Displacements of Prohead Protease Genes in the Late Operons of Double-Stranded-DNA Bacteriophages (original) (raw)
Abstract
Most of the known prohead maturation proteases in double-stranded-DNA bacteriophages are shown, by computational methods, to fall into two evolutionarily independent clans of serine proteases, herpesvirus assemblin-like and ClpP-like. Phylogenetic analysis suggests that these two types of phage prohead protease genes displaced each other multiple times while preserving their exact location within the late operons of the phage genomes.
Capsid maturation in double-stranded-DNA (dsDNA) bacteriophages requires proteolytic cleavage by a prohead protease. The MEROPS database (27; http://merops.sanger.ac.uk) currently places phage prohead proteases into four families, two of which exclusively consist of phage and prophage prohead proteases with uncharacterized catalytic mechanisms. They are family U9 (prohead proteases from T4-like Myoviridae) and family U35 (prohead proteases from HK97 and related phages of the Siphoviridae family). The other two, serine protease families S14 and S49, are typified by ClpP protease and Escherichia coli peptidase IV, respectively, and contain phage prohead proteases such as gpC protein of phage lambda and prohead protease of Pseudomonas aeruginosa phage D3 (both phages are members of the Siphoviridae).
Prohead protease is an essential factor in phage capsid morphogenesis, as first demonstrated for phage T4 (2). The maturational cleavage of the capsid protein precursors confers stability to the nucleic acid-free prohead and triggers a conformational switch of prohead, which allows phage genomic DNA to enter and initiate the DNA packaging process. A similar capsid maturation mechanism has been reported for various dsDNA phages from the Siphoviridae family, such as HK97 (8), PVL (7), and D3 (12), suggesting that the proteolytic cleavage of structural proteins during phage capsid assembly is common in many dsDNA phages and that the prohead protease is the key enzyme in the process. The prohead protease is generally encoded next to the gene for the capsid protein that it cleaves and may be fused to it in some cases (e.g., Siphoviridae psiM2 and psiM100). Although the capsid assembly and maturation processes in phages HK97, from the lambda-like Siphoviridae group, and T4, from the Myoviridae family, are tightly regulated and share many common features, no sequence similarity or evolutionary connection between their prohead proteases has been reported. In this study, we use computational analysis to show that virtually all known prohead proteases from dsDNA phages belong to one of the two superfamilies of serine proteases and are unrelated to each other but apparently functionally interchangeable.
To examine the evolutionary relationship between the prohead proteases from phages T4 and HK97, we searched the NCBI nonredundant protein database (7 November 2003; 1,537,641 sequences; 502,645,420 total letters) with the PSI-BLAST program (1) using members of the U9 and U35 families as queries, with the threshold for inclusion set at a BLAST E value of 0.02. Starting with the prohead protease from HK97 (gi 9634157; family U35) as a query, we collected the prohead proteases from HK97-related Siphoviridae and observed statistically significant matches to proteases from other phages. In the course of these iterative searches, members of the family U35 were found to share significant sequence similarity with herpesvirus proteases and UL26 assemblins from family S21, and they were all remote homologs of prohead proteases from T4-like Myoviridae, members of the family U9. For example, when the prohead protease from Salmonella enterica serovar Typhimurium phage ST64B (Podoviridae; gi 23505450; family U35), was used as a query after phage prohead proteases from family U9 and herpesvirus proteases from family S21 were detected, the prohead core scaffold protein and protease gp21 from phage Aeh1 (gi 33414841; family U9), a T4-like member of the Myoviridae, were found in the ninth iteration, with an E value of 10−17 and without any false positives. In the PSI-BLAST searches that were initiated with herpesvirus proteases, a phage head maturation protease from Novosphingobium aromaticivorans (gi 23107536; COG3740) was always just below the threshold. When that protease was submitted as a query, a UL26 capsid maturation protease from meleagrid herpesvirus 1 was found in the second iteration, with an E value of 10−3.
In Myovirus P2, the internal scaffolding protein gpO is indispensable for the cleavage of capsid protein gpN (29), but it is unknown whether gpO itself has protease activity or if the cleavage reaction is performed by a host protease. During sequence similarity searches, we found that P2 gpO shares significant sequence similarity with phage Mu putative head maturation protease gpI, and both of them are distantly related to herpesvirus assemblin UL26. For instance, when a conserved hypothetical protein from Chromobacterium violaceum ATCC 12472 (gi 34496934) served as a query in a PSI-BLAST search, gpO from P2-like Myoviridae was detected, followed by the putative head maturation protease gpI from Mu-like phages. If we included the UL26 protein from gallid herpesvirus 3 (gi 10834896), whose similarity was just below the threshold in the seventh iteration in the later round of the PSI-BLAST search, we found many UL26 proteases from herpesviruses without any false positives until no new sequences could be retrieved after the 10th iteration.
