fibrodysplasia ossificans progressiva. J Oral Biosciences. 2012;54:119–123. [Google Scholar]
- 3.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis PW, et al. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013 doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roujeau T, et al. Stereotactic biopsy of diffuse pontine lesions in children. J Neurosurg. 2007;107:1–4. doi: 10.3171/PED-07/07/001. [DOI] [PubMed] [Google Scholar]
- 6.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 7.Puget S, et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7:e30313. doi: 10.1371/journal.pone.0030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paugh BS, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shore EM, Kaplan FS. Inherited human diseases of heterotopic bone formation. Nat Rev Rheumatol. 2010;6:518–27. doi: 10.1038/nrrheum.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shore EM, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38:525–7. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 12.Bocciardi R, Bordo D, Di Duca M, Di Rocco M, Ravazzolo R. Mutational analysis of the ACVR1 gene in Italian patients affected with fibrodysplasia ossificans progressiva: confirmations and advancements. Eur J Hum Genet. 2009;17:311–8. doi: 10.1038/ejhg.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrie KA, et al. Novel mutations in ACVR1 result in atypical features in two fibrodysplasia ossificans progressiva patients. PLoS One. 2009;4:e5005. doi: 10.1371/journal.pone.0005005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda T, et al. A unique mutation of ALK2, G356D, found in a patient with fibrodysplasia ossificans progressiva is a moderately activated BMP type I receptor. Biochem Biophys Res Commun. 2008;377:905–9. doi: 10.1016/j.bbrc.2008.10.093. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan FS, et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum Mutat. 2009;30:379–90. doi: 10.1002/humu.20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaikuad A, et al. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J Biol Chem. 2012;287:36990–8. doi: 10.1074/jbc.M112.365932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu PB, et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat Med. 2008;14:1363–9. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tumolo M, Moscatelli A, Silvestri G. Anaesthetic management of a child with fibrodysplasia ossificans progressiva. Br J Anaesth. 2006;97:701–3. doi: 10.1093/bja/ael251. [DOI] [PubMed] [Google Scholar]
- 19.Kan L, et al. CNS demyelination in fibrodysplasia ossificans progressiva. J Neurol. 2012;259:2644–55. doi: 10.1007/s00415-012-6563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu PB, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-7 and Supplementary Tables 2-4
Supplementary Table 1