Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 1.
Published in final edited form as: Am J Transplant. 2014 Nov 17;15(1):200–209. doi: 10.1111/ajt.13031
Abstract
Few studies have examined the lung virome in health and disease. Outcomes of lung transplantation are known to be influenced by several recognized respiratory viruses, but global understanding of the virome of the transplanted lung is incomplete. To define the DNA virome within the respiratory tract following lung transplantation we carried out metagenomic analysis of allograft bronchoalveolar lavage (BAL), and compared to healthy and HIV+ subjects. Viral concentrates were purified from BAL and analyzed by shotgun DNA sequencing. All of the BAL samples contained reads mapping to anelloviruses, with high proportions in lung transplant samples. Anellovirus populations in transplant recipients were complex, with multiple concurrent variants. Q-PCR quantification revealed that anellovirus sequences were 56-fold more abundant in BAL from lung transplant recipients compared with healthy controls or HIV+ subjects (p<0.0001). Anellovirus sequences were also more abundant in upper respiratory tract specimens from lung transplant recipients than controls (p=0.006). Comparison to metagenomic data on bacterial populations showed that high anellovirus loads correlated with dysbiotic bacterial communities in allograft BAL (p=0.00816). Thus the respiratory tracts of lung transplant recipients contain high levels and complex populations of anelloviruses, warranting studies of anellovirus lung infection and transplant outcome.
Introduction
Little is known about the virome of the human respiratory tract as a whole, though infections by individual viruses are well characterized. For the case of lung transplantation, viral infection is a major complicating factor impacting graft survival rates (1–4). Respiratory infections with known viruses can cause direct lung injury or increase risk of graft failure, as in the case of cytomegalovirus and community acquired respiratory viruses (1, 5). Intense interest has thus focused on viruses in the respiratory tract and transplantation outcome.
Today it is possible to characterize large viral populations using high throughput metagenomic sequencing (6–8), which has identified both well-recognized and little-studied viruses living in association with humans. Only a few studies have applied metagenomic approaches to understand viruses of the lower respiratory tract (8, 9), and none in lung transplantation.
Anelloviruses are circular, nonenveloped, negative-sense, single-stranded DNA viruses that commonly colonize humans and show increased abundance in blood after hematopoietic and solid organ transplantation (10–12). The anellovirus family consists of Torque Teno viruses (TTVs), Torque Teno Midi Viruses (TTMDV), Torque Teno Mini Viruses (TTMV), and Small Anelloviruses (SAVs), each of which has multiple subtypes (12, 13). Their small genomes (2.3–3.8 kb) consist of three to four open reading frames and a highly conserved untranslated region (UTR) (12). These viruses are ubiquitous in the human population and have not yet been causally linked with any human disease (12, 13). Diverse types of anelloviruses have been found in various organs, tissues, and cell types (12, 14–16).
In the respiratory tract, TTV was recently identified in bronchoalveolar lavage fluid from 28% of individuals with acute exacerbations of idiopathic pulmonary fibrosis (IPF), but not those with stable IPF, and in a quarter of individuals with acute lung injury (17). In the upper respiratory tract, elevated levels of TTV have been found in nasal secretions from children with respiratory diseases, and correlated with disease severity (15, 16). In HIV-infected patients, plasma concentrations of anelloviruses increased during progression to AIDS (18) and decreased following therapy (19). TTV viremia was reported to increase following autologous hematopoietic stem cell transplantation, and then return to baseline levels following immune reconstitution (10). Recently, TTV levels in blood were shown to increase in association with immunosuppression following lung and heart transplantation (11). The association between TTV levels and immune status has led some authors to propose that anelloviruses genome copy numbers may serve as an empirical measure of successful immune suppression (10, 11, 19).
We report here the first study to apply viral metagenomics to understand the lung virome in lung transplantation. We first used Illumina metagenomic sequencing to characterize lung DNA viromes from lung transplant recipients, along with another immunologically impaired group, HIV-positive individuals. This showed that anellovirus sequences were prominent in BAL from transplant recipients, and revealed the presence of complex populations with multiple concurrent variants. Based on these metagenomic data, we then quantified anellovirus levels within the lungs and upper respiratory tracts of lung transplant recipients, HIV+ subjects, and healthy individuals using Q-PCR, demonstrating high levels of anellovirus DNA in BAL and OW of lung transplant recipients. Our findings demonstrate metagenomic detection and genetic characterization of the allograft virome, which is then followed by broader quantitative analysis. We also compared metagenomic data on bacterial populations, and found that high levels of anelloviruses correlated with dysbiotic bacterial communities, revealing covariation among microbiome constituents.
Methods
Sample Collection
Bronchoalveolar lavage (BAL) was carried out within the lung allograft on lung transplant recipients, most of whom were undergoing routine clinical surveillance bronchoscopy during the first year post-transplant (Table S1) as described previously (20). Oropharyngeal wash (OW) to sample the upper respiratory tract was collected as previously described (20). One group of control samples were obtained from healthy volunteers who underwent research bronchoscopy using the same procedure (Table S2). Samples from three HIV+ subjects not on antiretroviral therapy (CD4 T cell counts of 301, 321 and 682; Table S3) and a second set from healthy volunteers (Table S2) were obtained by bronchoscopy using a previously described two-scope procedure (21). The University of Pennsylvania IRB approved all procedures (protocols # 812748 and #810851), and subjects gave written informed consent.
Virus-Like Particle (VLP) Purification
Virus-like particles were purified from 1–5 milliliters of BAL or OW, depending upon availability. 10mM MgSO4 and 10mM dithiothreitol were added to the BAL or OW and filtered through a 0.22 µm filter (Millipore). The filtrate was concentrated using a 100kDa molecular weight cut-off filter (Amicon 20, Millipore), resuspended in 1 ml Buffer SM, and reconcentrated. The concentrate was treated with DNase I and RNase (Roche) at 37°C for 15 min to eliminate non-encapsidated nucleic acids, then the enzymes were deactivated at 70°C for 5 min.
Nucleic Acid Extraction and Metagenomic Sequencing
Nucleic acids were extracted from virus particle preps using Qiagen’s All Prep nucleic acid isolation kit. Six lung transplant and three HIV+ BAL samples that had large volumes of BAL and detailed patient metadata available were used for metagenomic sequencing. Whole genome amplification was performed on these samples using the GenomiPhi V2 Amplification Kit (GE Healthcare). Libraries for sequencing were made using Illumina’s Nextera XT DNA Sample Preparation Kit with 1 ng of input DNA, generating paired-end fragments. Metagenomic sequencing was performed on an Illumina MiSeq instrument.