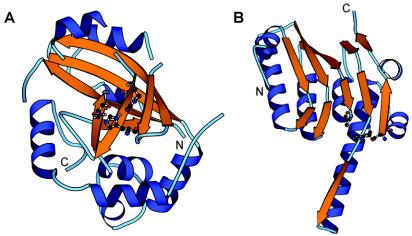
Herpesviruses are enveloped dsDNA viruses that can cause severe health problems in humans and other animals. As in dsDNA bacteriophages, the proteolytic processing of capsid protein precursor by a virus-encoded enzyme is essential for producing infectious virions (17, 20, 37-39), and several structural and mechanistic parallels between the maturation of phage heads and herpesvirus capsids have been noted (14, 31). So far, the UL26 gene products are the only known proteases encoded by herpesvirus genomes, and the three-dimensional structures of UL26 proteases from all three herpesvirus subfamilies have been determined (3, 6, 15, 25, 26, 28, 32, 35) (Fig. 1A). The central core of the enzyme is a β-barrel, consisting of two sheets held together at one edge by a long β-strand (β3) passing between the sheets and forming a triple-stranded corner and at the other edge by two hydrogen bonds between strands β5 and β7. All residues with a direct role in catalysis, including the three members of the catalytic triad and two residues predicted to form the oxyanion hole, are located within this core. The core is surrounded by eight α-helices, four of which play a role in the formation of the homodimer. The similarity between phage proteases and UL26 proteases extends along most of the β-strand's rich central region of the UL26 sequence, except for the middle region of the herpesvirus sequences, which consists of three α-helices that appear to be lacking in phage proteases.
FIG. 1.
Structures of HCMV protease (PDB code 1CMV) and ClpP protease from E. coli (PDB code 1TYF). The α-helices are blue, and the β-strands are orange. The residues in the catalytic triad are shown with balls and sticks. They are His63-Ser132-His157 in HCMV protease and Ser97-His122-Asp171 in E. coli ClpP protease. The structures were drawn by using the program MOLSCRIPT (19).
To further validate the sequence and structure similarities between proteases from phages and herpesviruses, we queried the global network of independent structure prediction servers via the 3D-Jury Meta predictor (13; http://bioinfo.pl/Meta/) with prohead proteases from HK97 and ST64B. Herpesvirus proteases were ranked as the top-scoring matches, with 3D-Jury scores greater than 90, which are typical of true positives (13). We concluded that the three families are probably derived from a common ancestor and propose to group them into a single protease clan, SH.
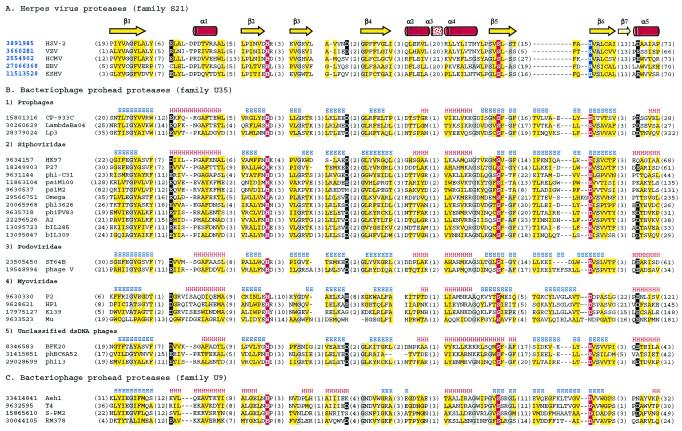
A multiple-sequence alignment of proteases from the newly defined clan SH is shown in Fig. 2. The fully conserved serine residue in the middle of the fifth β-strand suggests that the phage prohead proteases from families U9 and U35 and the internal scaffolding protein gpO from P2-like phages are serine proteases. The second member of the catalytic triad, histidine, located near the C terminus of strand 2, is also universally conserved. In UL26 proteases, a histidine residue (His157 in human cytomegalovirus [HCMV] protease) at the N terminus of strand 6 is predicted to be the third triad member according to its position in the three-dimensional structure (6, 25, 32, 35). This position aligns with a well-conserved acidic residue in all phage proteases, which therefore appear to have a more conventional His-Ser-Asp(Glu) catalytic triad. Kinetic studies of HCMV protease with the His157 residue mutated to alanine (H157A) showed about a 10-fold loss in activity relative to that of the wild-type enzyme, while the H157D and H157E mutants were about three times less active than the wild type (16). It would be interesting to see whether phage prohead proteases from families U9 and U35, which all have an aspartate or glutamate in the third position, display lower activities than their herpesvirus homologs or whether evolutionary changes elsewhere in the molecule have compensated for that.
