Monoclonal 1- and 3-Phosphohistidine Antibodies: New Tools to Study Histidine Phosphorylation (original) (raw)
. Author manuscript; available in PMC: 2016 Jul 2.
Summary
Histidine phosphorylation (pHis) is well studied in bacteria; however, its role in mammalian signaling remains largely unexplored due to the lack of pHis-specific antibodies and the lability of the phosphoramidate (P-N) bond. Both imidazole nitrogens can be phosphorylated, forming 1-phosphohistidine (1-pHis) or 3-phosphohistidine (3-pHis). We have developed monoclonal antibodies (mAbs) that specifically recognize 1-pHis or 3-pHis; they do not cross-react with phosphotyrosine or the other pHis isomer. Assays based on the isomer-specific auto-phosphorylation of NME1 and phosphoglycerate mutase were used with immunoblotting and sequencing IgG variable domains to screen, select and characterize anti-1-pHis and anti-3-pHis mAbs. Their sequence independence was determined by blotting synthetic peptide arrays, and they have been tested for immunofluorescence staining and immunoaffinity purification, leading to putative identification of pHis-containing proteins. These reagents should be broadly useful for identification of pHis substrates and functional study of pHis using a variety of immunological, proteomic and biological assays.
Introduction
The majority of intracellular proteins are phosphorylated at any given time, and, while 9 of the 20 amino acids can be phosphorylated, the current focus has been on serine (Ser), threonine (Thr), and tyrosine (Tyr) phosphorylation, despite pHis having been first identified over 50 years ago (Boyer, 1962). Ser, Thr, and Tyr all form acid-stable, phosphoester (P-O) bonds upon phosphorylation (Attwood et al., 2007), whereas His forms heat and acid-labile phosphoramidate (P-N) bonds. Phosphospecific antibodies have enabled routine study of phosphoester protein phosphorylation, and the use of MS-based phosphoproteomics has identified thousands of phosphorylation sites in human cells, tissues and tumors. The lack of specific antibodies to study pHis and the relative instability of the P-N bond under typical conditions used for proteomics have made it impossible to determine the prevalence of pHis. Early estimates suggest that pHis could be as abundant as pTyr (Matthews, 1995; Pesis et al., 1988), which comprises ~1% of all known phosphorylation in cells (Hunter and Sefton, 1980; Olsen et al., 2006). Since current biochemical and proteomic technologies have been optimized for preservation, enrichment and detection of the phosphoester amino acids, pHis has remained largely invisible and its importance has likely been underestimated.
A large family of His kinases and downstream signaling proteins, known as two-component regulatory systems, are widely employed by bacteria to link extracellular signals with transcription and chemotaxis. Similar phosphotransfer cascades function in plants to regulate processes such as ripening and circadian rhythms (Matthews, 1995). Its importance in these systems notwithstanding, whether or not pHis plays important roles in vertebrate cell signaling remains unresolved. NME1 and NME2 are the only mammalian protein-His kinases reported to date (Cai et al., 2014; Hartsough et al., 2002; Wagner, 1995) and there is growing evidence implicating these two closely related proteins in cancer and tumor metastasis (Thakur et al., 2011; Tso et al., 2013). Indeed, NME1 (AKA Nm23-H1 or nucleoside diphosphate kinase [NDPK]) was the first candidate metastasis suppressor gene identified (Steeg et al., 1988). NME family members are involved in intracellular nucleotide triphosphate homeostasis as well as in both physiological and pathophysiological cellular processes such as proliferation, differentiation, development, apoptosis, cytokinesis and dynamin-mediated endocytosis (Boissan et al., 2014; Conery et al., 2010). pHis is unique among phosphoamino acids in that two biologically relevant isomers occur. Both imidazole nitrogen atoms (N1 and N3) can be phosphorylated to generate 1-pHis or 3-pHis (Figure 1A). NME family members catalyze transfer of phosphate from ATP onto NDPs through a 1-pHis enzyme intermediate. 3-pHis is used by bacterial His kinases to initiate phosphotransfer cascades and plays a role as an enzyme intermediate for phospholipase D as well as several metabolic enzymes, including phosphoglycerate mutase (PGAM), succinyl-CoA synthetase (SCS) and ATP-citrate lyase (ACLY) (Kee and Muir, 2012). pHis regulatory sites have also been identified in a number of proteins with non-enzymatic functions. For example, phosphorylation of KCa3.1 (His358) and TRPV5 (His711) by NME2 promotes channel activation that is negatively regulated by a pHis-specific phosphatase (PHPT1) (Cai et al., 2014; Srivastava et al., 2006). Phosphorylation of GNB1 (His266) by NME2 activates Gs and regulates basal cAMP accumulation (Wieland et al., 2010). Histone H4 phosphorylation (His18) is highly conserved, and was first observed in eukaryotes over 40 years ago (Besant and Attwood, 2012).