Quantitative PCR
Q-PCR was carried out on non-Genomiphi amplified DNA using primers targeting the UTR of anelloviruses from a previously-described assay (22) that we adapted to a SYBR green-based method as described in Supplemental Methods. All samples were run in duplicate or triplicate and values averaged. The detection limit was 1.4 copies per reaction.
Bioinformatics Pipeline
Paired-end reads from the MiSeq instrument were quality-trimmed and processed through BMTagger to remove human sequences. Non-human reads were then analyzed using BLAST against the NCBI viral database. Reads were assembled into contigs by iterative deBruijn graph assembly using IDBA-UD (23). To verify contig assembly, contig-specific PCR primers were synthesized and amplification products subjected to Sanger sequencing. Dot plots were generated using Gepard with a word size of 10 nucleotides (24). Anellovirus ORF1 amino acid sequences were identified from our contigs and aligned along with 49 reference TTV, TTMDV, and TTMV sequences from Genbank. The phylogenetic tree was built using FastTree v2.1.3 (25). Further details of bioinformatic analysis are provided in Supplemental Methods.
Bacterial 16S rRNA gene sequence data generated from whole BAL have been previously reported (20). Sequences were clustered into operational taxonomic units based on 97% sequence similarity, aligned with 16S reference databases for taxonomic assignment, and pairwise UniFrac distances calculated using QIIME v1.8 (26).
Statistical Analyses
Mann-Whitney tests, paired Wilcoxon signed rank tests, t-tests, and Spearman correlation tests were performed in R (27).
Results
BAL and OW samples were collected from 1) lung transplant recipients undergoing post-transplant bronchoscopy, 2) HIV+ individuals without lung disease or respiratory symptoms, and 3) healthy control subjects (20, 21) (Tables S1, S2, and S3). VLPs were purified from BAL and OW, followed by concentration and treatment with nucleases to eliminate non-encapsidated nucleic acids (Figure 1). DNA was then purified from each sample. To verify selective reduction in human and bacterial cells, two BAL samples were analyzed using Q-PCR to quantify bacterial 16S rRNA gene copies or human beta-tubulin gene copies. Copies of bacterial 16S DNA decreased from 3000 copies/ng in unfractionated BAL to 80 copies/ng after viral particle purification (38-fold reduction). Human beta-tubulin DNA was detected at 925 copies/ng in unfractionated BAL but became undetectable after purification (>900-fold reduction).
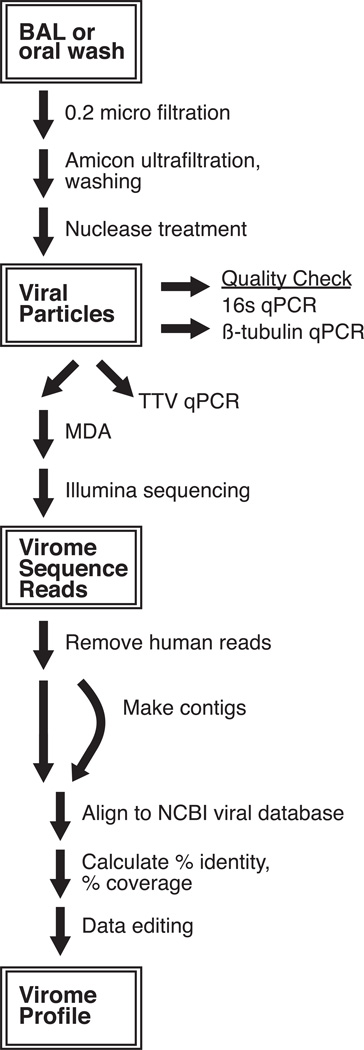
Figure 1. Overview of experimental design.
BAL and OW samples (1–5 ml) were filtered through a 0.22 µM filter, concentrated on an Amicon 10 kDa MWCO filter, washed, and treated with DNase and RNase to yield purified virus-like particles. Select samples were checked for viral purity by quantifying the bacterial 16S rRNA and human β-tubulin genes using Q-PCR. Anelloviruses were quantified in all samples by Q-PCR. Select lung transplant and HIV+ samples were whole-genome amplified by multiple displacement amplification (MDA) and shotgun sequenced using the Illumina MiSeq platform. Bioinformatic analysis of the virome reads consisted of removing human reads (BMTagger) and aligning to the NCBI viral database. Reads were assembled into contigs, aligned to the NCBI viral database and analyzed.
Metagenomic Sequencing of Lung Transplant and HIV+ Samples
We first applied metagenomic analysis to viral DNA from six lung transplant and three HIV+ BAL samples. DNA from viral preps was subjected to whole genome amplification and then shotgun sequenced using the Illumina MiSeq platform (2 × 250 bp reads) (Table 1). Up to 3.4 million reads per sample were generated, for a total of 5.42 × 109 bp of sequence data. Reads were filtered and trimmed to remove low quality sequences, and human reads were identified and removed. Reads were then aligned using BLAST to the NCBI viral database (Table 1 and Table S4).
Table 1.
Anelloviruses identified through metagenomic sequencing of BAL samples from lung transplant recipients and HIV+ individuals.