FIG. 2.
Multiple-sequence alignment of protease clan SH members. The left column is the gi number for each sequence followed by the name of the virus or phage. The identifiers of herpesvirus proteases with known three-dimensional structures are shown in bold blue font. Distances, in amino acid residues, from the ends of each sequence and between the blocks with highest sequence similarities are shown in parentheses. Consensus positions of the structural elements are shown above the alignment. Yellow shading indicates the conservation of hydrophobic residues, gray shading indicates the conservation of residues with small side chains (A, G, and S), and a white font on a black background indicates the conservation of negatively charged residues (D and E). The catalytic Ser, His, and Asp/Glu residues in the catalytic triad are in white font on a red background, except that the His in the third position of the triad in all herpesvirus proteases is on a blue background. (A) Representatives from family S21 with known three-dimensional structures. HSV-2, herpes simplex virus 2; VZV, varicella-zoster virus; EBV, Epstein-Barr virus; KSHV, Kaposi's sarcoma-associated herpesvirus. (B) Representatives from family U35. (C) Representatives from family U9.
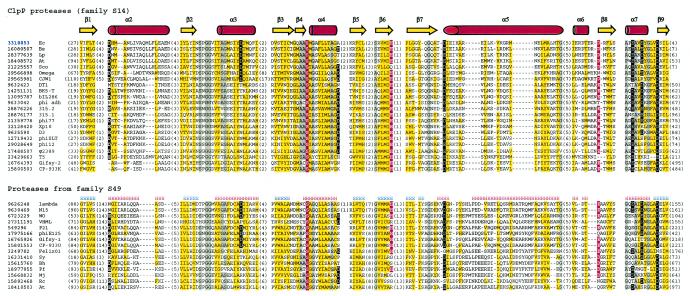
Two other recognized families of phage prohead proteases are families S14 and S49, which include proteases widely distributed in bacteria, archaea, and eukarya. The founding members of the S14 and S49 families are, respectively, ClpP protease and E. coli peptidase IV. Using lambda prohead protease gpC protein (gi 9626248) as a query and after detecting many S49 family proteases, we also retrieved ClpP proteases at the second iteration. Statistically significant matches to many members of the large crotonase superfamily were detected in later iterations, which is compatible with the observation of a distant structural similarity between ClpP protease and crotonases (11, 22). The computational fold recognition experiments further validated the sequence and structure similarities between proteases from families S14 and S49. As expected, the best match from the 3D-Jury consensus prediction (13) for lambda gpC protein is ClpP protease (PDB code 1TYF), with a score of 126.33. With an α/β fold composed of six repeats of the β-β-α unit (Fig. 1B) (36), ClpP is distinct from the herpesvirus protease fold. An analysis of a multiple-sequence alignment (Fig. 3) indicates that most of the enzyme catalytic core is well conserved among proteases from families S14 and S49; therefore, proteases from the two families are expected to adopt a ClpP/crotonase fold and constitute clan SK.
FIG. 3.
Multiple-sequence alignment of protease clan SK members. Designations are as described in the legend to Fig. 2. Species abbreviations: Ec, E. coli; Bs, Bacillus subtilis; Lp, Lactobacillus plantarum; At, Arabidopsis thaliana; Sco, Streptomyces coelicolor A3(2); Sy, Synechocystis sp. strain PCC 6803; Bh, Bacillus halodurans; Pf, Pyrococcus furiosus DSM 3638; Mj, Methanococcus jannaschii; Rc, Rickettsia conorii.