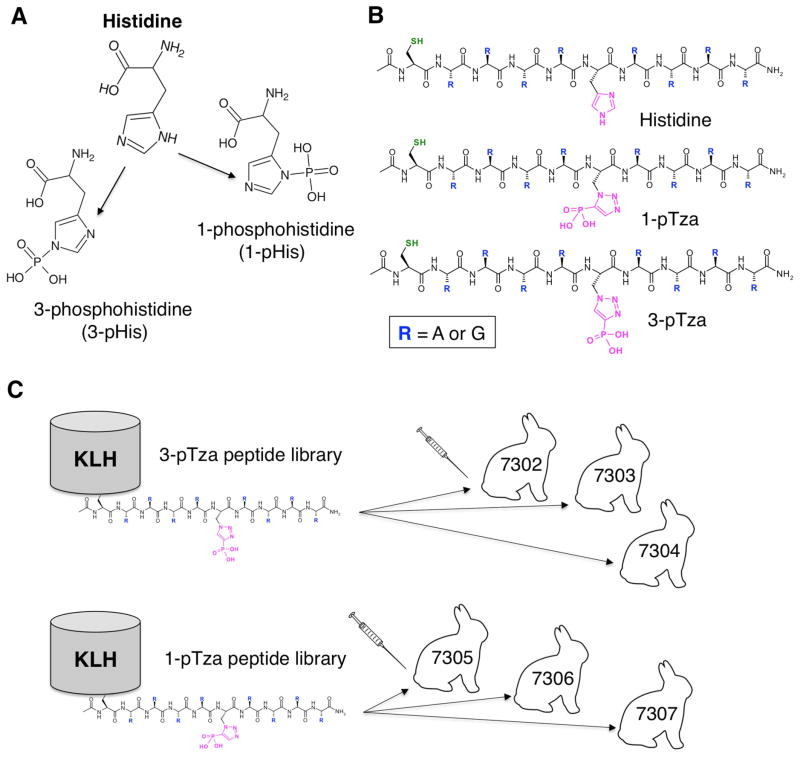
Figure 1. Incorporation of Non-Hydrolyzable Phosphohistidine Analogues into Degenerate Peptide Libraries for Use as Immunogens.
(A) Structure of histidine and the two pHis isomers; 1-phosphohistidine (1-pHis) and 3-phosphohistidine (3-pHis). (B) Structures of the three synthetic peptide libraries used in this study in which either His or a stable pHis mimetic (1-pTza or 3-pTza) is flanked by randomized, neutral amino acids (alanine [A] and glycine [G)]). Each library is composed of 28 = 256 unique peptides (Figure S1A–B), acetylated at the N-terminus and containing L-cysteine (Cys) for chemical ligation to KLH (Ac-Cys.G/A.G/A.G/A.G/A.X.G/A.G/A.G/A.G/A-CONH2). (C) The peptide libraries were conjugated to the carrier protein keyhole limpet hemocyanin (KLH). Three rabbits were immunized with the 3-pTza library (7302, 7303 and 7304) and three rabbits were immunized with the 1-pTza library (7305, 7306 and 7307). See also Figure S1.
Recently, sequence-specific pHis polyclonal antibodies towards pHis18 in histone H4 have been generated (Kee et al., 2010). First and second-generation “pan-pHis” polyclonal antibodies against 3-pHis have also been reported (Kee et al., 2015; Kee et al., 2013). However, these antibodies appear to be limited in their usefulness by their cross-reactivity with pTyr. Isoform-specific pHis mAbs have not yet been developed. We used non-hydrolyzable phosphoryl-triazolylalanine (pTza) pHis analogues (Kee et al., 2010; McAllister et al., 2011) incorporated into degenerate peptide libraries to immunize rabbits and develop selective anti-1-pHis and anti-3-pHis mAbs. We demonstrate that these mAbs do not cross-react with pTyr, appear to detect pHis in a sequence-independent manner and can be used in a variety of immunological assays.