| HIV+ | Lung Transplant Recipients | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HUP1A03 | HUP1B03 | HUP1B07 | Tx-24 | Tx-34 | Tx-38 | Tx-49 | Tx-51 | Tx-52 | |
| Total Reads (Unfiltered) | 1,134,270 | 3,480,764 | 649,864 | 644,536 | 869,518 | 1,236,626 | 1,169,756 | 171,892 | 71,018 |
| Total Reads after Quality Trimming | 1,134,256 | 3,480,750 | 649,852 | 644,526 | 869,504 | 1,236,620 | 1,169,742 | 171,884 | 71,010 |
| % Human Reads | 0% | 0% | 0% | 1% | 0% | 26% | 29% | 21% | 14% |
| % Reads Matching to Viral DB | 4% | 4% | 5% | 21% | 23% | 1% | 10% | 7% | 1% |
| Small anellovirus 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| Small anellovirus 2 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 |
| Torque teno felis virus | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Torque teno midi virus 1 | 0 | 0 | 0 | 20 | 1 | 0 | 0 | 0 | 0 |
| Torque teno midi virus 2 | 0 | 0 | 0 | 13 | 1 | 0 | 0 | 0 | 0 |
| Torque teno mini virus 2 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| Torque teno mini virus 3 | 0 | 0 | 0 | 0 | 0 | 5 | 2 | 0 | 0 |
| Torque teno mini virus 4 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 |
| Torque teno mini virus 5 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 18 |
| Torque teno mini virus 7 | 0 | 0 | 0 | 5 | 0 | 9 | 0 | 0 | 6 |
| Torque teno mini virus 8 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 |
| Torque teno virus 1 | 4 | 6 | 3 | 3124 | 10901 | 44 | 1051 | 128 | 2 |
| Torque teno virus 10 | 8 | 10 | 3 | 6984 | 10537 | 97 | 4828 | 513 | 1 |
| Torque teno virus 12 | 4 | 13 | 0 | 9946 | 222 | 21 | 2695 | 98 | 4 |
| Torque teno virus 14 | 1 | 4 | 0 | 480 | 15 | 290 | 10819 | 4235 | 0 |
| Torque teno virus 15 | 19 | 46 | 19 | 44356 | 75859 | 115 | 7173 | 564 | 3 |
| Torque teno virus 16 | 7 | 18 | 5 | 10860 | 5918 | 166 | 19424 | 175 | 0 |
| Torque teno virus 19 | 7 | 12 | 5 | 3102 | 12143 | 78 | 14768 | 233 | 2 |
| Torque teno virus 2 | 1 | 6 | 0 | 262 | 7 | 82 | 66 | 501 | 0 |
| Torque teno virus 27 | 2 | 0 | 1 | 1381 | 1046 | 46 | 1036 | 57 | 1 |
| Torque teno virus 28 | 3 | 3 | 1 | 211 | 80 | 11 | 161 | 6 | 0 |
| Torque teno virus 3 | 5 | 33 | 1 | 15693 | 59 | 79 | 19995 | 1202 | 0 |
| Torque teno virus 4 | 2 | 1 | 0 | 853 | 9 | 0 | 1114 | 661 | 0 |
| Torque teno virus 6 | 19 | 24 | 5 | 22153 | 13397 | 184 | 10487 | 127 | 6 |
| Torque teno virus 7 | 4 | 48 | 2 | 8714 | 28 | 20 | 1950 | 22 | 0 |
| Torque teno virus 8 | 20 | 28 | 79 | 6834 | 69854 | 868 | 17068 | 2440 | 24 |
| Torque teno virus | 0 | 1 | 0 | 1076 | 3 | 24 | 970 | 0 | 0 |
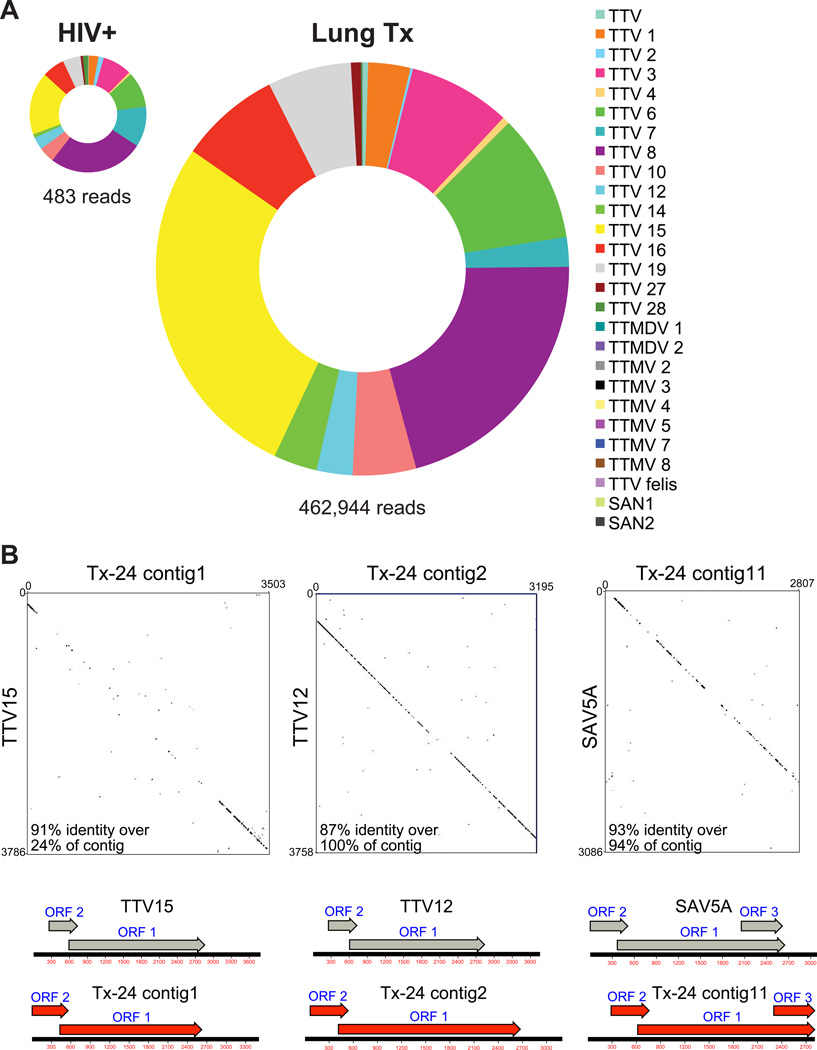
Of reads assigned to reference viruses, the majority (>68%) mapped to anelloviruses, which included Torque Teno viruses, Torque Teno Mini viruses, Torque Teno Midi viruses, and Small Anelloviruses (Table 1, Figure 2A). Based on the closest match to reference viruses, a wide variety of anelloviruses were present within the lungs of lung transplant subjects, while substantially fewer anellovirus reads were detected in lungs of HIV+ individuals (Figure 2A). Furthermore, diverse anelloviruses were present in single individuals, especially in the lung transplant recipients, as evidenced by reads aligning to many different TTVs, TTMVs, TTMVDs, and SAVs within single samples (Table 1).
Figure 2. Anelloviruses in BAL of HIV+ individuals and lung transplant recipients.
A) Distribution of reads aligning to anelloviruses. Metagenomic sequencing reads were searched against the NCBI viral database and the number of combined reads aligning to each anellovirus calculated for lung transplant recipients (n=6) and HIV+ subjects (n=3). B) Presence of multiple anelloviruses present within the lungs of lung transplant recipient Tx-24 verified by Sanger sequencing. For each of the three analyzed by Sanger sequencing (x-axis), similarity was scored versus their closest matching NCBI reference sequence (y-axis). Open reading frames were assigned using MacVector and are illustrated on the genetic maps under the dot plots.