Herpesvirus-like proteases and ClpP-like proteases represent two distinct serine protease folds that are most likely evolutionarily independent but appear to play the same essential function in the phage life cycle. The only phage prohead protease with experimentally demonstrated activity that does not seem to belong to these two classes and lacks similarity to any other proteins is the product of orf13 in pneumococcal phage Cp-1 (21). While ClpP-like proteases are found in both prokaryotic and eukaryotic organisms, a striking feature of clan SH is that its members are found only in the dsDNA viruses of bacteria and eukaryotes. Sensitive probabilistic searches of sequence databases, such as with the HMMER package (9) and Wise2 package (http://www.ebi.ac.uk/Wise2/), as well as searches of the databases of unfinished prokaryotic and eukaryotic genomes and expressed sequence tags, failed to reveal clan SH members in any other viruses or cellular organisms. All significant matches to bacterial genomes turned out to be in the recently integrated prophages, which typically possess the full complement of structural components of the virions (data not shown).
Mechanistic similarities between the maturation of phage heads and herpesvirus capsids are quite remarkable. Both procapsids are assembled on an asymmetric α-helical scaffold, which is cut into fragments by viral protease and leaves behind an empty head shell. In both cases, replicated virus genome is packaged into this shell until the associated virus-encoded terminase enzyme cleaves genomic DNA. Concomitant with DNA packaging, the capsid changes from nearly spherical to a much more stable icosahedral form. These parallel relationships remained an evolutionary puzzle without direct evidence for sequence relationships between individual components. It has been noted, however, that the orders of structural components and factors of their assembly in virus genomes were similar and that large subunits of phage and herpesvirus terminases are related ATP-hydrolyzing enzymes (5, 14, 31). Statistically significant sequence similarity between the SH-type proteases from phages and herpesviruses strengthens the argument that the complement of genes encoding capsid components and maturation factors in bacteriophages and herpesviruses has evolved from a common ancestor.
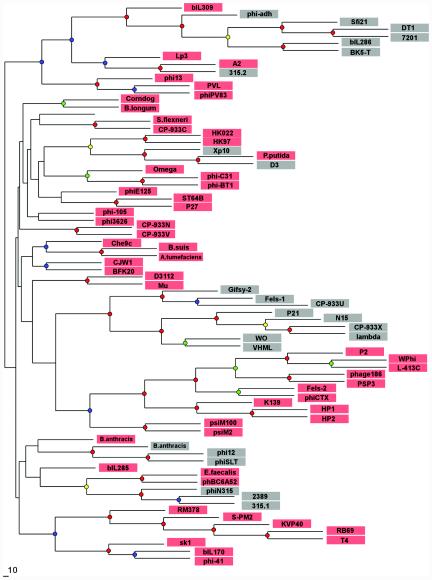
In phages, the prohead protease gene is always located between genes that encode the portal protein and the major capsid protein. Since these three gene products interact intimately with each other during the phage capsid assembly and maturation process, the tandem arrangement appears to be under a selective constraint. Strikingly, however, the type of the maturation protease is not correlated with the extent of sequence similarity between the neighboring structural proteins. One such example is presented by phages Xp10, D3, and Pseudomonas putida prophage; their genes coding for the portal protein and the major capsid protein are closely related and must have diverged very recently (Fig. 4 and data not shown), yet in phages Xp10 and D3, the protease belongs to clan SK, and the gene in exactly the same location in P. putida prophage codes for a clan SH member (40).
FIG. 4.
Phylogenetic tree constructed by using the sequence alignment of phage portal proteins. Sequences of the portal proteins from phages and prophages were aligned by using the T-Coffee (23) and CLUSTAL_X (34) programs, followed by manual validation and the removal of poorly aligned regions. The phylogenetic tree was constructed by using the neighbor-joining method as implemented in the NEIGHBOR program, and subsets of the data were used to rebuild the tree multiple times. The root was set to midpoint by the RETREE program of the PHYLIP package (10), and the consensus tree was subsequently reviewed by TreeView (24). Bootstrap values were estimated by resampling the set of the alignment 100 times (see the supplemental material). The level of bootstrap support is marked by small circles in the following colors: red (90 to 100%), yellow (80 to 90%), green (70 to 80%), and blue (50 to 70%). The nodes with <40% support are unlabeled. Phages that encode prohead protease belonging to clan SH are shaded in pink, and those that encode prohead protease belonging to clan SK are shaded in gray.