Results
Design of pHis mAb Immunogens: Incorporation of Non-Hydrolyzable pHis Analogues into Degenerate Peptide Libraries
Previous attempts to make pHis antibodies using pHis itself as the antigen have been unsuccessful, presumably because the labile phosphoramidate bond is hydrolyzed too rapidly after immunization to elicit an immune response. The development of non-hydrolyzable, phosphonate analogues of both isomers (1-phosphoryl-triazolylalanine [1-pTza] and 3-phosphoryl-triazolylalanine [3-pTza]) allowed us to develop a strategy for generation of both 1-pHis- and 3-pHis-specific mAbs. We synthesized two peptide libraries consisting of 1-pTza or 3-pTza flanked by randomized sets of alanine (Ala) and glycine (Gly) to serve as immunogens. An unphosphorylated version with His in place of pTza was also synthesized (Figure 1B). MS analysis of the peptide libraries confirmed that the random incorporation of Ala and Gly matched the expected distribution of peptides sharing the same composition of 0–8 Ala and/or Gly residues (Figure S1A–B). The N-terminal Cys was used to couple the pTza libraries to KLH (keyhole limpet hemocyanin) and three rabbits were immunized for each pHis isomer (Figure 1C).
Development of Phospho-NME1 Screening Assay Demonstrates 1-pHis Antibody Specificity
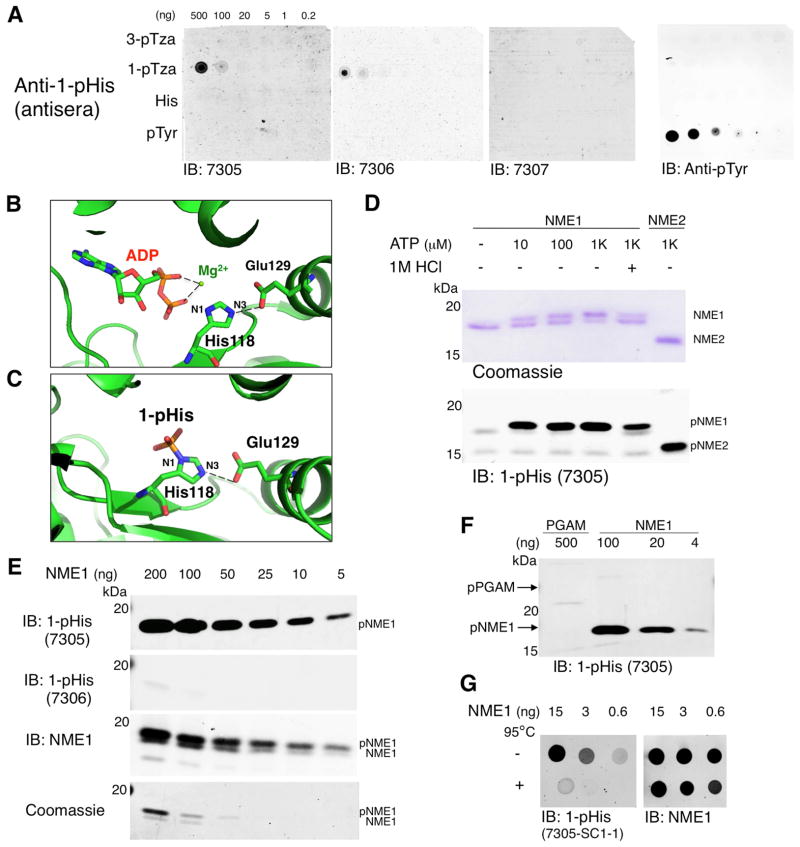
Bleeds from immunized rabbits (7305, 7306 and 7307) were screened by dot blot using the 1-pTza library. To define pHis isoform specificity and control for cross-reactivity, the 3-pTza peptide library, a His control library and a pTyr peptide (Nck pY105) were also included (Figure 2A). Antisera from two rabbits (7305 and 7306) detected only the 1-pTza immunizing library and did not cross-react with pTyr. While the synthetic pTza immunogen peptides were useful for preliminary antisera screening, protein samples with pure 1-pHis or 3-pHis were needed for screening antibodies for isomer specificity. Two in vitro phosphorylation assays capable of rapidly generating either pure 1-pHis or 3-pHis on proteins were developed using NME1/NME2 and PGAM respectively. To generate 1-pHis, we utilized the unique properties of NME1/2, which autophosphorylate on a conserved catalytic His (H118) when incubated with ATP and Mg2+. This occurs solely at the N1 position due to a conserved Glu (E129) that H-bonds with the protonated N3 nitrogen, leaving N1 able to accept the γ-phosphate from ATP (PBD entry 1nsp and 4dy9, Figure 2B–C) (Moréra et al., 1994). Autophosphorylation of recombinant NME1/2 occurred in an ATP dose-dependent fashion up to 1 mM ATP (Figure 2D). Reactions were stopped by addition of 5X pH 8.8 sample buffer and immediately analyzed by a modified SDS-PAGE method without heating (Extended Experimental Procedures). Autophosphorylation on H118 was confirmed by LC-MS/MS with modifications to preserve and detect pHis (Figure S1C and Extended Experimental Procedures). Analysis by immunoblotting and Coomassie staining revealed a phosphorylation-dependent doublet (for NME1 though not NME2) with the slower migrating band representing phospho-NME1 (pNME1). 1 M HCl acid treatment at 37°C or omission of ATP reduced or abolished phosphorylation of NME1 (Figure 2D). Mutagenesis of the catalytic residue H118 prevented phosphorylation of NME1/2 (data not shown).