Other eukaryotic viruses were detected within the samples, but at much lower levels. These included Epstein-Barr virus (Human Herpesvirus 4) (2 reads in one sample), Human Herpesvirus 7 (2 reads in one sample, 36 reads in another sample), and Human Papillomavirus (1 read) (Table S4). To determine whether use of the 0.22 micron filter step during virus purification may have resulted in depletion of some of the larger viruses, we compared filtration and centrifugation (Table S6). This showed that filtration resulted in reduced recovery of herpesviruses, but was more efficient at capturing small viruses such as human papillomaviruses and anelloviruses.
A small percentage of reads (0.75% of total reads for all samples combined) aligned to bacteriophage genomes, predominantly phages of Enterobacteria, Salmonella, Pseudomonas, Streptococcus and Yersinia. When bacterophages were compared with bacterial 16S sequences from these subjects that were previously analyzed (28), we found representation of several of these bacterial lineages (Streptococcus, Pseudomonas, Staphylococcus, and Enterobacteria), consistent with possible phage/host pairs in our samples.
An average of 81% of the reads did not match reference viruses in the NCBI viral database, a high proportion similar to results of other viral metagenomic studies (9, 29). We surmise that many of these may correspond to DNA phage sequences, because phage are hyperabundant globally and poorly represented in sequence databases (30). Unidentified reads could also correspond to novel mammalian viruses or low quality reads not removed by our filtering strategy.
Genetic structure of lung anellovirus populations
Anellovirus reads were assembled into contigs for more complete analysis of anelloviruses community structure, using iterative deBruijn graph assembly (23). To validate correct contig assembly, we designed specific primer sets for three of the contigs, then PCR amplified 500–700 base pair regions, sequenced using the Sanger method, and assembled using the conventional overlap method. Assembled contigs were then aligned to the NCBI nt database (Figure 2B). Selected regions within the anellovirus contigs were 87–93% identical to the reference sequences, though highly divergent regions were also detected. Anellovirus open reading frames (ORF) 1 and 2 were identified in the three anellovirus contigs. ORF1 was previously shown to encode the capsid gene and contains a hypervariable region, which may evolve rapidly to evade the host immune response (12). The ORF2 encoded protein was reported to suppress the NF-ΚB pathway, thereby potentially regulating the host innate and adaptive immune response (12, 31).
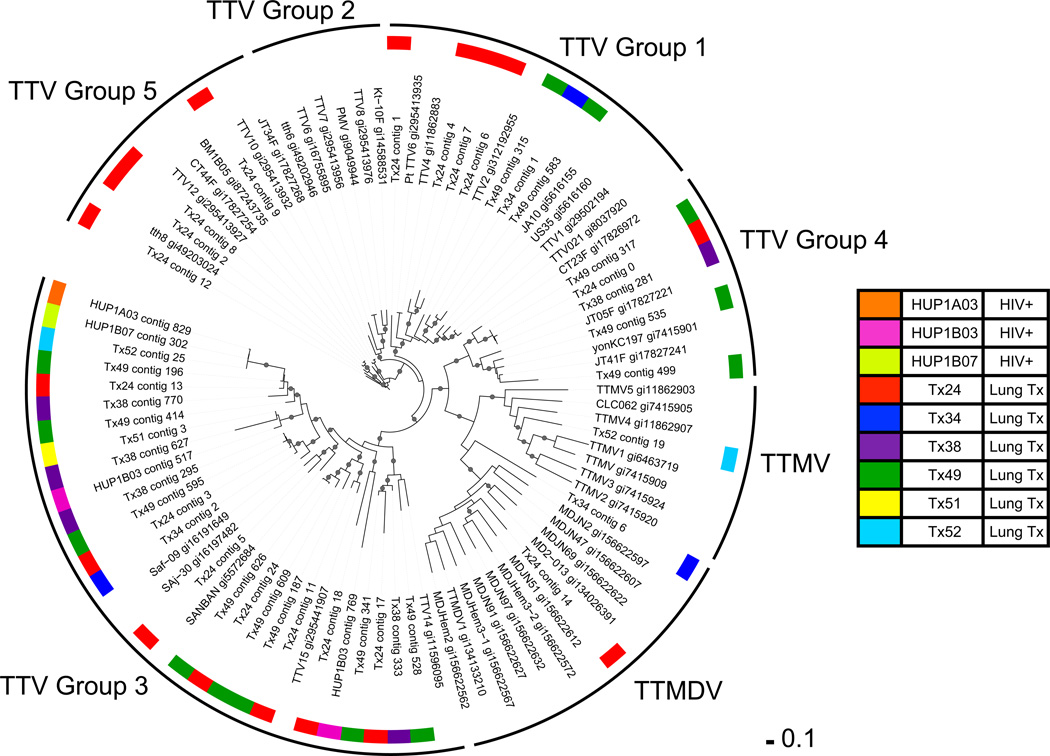
To compare anelloviruses populations within and between individuals, a phylogenetic tree was generated using ORF1 sequences from reference genomes and contigs assembled from the Illumina and Sanger sequence data. As shown in Figure 3, multiple different anellovirus ORF1s were found within individual lung transplant patient allografts, with subjects exhibiting as many as 17 distinct contigs. In contrast, richness of anelloviruses was lower in BAL of HIV+ individuals, even though sequences were detected in all of them. Anellovirus contigs were mostly different between individuals (Figure 3).
Figure 3. Diversity of anellovirus ORF1 sequences in samples from each individual studied.
ORF1 amino acid sequences from anellovirus contigs and Genbank reference sequences were aligned and trimmed to generate the phylogenetic tree shown. The names of the reference sequences include the TTV strain name followed by the Genbank identification (gi) number. Clades are designated as groups, in the outer ring, as described by Okamoto et al (12, 22). The BAL sample of origin of each ORF1 sequence is designated by the color code (key at right). The Shimodaira-Hasegawa (SH) score was calculated using FastTree to estimate the reliability of each split compared to alternate topologies (25). Local SH support values over 0.9 are labeled as circles on the nodes of the tree. The scale bar represents the number of amino acid substitutions per position.