A prohead protease gene can thus be replaced in evolution by a functionally equivalent gene coding for a structurally unrelated protease. Such nonorthologous gene displacement is thought to be common in the genomes of cellular life forms (18) and has been noted to occur in some other phage genes, such as integrase (33) and lysozyme (4). In order to trace the history of the mutual displacement of prohead proteases, we built a phylogenetic tree of dsDNA phages based on the similarity of their portal proteins and labeled the leaves of the resulting consensus tree by the type of prohead protease observed in each lineage. Analysis of the resulting tree (Fig. 4) indicates that the SK-type protease may be a derived characteristic, with the possessors of this protease typically being nested within a group of phages with the SH-type protease. All of the deepest branches of the tree, or all clades but one with bootstrap support of more than 50%, are inferred to have the SH-type protease at the root (Fig. 4 and data not shown). Thus, the SH-type protease may be evolutionarily ancient, predating the common ancestor of present-day DNA phages, and several in situ gene replacements by the ClpP-like protease may have occurred in different evolutionary lineages. Interestingly, at lease in one case, such SH-to-SK displacement appears to have been followed by a reverse displacement, from SK to SH, in the P. putida prophage (Fig. 4).
The phage phylogeny shown in Fig. 4 is inferred on the basis of a single gene, albeit one that has strong functional links to prohead protease. We also examined the distribution of two classes of portal proteases in a phylogenetic tree built on the basis of a whole-genome approach (30). There are topological differences between that phage proteomic tree and our single-gene tree, but remarkably, the pattern of incongruity between the protease type and the whole-genome phylogeny still holds, with phages with the SK-type protease typically nested within a group of phages with the SH-type protease (details are available from the authors upon request).
To conclude, the proteolytic cleavage of the scaffold during the assembly of capsids in dsDNA bacteriophages appears to be a virus-specific function provided by the proteolytic enzymes from at least two distinct families and folds. One of these folds, which is perhaps evolutionarily more ancient, is shared with the functionally very similar protease of herpesviruses. Phylogenetic analysis suggests that the two types of head maturation proteases in dsDNA bacteriophages have displaced each other on multiple occasions.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Galina Glazko and Helen Piontkivska for assistance with the phylogenetic analysis and two anonymous reviewers for many suggestions for improving the manuscript.
Footnotes
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, L. W., and M. K. Showe. 1983. Morphogenesis of the T4 head, p. 219-245. In C. K. Mathews, E. M. Kutter, G. Mosig, and P. B. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 3.Buisson, M., J. F. Hernandez, D. Lascoux, G. Schoehn, E. Forest, G. Arlaud, J. M. Seigneurin, R. W. Ruigrok, and W. P. Burmeister. 2002. The crystal structure of the Epstein-Barr virus protease shows rearrangement of the processed C terminus. J. Mol. Biol. 324**:**89-103. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of the dsDNA tailed-bacteriophage genomes. Semin. Virol. 3**:**383-397. [Google Scholar]
- 5.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57**:**128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, P., H. Tsuge, R. J. Almassy, C. L. Gribskov, S. Katoh, D. L. Vanderpool, S. A. Margosiak, C. Pinko, D. A. Matthews, and C. C. Kan. 1996. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86**:**835-843. [DOI] [PubMed] [Google Scholar]
- 7.Desiere, F., S. Lucchini, and H. Brussow. 1999. Comparative sequence analysis of the DNA packaging, head, and tail morphogenesis modules in the temperate cos-site Streptococcus thermophilus bacteriophage Sfi21. Virology 260**:**244-253. [DOI] [PubMed] [Google Scholar]
- 8.Duda, R. L., K. Martincic, and R. W. Hendrix. 1995. Genetic basis of bacteriophage HK97 prohead assembly. J. Mol. Biol. 247**:**636-647. [DOI] [PubMed] [Google Scholar]
- 9.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14**:**755-763. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5**:**164-166. [Google Scholar]
- 11.Gerlt, J. A., and P. C. Babbitt. 2001. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 70**:**209-246. [DOI] [PubMed] [Google Scholar]