Figure 2. Screening of 1-pHis Antisera using NME1 in vitro Phosphorylation Assays.
(A) Dot blot screening of 1-pHis antisera. The 1-pTza, 3-pTza and His libraries (Figure 1B) were dissolved in water and 1 μl of 1:5 serial dilutions were spotted directly on nitrocellulose and probed with antisera from rabbits 7305, 7306 and 7307. A synthetic pTyr peptide was included to control for pTyr cross-reactivity and membranes were probed with pTyr mAb 4G10 as a positive control. (B) Ribbon representation of PDB entry 1nsp. NME1 was co-crystallized with ADP and Mg2+ and the hydrogen bond between NME1 E129 with the N3 nitrogen on the catalytic H118 is shown as a dashed line. (C) Phospho-NME1 (PDB entry 4dy9) is shown to highlight that 1-pHis (but not 3-pHis) is formed on the catalytic His residue H118. (D) NME1 and NME2 in vitro autophosphorylation assay. Recombinant NME1 and NME2 were incubated with up to 1 mM ATP at RT for 10 min. Negative controls were performed by omitting ATP or treating with 1M HCl for 15 min at 37°C. Immunoblotting was performed with antiserum from rabbit 7305 and an identical gel was run in parallel and stained with Coomassie blue. (E) 1-pHis detection limit assay. In vitro phosphorylated NME1 was diluted from 200 ng to 5 ng and blotted with 1-pHis antisera from rabbits 7305 and 7306 as well as NME1 antibodies. Identical gels were stained with Coomassie blue. (F) 1-pHis isoform specificity. Recombinant PGAM (Figure 3E) and NME1 were auto-phosphorylated in vitro and blotted with 1-pHis antisera (rabbit 7305). (G) Phospho-NME1 spot blots. In vitro phosphorylation of NME1 was performed as in Figure 2D–E except reactions were stopped with addition of 2% SDS (rather than sample buffer), treated with or without heating to 95°C for 10 min, diluted 1:5 and spotted directly on nitrocellulose. Representative immunoblots with mAb SC1-1 and NME1 antibodies are shown. See also Figure S2.
In vitro phosphorylated NME1 was used to measure the sensitivity of 1-pHis antisera (Figure 2E) and gauge titer as rabbits were boosted with antigen until titers peaked and stabilized. Identical samples were blotted with NME1 antibodies or stained by Coomassie as a loading control. Interestingly, 1-pHis was detected by rabbit 7305 antisera, but not 7306 (Figure 2E), despite both binding 1-pTza peptides (Figure 2A). We also confirmed that 1-pHis antisera did not cross-react with 3-pHis. In vitro phosphorylated PGAM (pPGAM) was analyzed alongside 5- to 125-fold less NME1 (Figure 2F) and no 3-pHis signal was observed. Identical samples blotted with 3-pHis antisera (Figure 3F) served as a positive control for pPGAM detection. A method for rapid analysis of pHis was also developed for screening. pNME1 or pPGAM were directly spotted onto nitrocellulose to avoid potential loss of signal that may occur during SDS-PAGE. This allowed for comparison of mAb sensitivity by spotting limiting dilutions of the phosphoproteins; 1-pHis mAb SC1-1 was able to detect less than 600 pg of pNME1 and signal was abolished when samples were heated at 95°C for 10 min to induce dephosphorylation, whereas NME1 detection was unaffected (Figure 2G).