Quantification of Anellovirus DNA in Lung and Upper Respiratory Tract Samples Using Quantitative PCR
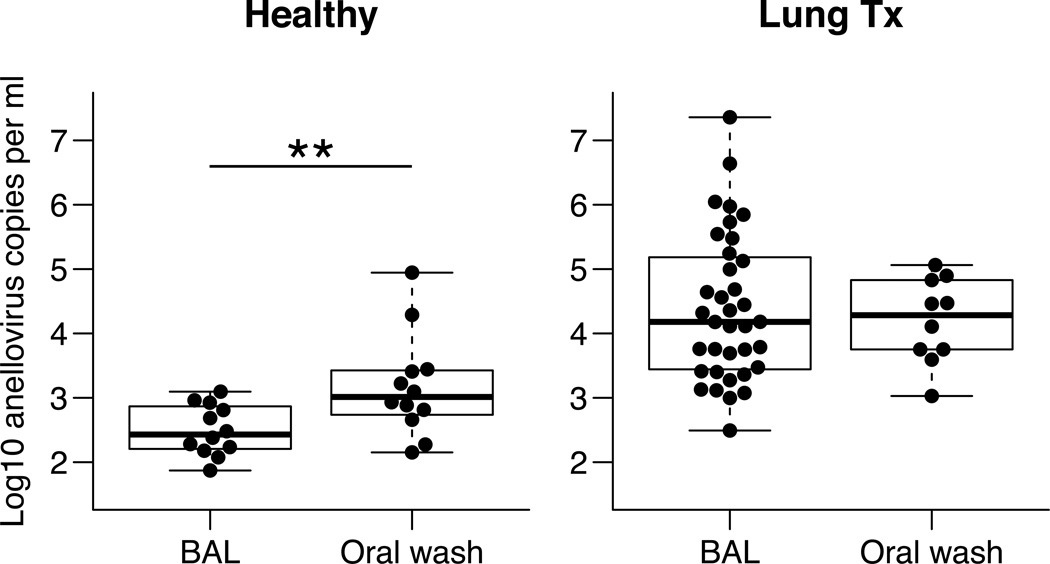
Given diverse anellovirus populations found in some of the transplant samples, we sought to quantify absolute anellovirus DNA levels in a larger group of subjects using Q-PCR. We used the universal PCR primers targeting the anellovirus highly conserved UTR (22) to generate a Q-PCR assay. These primers recognize TTVs, TTMVs, TTMVDs, and SAVs. We analyzed a total of 53 BAL samples from lung transplant recipients (n=38), HIV+ subjects (n=3), and healthy controls (n=12). To assess the upper respiratory tract, we also analyzed 24 OW samples from lung transplant recipients (n=12) and healthy controls (n=12).
As shown in Figure 4, anellovirus copy numbers in lung ranged widely among transplant recipients, with a median of 15,133 copies/ml (range=314 to 2.3 × 107 copies/ml). These levels were far higher than in lungs of healthy or HIV+ subjects (medians of 271 and 191 copies/ml, respectively; p<0.0002 for comparison to lung transplant samples; Mann-Whitney test).
Figure 4. Abundance of anelloviruses in BAL samples.
Anelloviruses were quantified by Q-PCR in BAL from lung transplant recipients, healthy individuals, and HIV+ individuals. Boxes represent the middle two quartiles for each group and whiskers are placed at the minimum and maximum values. Quantities were higher in lung transplant recipients compared with healthy and HIV+ individuals as determined by the Mann-Whitney test with Bonferroni correction: p<0.0002. All samples quantified were above the Q-PCR detection limit (1.4 copies per reaction).
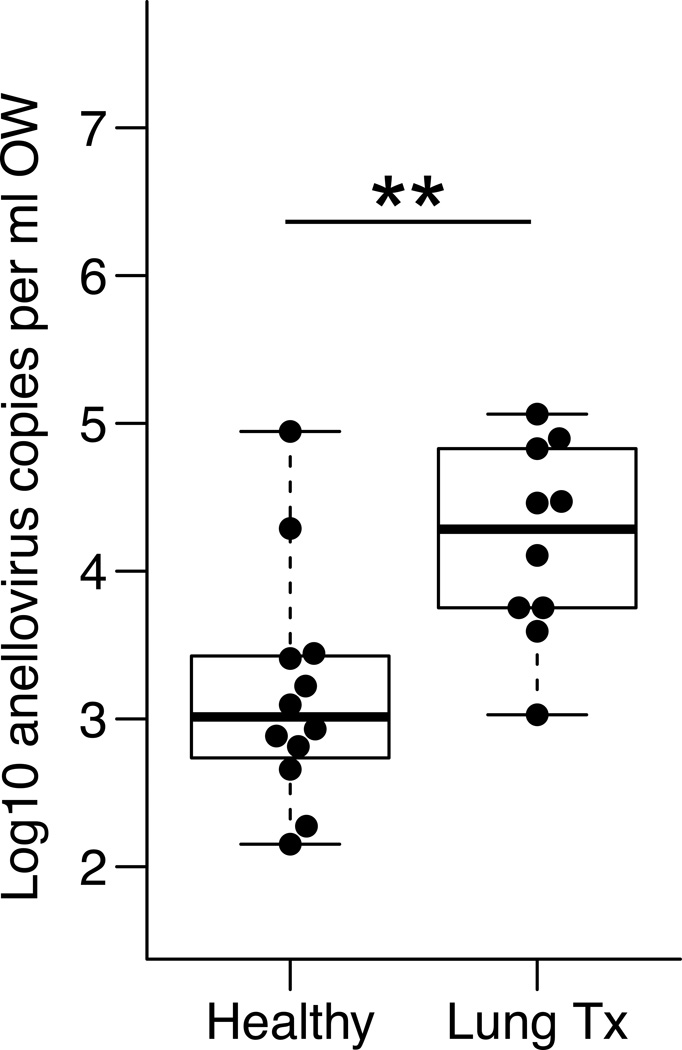
Quantities of anelloviruses in the upper respiratory tract were also measured in OW collected from lung transplant recipients and healthy subjects. As was seen in lung, anellovirus genome copies were significantly more abundant in the upper respiratory tract of transplant recipients compared with healthy controls (Figure 5) (median=16,700 in transplant recipients, range=634 to 632,759 copies/ml compared to median=1,033 in healthy; p=0.0055). When quantities of anellovirus were compared between lung and upper respiratory tract within the same treatment group, healthy subjects had significantly lower titers in their BAL compared to oropharyngeal wash (p=0.0068, Mann-Whitney test). However, in lung transplant recipients, anellovirus genome copies were higher in both sample types and not significantly different in lung compared to the upper respiratory tract (p=0.23, Figure 6).
Figure 5. Abundance of anelloviruses in OW samples.
Anelloviruses were quantified by Q-PCR in oropharyngeal wash from healthy individuals and lung transplant recipients. Anellovirus quantities were higher in the upper respiratory tract of lung transplant recipients compared with healthy individuals. Mann-Whitney test: p=0.0055. All samples quantified were above the Q-PCR detection limit.
Figure 6. Comparison of anellovirus quantities in lung and upper respiratory tract within individuals.