- 12.Gilakjan, Z. A., and A. M. Kropinski. 1999. Cloning and analysis of the capsid morphogenesis genes of Pseudomonas aeruginosa bacteriophage D3: another example of protein chain mail? J. Bacteriol. 181**:**7221-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginalski, K., A. Elofsson, D. Fischer, and L. Rychlewski. 2003. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19**:**1015-1018. [DOI] [PubMed] [Google Scholar]
- 14.Hendrix, R. W. 1999. Evolution: the long evolutionary reach of viruses. Curr. Biol. 9**:**R914-R917. [DOI] [PubMed] [Google Scholar]
- 15.Hoog, S. S., W. W. Smith, X. Qiu, C. A. Janson, B. Hellmig, M. S. McQueney, K. O'Donnell, D. O'Shannessy, A. G. DiLella, C. Debouck, and S. S. Abdel-Meguid. 1997. Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry 36**:**14023-14029. [DOI] [PubMed] [Google Scholar]
- 16.Khayat, R., R. Batra, M. J. Massariol, L. Lagace, and L. Tong. 2001. Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease. Biochemistry 40**:**6344-6351. [DOI] [PubMed] [Google Scholar]
- 17.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85**:**4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin, E. V., A. R. Mushegian, and P. Bork. 1996. Non-orthologous gene displacement. Trends Genet. 12**:**334-336. [PubMed] [Google Scholar]
- 19.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24**:**946-950. [Google Scholar]
- 20.Liu, F., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65**:**5149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martín, A. C., R. López, and P. García. 1998. Pneumococcal bacteriophage Cp-1 encodes its own protease essential for phage maturation. J. Virol. 72**:**3491-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murzin, A. G. 1998. How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol. 8**:**380-387. [DOI] [PubMed] [Google Scholar]
- 23.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302**:**205-217. [DOI] [PubMed] [Google Scholar]
- 24.Page, R. D. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12**:**357-358. [DOI] [PubMed] [Google Scholar]
- 25.Qiu, X., J. S. Culp, A. G. DiLella, B. Hellmig, S. S. Hoog, C. A. Janson, W. W. Smith, and S. S. Abdel-Meguid. 1996. Unique fold and active site in cytomegalovirus protease. Nature 383**:**275-279. [DOI] [PubMed] [Google Scholar]
- 26.Qiu, X., C. A. Janson, J. S. Culp, S. B. Richardson, C. Debouck, W. W. Smith, and S. S. Abdel-Meguid. 1997. Crystal structure of varicella-zoster virus protease. Proc. Natl. Acad. Sci. USA 94**:**2874-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawlings, N. D., E. A. O'Brien, and A. J. Barrett. 2002. MEROPS: the protease database. Nucleic Acids Res. 30**:**343-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiling, K. K., T. R. Pray, C. S. Craik, and R. M. Stroud. 2000. Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements. Biochemistry 39**:**12796-12803. [DOI] [PubMed] [Google Scholar]
- 29.Rishovd, S., and B. Lindqvist. 1992. Bacteriophage P2 and P4 morphogenesis: protein processing and capsid size determination. Virology 187**:**548-554. [DOI] [PubMed] [Google Scholar]
- 30.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184**:**4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75**:**687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh, H. S., R. G. Kurumbail, A. M. Stevens, R. A. Stegeman, E. J. Sturman, J. Y. Pak, A. J. Wittwer, M. O. Palmier, R. C. Wiegand, B. C. Holwerda, and W. C. Stallings. 1996. Three-dimensional structure of human cytomegalovirus protease. Nature 383**:**279-282. [DOI] [PubMed] [Google Scholar]
- 33.Smith, M. C., and H. M. Thorpe. 2002. Diversity in the serine recombinases. Mol. Microbiol. 44**:**299-307. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25**:**4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong, L., C. Qian, M. J. Massariol, P. R. Bonneau, M. G. Cordingley, and L. Lagace. 1996. A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature 383**:**272-275. [DOI] [PubMed] [Google Scholar]
- 36.Wang, J., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91**:**447-456. [DOI] [PubMed] [Google Scholar]
- 37.Weber, J. M., and K. Tihanyi. 1994. Adenovirus endopeptidases. Methods Enzymol. 244**:**595-604. [DOI] [PubMed] [Google Scholar]
- 38.Weber, J. M. 1995. Adenovirus endopeptidase and its role in virus infection. Curr. Top. Microbiol. Immunol. 199**:**227-235. [DOI] [PubMed] [Google Scholar]
- 39.Welch, A. R., A. S. Woods, L. M. McNally, R. J. Cotter, and W. Gibson. 1991. A herpesvirus maturational proteinase, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc. Natl. Acad. Sci. USA 88**:**10792-10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuzenkova, J., S. Nechaev, J. Berlin, D. Rogulja, K. Kuznedelov, R. Inman, A. Mushegian, and K. Severinov. 2003. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J. Mol. Biol. 330**:**735-748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]