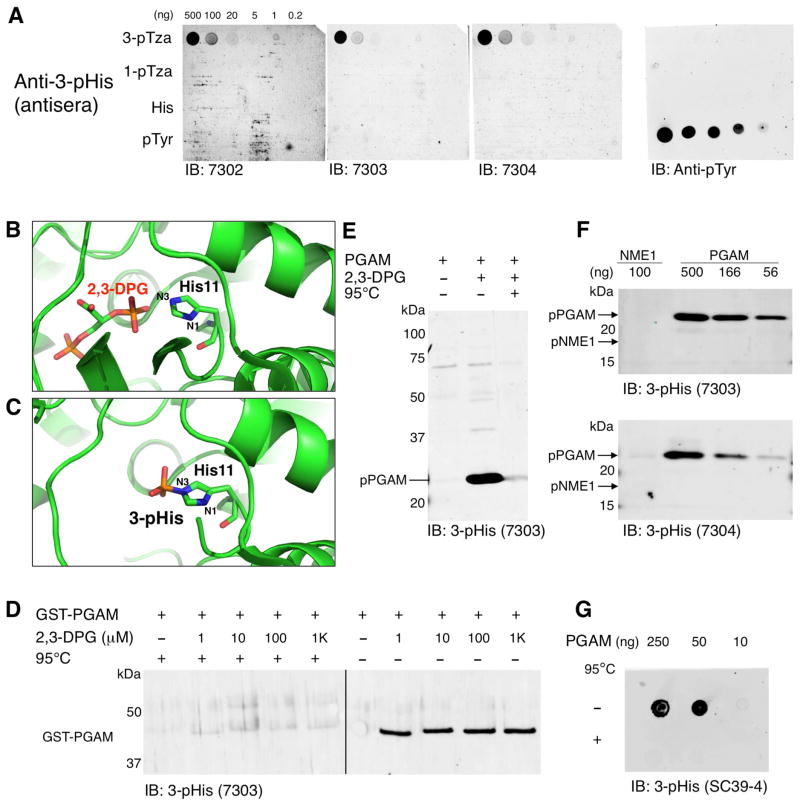
Figure 3. Screening of 3-pHis Antisera by PGAM in vitro Phosphorylation Assays.
(A) Dot blot screening of 3-pHis antisera from rabbits 7302, 7303 and 7304 was performed as described in Figure 2A. (B) Ribbon representation of PDB entry 4hze shows the relative positions of N1 and N3 in H11 of PGAM and its phosphate donor 2,3-DPG. (C) Structure of PGAM (PDB entry 1e58) highlighting 3-pHis formation on the catalytic His residue H11. (D) GST-PGAM fusion protein was auto-phosphorylated in vitro by addition of increasing concentrations of 2,3-DPG. Reactions were stopped by addition of 5X pH 8.8 sample buffer and treated with or without heating to 95°C for 10 min. (E) Purified PGAM was auto-phosphorylated in vitro by incubation with 2,3-DPG for 10 min at 30°C. Reactions were stopped by addition of 5X pH 8.8 sample buffer and treated with or without heat. (F) 3-pHis isoform specificity. Recombinant NME1 and PGAM were auto-phosphorylated in vitro by incubation with ATP or 2,3-DPG respectively and blotted with 3-pHis antisera from rabbits 7303 and 7304. (G) Phospho-PGAM spot blots. In vitro phosphorylation of PGAM was performed as in Figure 3E except reactions were stopped by addition of 2% SDS. Reactions were treated with or without heat, diluted 1:5 and spotted directly on nitrocellulose. A representative immunoblot with 3-pHis mAb SC39-4 is shown. See also Figure S2.