Paired Wilcoxon signed rank tests were performed on anellovirus copies from BAL and OW in A) healthy control subjects and B) lung transplant recipients. Anellovirus DNA copies were lower in the lung compared with the upper respiratory tract of healthy controls but not in lung transplant recipients (p-values: 0.0068 and 0.23, respectively).
Comparison of anellovirus DNA levels to transplant subject metadata
We then asked whether anellovirus levels in the transplant recipients’ lungs were correlated with clinical variables that might explain the high variability. We queried patient age, gender, time since transplant, number of immunosuppressive drugs, tacrolimus levels, prednisone dosing, target immunosuppression range, surgical type (single vs. bilateral lung transplant), use of azithromycin, cytomegalovirus status (donor and recipient), BAL bacterial culture results and bacterial load in BAL based on 16S rRNA gene Q-PCR, acute rejection grade, and pre-transplant diagnosis (suppurative vs. nonsuppurative lung disease). No significant associations were detected (Table S5). Though power was limited by the relatively small sample sizes, these data suggest that anellovirus replication in the lung allograft is linked to factors other than these clinical variables.
Comparison of lung anellovirus DNA levels to the bacterial microbiome
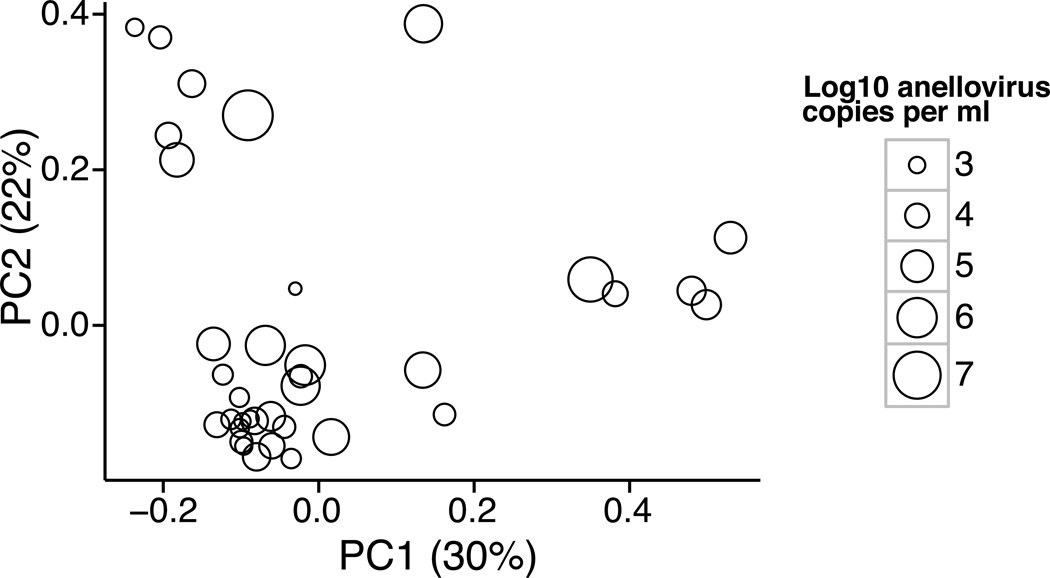
Finally, we asked whether there was a relationship between anellovirus DNA levels and composition of the bacterial microbiome in allografts. Previously, DNA was purified from whole BAL, PCR ampified using primers that recognize conserved regions of the bacterial 16S rRNA gene, and products analyzed by deep sequencing (20). That analysis showed that lung transplant bacterial communities differed in composition from healthy subjects, and transplant recipients were commonly colonized by oral bacteria, known pathogenic bacteria, and sometimes by unexpected bacterial lineages. Absolute levels of bacteria were also higher than in healthy controls (40–100-fold). Here we compared the anellovirus titers with bacterial community composition among transplant recipients, and found a significant correlation (Figure 7; weighted UniFrac, p=0.032). However, no specific bacterial lineages were significantly linked to anellovirus levels among transplant subjects. We then asked if the relationship between anellovirus and 16S bacterial composition might be linked to the degree to which the bacterial communities diverged from healthy controls. This analysis revealed a significant correlation between transplant BAL anellovirus levels and a metric measuring the divergence between the corresponding bacterial community and those of healthy subjects (Figure S1; p=0.008). Thus, high anellovirus levels in transplant recipients are also associated with aberrant bacterial microbiota.
Figure 7. Ordination based on composition of bacterial 16S sequence analysis, showing the relationship to anellovirus DNA copy numbers.
Each circle on the plot shows data for a single transplant recipient BAL sample. For each sample, bacterial 16S rRNA gene tags were subject to deep sequencing (median 7,925 reads per sample; data published in (20)) and analyzed using Weighted UniFrac, which generated a set of pairwise distances among samples. The distances were then analyzed using Principal Coordinate Analysis (PCoA), and the samples plotted along the first two principal coordinates. For each sample, the anellovirus titer is summarized by the size of the open circle. Among these transplant recipient BALs there is a significant relationship between anellovirus DNA level and 16S bacterial community composition, which is reflected in the observation that samples with low anellovirus titers tend to appear on the lower left side of the PCoA plot (p=0.032; ADONIS test).
Discussion
Here we used metagenomic sequencing to analyze DNA viruses present in the respiratory tract of lung transplant recipients and HIV-infected individuals. The metagenomic data then enabled targeted analysis of anellovirus levels in a larger cohort of lung transplant and control subjects. This is the first study to apply viral metagenomics to the lung transplant allograft. Multiple viruses infecting animal cells were detected by sequencing, including a remarkable abundance and complexity of anelloviruses, as well as much lower levels of papilloma viruses and herpes viruses. Bacteriophage were also detectable, as were unidentified sequences likely corresponding at least in part to phages. Our most notable finding was that the anelloviruses, including TTVs, TTMVDs, TTMVs, and SAVs, were greatly increased in abundance in the lungs and the upper respiratory tract of lung transplant recipients compared with healthy and HIV+ subjects. As this work was being completed, Quake and colleagues reported that anellovirus sequences were increased in abundance in blood in organ transplant recipients (11). Our data reported here show that after lung transplantation, anellovirus sequences are abundant in the lung allograft itself.
The shotgun metagenomic sequence data enabled assembly of near-complete genome sequences for some of the most abundant anelloviruses, allowing the genetic nature of anelloviruses present to be analyzed in detail. A notable feature of transplant recipients was the great diversity of anelloviruses represented within single individuals. Quantifying the exact number of different forms is challenging and dependent on the metric chosen. Taking each independent contig into account, there were up to 247 contigs identified as anelloviruses within one sample (Tx-24). However, this may be an overestimate because some contigs are only partial genomes (sizes ranged from 250–3600 base pairs), though this is hard to assess definitively because replication-competent subviral molecules have been reported (32). Analysis based on viral ORF1 regions showed that up to 17 different anelloviruses were present within a single transplant recipient (Tx-24), indicating remarkable diversity. Some of the anelloviruses identified in these subjects were distant in sequence from database genomes. Such high divergence was confirmed in the three genomes reconstructed by Sanger sequencing, and so is not a result of error in Illumina sequence determination or assembly. The origin of this extreme diversity is unclear, but may be due in part to the known high mutation rates of single-stranded DNA viruses (33) combined with high virus levels likely reflecting high rates of virus replication in lung.
We found large numbers of bacteriophage sequences in our shotgun metagenomic data, likely due to the abundant bacterial populations in lung transplant subjects (20), but relatively few mammalian DNA viruses besides anelloviruses. This is likely in large part related to the near-universal use of herpesviruses prophylaxis in transplant subjects, and relatively modest level of immune dysfunction based on CD4+ T cell counts in our HIV+ subjects. In addition, samples for metagenomic analysis were subject to multiple displacement amplification, which favors small circular genomes such as anelloviruses. An additional technical factor may be that the filtration step used could have depleted large viruses or viral particles bound to other macromolecules, although reconstruction experiments suggest that the losses were modest. In addition, a large proportion of sequences did not match any database genomes, which is concordant with findings in other metagenomic virome studies (6, 9, 29). While many of these are likely novel bacterophages, it is possible that novel mammalian viruses may exist within those sequences as well. One limitation of our sample set is the relatively short elapsed time since transplant (median of 5 months), and it will be useful to investigate whether additional viral lineages emerge over longer times.
Anellovirus genome copy numbers in lung from Q-PCR data were remarkably different among transplant subjects, varying across nearly 5 orders of magnitude. However, comparison to standard patient metadata did not show significant correlations with any of the clinical parameters queried. A recent study of anellovirus in serum following organ transplantation reported that anellovirus genome levels correlated with the extent of immunosuppression, suggesting that anellovirus DNA in blood might serve as an assay for the overall level of immunosuppression (11). In contrast, our subjects were receiving relatively homogenous immunosuppression regimens (Table S1), and we found no association with any measure of immunosuppression, nor with time since transplant (Table S5). Thus, the absence of correlations seen here between lung anellovirus levels and clinical variables suggests that anellovirus replication in lung allografts may be regulated by factors that have yet to be identified.
We compared anellovirus DNA quantification to previously determined data on the structure of bacterial communities, and identified a novel form of covariation. Among transplant recipients, BAL samples with high anellovirus levels differed from those with lower levels based on the bacterial 16S data. Furthermore, samples with highest levels of anellovirus were more divergent from the bacterial communities of healthy controls, indicating bacterial dysbiosis. This suggests that the anellovirus and bacterial communities may both be responding to some common property of the host, such as loss of immune control. If so, the immune impairment would not be reflected by standard clinical measures, which did not correlate with features of anelloviral DNA levels. The correlation between anellovirus titers and bacterial dysbiosis was imperfect, however, indicating that there are likely both shared and unique factors influencing viral and bacterial microbiota, respectively.
In summary, our results employing shotgun metagenomic sequencing revealed robust anellovirus populations in lungs of lung transplant recipients, which was then confirmed and quantified by targeted Q-PCR. Long-term lung transplant outcomes have been linked to prior infection with several other viruses (1, 2, 4). While our subjects were sampled at relatively early time points (median 5 months post-transplant), early post-transplant colonization with other microbial agents has been associated with BOS at later time points (34, 35). Thus, the finding of unexpected high levels of anellovirus replication within lung allografts and marked inter-subject variability suggest that longitudinal studies are warranted to determine whether levels of anelloviruses in the lung allograft are associated with, and may play a role in, transplant outcomes.
Supplementary Material
Supp MaterialS1
Supp TableS1
Supp TableS2
Supp TableS3
Supp TableS4
Supp TableS5
Supp TableS6
Acknowledgments
We are grateful to members of the Bushman and Collman laboratory for help and suggestions. We are grateful to the research subjects who volunteered for this study and the clinicians who assisted with specimen collection; A. Fitzgerald for project management; W. Russell and D. Frame for critical study assistance. This work was supported by NIH awards U01 HL098957 and R01 HL113252. JCY was supported by NIH T32 AI007632. We also acknowledge assistance and support from the Penn Center for AIDS Research (P30-AI045008), and the Penn DNA Sequencing Facility.
Abbreviations
BAL
bronchoalveolar lavage
OW
oropharyngeal wash
TTV
torque teno virus
TTMDV
torque teno midi virus
TTMV
torque teno mini virus
SAV
small anellovirus
Q-PCR
quantitative PCR
UTR
untranslated region
VLP
virus-like particle
ORF
open reading frame
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supplementary Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Methods
Table S1: Lung transplant recipients
Table S2: Healthy Human subjects
Table S3: HIV+ subjects
Table S4: Summary of viruses detected
Table S5: Metadata p-values
Table S6: Impact of filtration on read distribution
Figure S1. The bacterial communities of lung transplant BAL samples are more divergent from healthy controls.
(A) PCoA ordination of BAL samples from healthy subjects and lung transplant recipients according to weighted UniFrac distance. The anellovirus titer is indicated by the size of the open circle. For each transplant subject, a line is drawn to the centroid (center-of-gravity) position for healthy subjects. (B) For lung transplant recipients, the anellovirus titer was positively correlated with weighted UniFrac distance from the centroid position of healthy subjects (Spearman correlation test, ρ=0.4, P=0.008).
References
- 1.Burguete SR, Maselli DJ, Fernandez JF, Levine SM. Lung transplant infection. Respirology. 2013;18:22–38. doi: 10.1111/j.1440-1843.2012.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vu DL, Bridevaux PO, Aubert JD, Soccal PM, Kaiser L. Respiratory viruses in lung transplant recipients: A critical review and pooled analysis of clinical studies. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:1071–1078. doi: 10.1111/j.1600-6143.2011.03490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbino J, Gerbase MW, Wunderli W, Kolarova L, Nicod LP, Rochat T, Kaiser L. Respiratory viruses and severe lower respiratory tract complications in hospitalized patients. Chest. 2004;125:1033–1039. doi: 10.1378/chest.125.3.1033. [DOI] [PubMed] [Google Scholar]
- 4.Clark NM, Lynch JP, 3rd, Sayah D, Belperio JA, Fishbein MC, Weigt SS. DNA viral infections complicating lung transplantation. Seminars in respiratory and critical care medicine. 2013;34:380–404. doi: 10.1055/s-0033-1348473. [DOI] [PubMed] [Google Scholar]
- 5.Speich R, van der Bij W. Epidemiology and management of infections after lung transplantation. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2001;33(Suppl 1):S58–S65. doi: 10.1086/320906. [DOI] [PubMed] [Google Scholar]
- 6.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, Rodriguez-Brito B, Salamon P, Rohwer F. Viral diversity and dynamics in an infant gut. Research in microbiology. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PloS one. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, Rainey PB, Schmieder R, Youle M, Conrad D, Rohwer F. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. American journal of respiratory cell and molecular biology. 2012;46:127–131. doi: 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Focosi D, Maggi F, Albani M, Macera L, Ricci V, Gragnani S, Di Beo S, Ghimenti M, Antonelli G, Bendinelli M, Pistello M, Ceccherini-Nelli L, Petrini M. Torquetenovirus viremia kinetics after autologous stem cell transplantation are predictable and may serve as a surrogate marker of functional immune reconstitution. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2010;47:189–192. doi: 10.1016/j.jcv.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 11.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, Weill D, Bernstein D, Valantine HA, Quake SR. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Villiers E-M, Zur Hausen H. Tt viruses: The still elusive human pathogens. Springer; 2009. [PubMed] [Google Scholar]
- 13.Hino S, Miyata H. Torque teno virus (ttv): Current status. Reviews in medical virology. 2007;17:45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- 14.Maggi F, Fornai C, Zaccaro L, Morrica A, Vatteroni ML, Isola P, Marchi S, Ricchiuti A, Pistello M, Bendinelli M. Tt virus (ttv) loads associated with different peripheral blood cell types and evidence for ttv replication in activated mononuclear cells. Journal of medical virology. 2001;64:190–194. doi: 10.1002/jmv.1035. [DOI] [PubMed] [Google Scholar]
- 15.Maggi F, Pifferi M, Fornai C, Andreoli E, Tempestini E, Vatteroni M, Presciuttini S, Marchi S, Pietrobelli A, Boner A, Pistello M, Bendinelli M. Tt virus in the nasal secretions of children with acute respiratory diseases: Relations to viremia and disease severity. Journal of virology. 2003;77:2418–2425. doi: 10.1128/JVI.77.4.2418-2425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maggi F, Pifferi M, Tempestini E, Fornai C, Lanini L, Andreoli E, Vatteroni M, Presciuttini S, Pietrobelli A, Boner A, Pistello M, Bendinelli M. Tt virus loads and lymphocyte subpopulations in children with acute respiratory diseases. Journal of virology. 2003;77:9081–9083. doi: 10.1128/JVI.77.16.9081-9083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wootton SC, Kim DS, Kondoh Y, Chen E, Lee JS, Song JW, Huh JW, Taniguchi H, Chiu C, Boushey H, Lancaster LH, Wolters PJ, DeRisi J, Ganem D, Collard HR. Viral infection in acute exacerbation of idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2011;183:1698–1702. doi: 10.1164/rccm.201010-1752OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thom K, Petrik J. Progression towards aids leads to increased torque teno virus and torque teno minivirus titers in tissues of hiv infected individuals. Journal of medical virology. 2007;79:1–7. doi: 10.1002/jmv.20756. [DOI] [PubMed] [Google Scholar]
- 19.Madsen CD, Eugen-Olsen J, Kirk O, Parner J, Kaae Christensen J, Brasholt MS, Ole Nielsen J, Krogsgaard K. Ttv viral load as a marker for immune reconstitution after initiation of haart in hiv-infected patients. HIV clinical trials. 2002;3:287–295. doi: 10.1310/8c94-vypq-ng1h-4cnw. [DOI] [PubMed] [Google Scholar]
- 20.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, Bushman FD, Collman RG. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. American journal of respiratory and critical care medicine. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. American journal of respiratory and critical care medicine. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. Development of pcr assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. Journal of clinical microbiology. 2008;46:507–514. doi: 10.1128/JCM.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Y, Leung HC, Yiu SM, Chin FY. Idba-ud: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 24.Krumsiek J, Arnold R, Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 25.Price MN, Dehal PS, Arkin AP. Fasttree 2--approximately maximum-likelihood trees for large alignments. PloS one. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. Qiime allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria.: R Foundation for Statistical Computing. 2011 [Google Scholar]
- 28.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, Bushman FD. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PloS one. 2012;7:e42786. doi: 10.1371/journal.pone.0042786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogilvie LA, Bowler LD, Caplin J, Dedi C, Diston D, Cheek E, Taylor H, Ebdon JE, Jones BV. Genome signature-based dissection of human gut metagenomes to extract subliminal viral sequences. Nature communications. 2013;4:2420. doi: 10.1038/ncomms3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: Next-generation sequencing applied to phage populations in the human gut. Nature reviews Microbiology. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng H, Ye L, Fang X, Li B, Wang Y, Xiang X, Kong L, Wang W, Zeng Y, Ye L, Wu Z, She Y, Zhou X. Torque teno virus (sanban isolate) orf2 protein suppresses nf-kappab pathways via interaction with ikappab kinases. Journal of virology. 2007;81:11917–11924. doi: 10.1128/JVI.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Villiers EM, Borkosky SS, Kimmel R, Gunst K, Fei JW. The diversity of torque teno viruses: In vitro replication leads to the formation of additional replication-competent subviral molecules. Journal of virology. 2011;85:7284–7295. doi: 10.1128/JVI.02472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: Patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 34.Weigt SS, Elashoff RM, Huang C, Ardehali A, Gregson AL, Kubak B, Fishbein MC, Saggar R, Keane MP, Saggar R, Lynch JP, 3rd, Zisman DA, Ross DJ, Belperio JA. Aspergillus colonization of the lung allograft is a risk factor for bronchiolitis obliterans syndrome. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2009;9:1903–1911. doi: 10.1111/j.1600-6143.2009.02635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vos R, Vanaudenaerde BM, Geudens N, Dupont LJ, Van Raemdonck DE, Verleden GM. Pseudomonal airway colonisation: Risk factor for bronchiolitis obliterans syndrome after lung transplantation? The European respiratory journal. 2008;31:1037–1045. doi: 10.1183/09031936.00128607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp MaterialS1
Supp TableS1
Supp TableS2
Supp TableS3
Supp TableS4
Supp TableS5
Supp TableS6