Development of Phospho-PGAM Screening Assay Demonstrates 3-pHis Antibody Specificity
Bleeds from 3-pTza-immunized rabbits (7302, 7303 and 7304) were screened by dot blot (as described for 1-pTza antisera [Figure 2A]) and only the 3-pTza immunizing library was detected (Figure 3A). PGAM is a glycolytic enzyme that converts 3-phosphoglycerate to 2-phosphoglycerate through a 3-pHis phosphoenzyme intermediate (Vander Heiden et al., 2010). Available crystal structures of pPGAM and PGAM co-crystallized with its phosphate donor (2,3-diphosphoglycerate [2,3-DPG]) show that only N3 of H11 is positioned to accept the phosphate from 2,3-DPG (PDB entries 1e58 and 2h4z, Figure 3B–C). To determine if PGAM could be phosphorylated in vitro, GST-PGAM was incubated with 2,3-DPG [1 μM to 1 mM] (Figure 3D). Identical samples were heated at 95°C for 10 min and immunoblotting with 3-pHis antisera revealed a heat-sensitive, 45 kDa band that was absent when 2,3-DPG was omitted. PGAM was subsequently cloned into a bacterial expression vector that allowed cleavage of the GST for analysis of untagged protein. PGAM was purified from E. coli and incubated with or without 2,3-DPG. Autophosphorylation on H11 was confirmed by LC-MS/MS (Figure S1D) and immunoblotting with 3-pHis antisera revealed a heat-sensitive band at 25 kDa (Figure 3E) that was abolished by mutagenesis of H11 (data not shown).
To confirm that 3-pHis antisera did not cross-react with 1-pHis, pNME1 was analyzed alongside pPGAM and no 1-pHis signal was detected (Figure 3F). As we observed for 1-pHis antisera, not all 3-pHis antisera that recognized the pTza analogues could bind pHis. Antisera from rabbits 7303 and 7304 (Figure 3F), but not 7302 detected pPGAM. For this reason we used splenocytes from rabbits 7303 and 7304 to generate hybridomas expressing 3-pHis mAbs in collaboration with Epitomics (Burlingame, CA). To determine 3-pHis mAb sensitivity, in vitro phosphorylated PGAM was spotted directly on nitrocellulose. A representative immunoblot with 3-pHis mAb SC39-4 showed phospho-PGAM was detected down to ~10 ng in a heat-sensitive manner (Figure 3G).
Screening and Sequencing of 1-pHis mAb Hybridomas Yielded Three Unique, Sequence-Independent mAbs
Splenocytes from rabbit 7305 were used for generation of 1-pHis hybridomas, as described for 3-pHis hybridomas. Several thousand hybridoma cell lines generated from rabbit 7305 were initially screened by ELISA in 96-well plates coated with the immunizing 1-pTza peptide library (data not shown). 48 ELISA-positive multiclonal cell lines were selected for secondary screening using several assays, including the pNME1 assays described above. Since pHis is known to be involved in bacterial two-component signaling, we also developed a bacterial based screening assay in which E. coli transformed with a pGEX-NME1 plasmid were induced with IPTG for 3 hr to overexpress the 1-pHis positive control protein, GST-NME1. Crude lysates were loaded on preparative minigels (i.e. a single sample well) and transferred to PVDF membranes, which were clamped into a slot blotting apparatus (Extended Experimental Procedures). Membranes were probed with cell supernatants from the 48 ELISA-positive multiclonal lines (Figure S2A). The top 3 multiclonal cell cultures (MC1, MC50 and MC77) were selected for subcloning based on multiple assays (ELISA, immunoblotting and peptide dot blots, etc.). Similar assays were used to screen subclones for final selection of 1-pHis mAb hybridoma cell lines. For example, small-scale screens were performed using E. coli lysates treated with or without heat (Figure S2B). Identical membranes were probed with crude 1-pHis antisera and the 1-pHis mAbs exhibited significantly decreased background. Since GST-NME1 is highly overexpressed in these cells, it is not surprising that it was the strongest signal detected; however, many other heat-sensitive bands were also detected (Figure S2B). We next sought to determine the extent to which these mAbs could detect 1-pHis in mammalian cells, by performing a similar screen using lysates from a stably transfected FLAG-NME1 293 cell line (Figure S2C). Membranes were probed simultaneously with FLAG-M2 and 1-pHis mAbs (Figure S2D). Multiple heat-sensitive bands were detected, further suggesting that 1-pHis mAbs are able to detect 1-pHis regardless of sequence context. Sequencing of the IgG variable domains (VH and VL) revealed that three unique sequences were encoded by subclones SC1-1, SC50-3 and SC77-11.
Screening and Sequencing of 3-pHis mAb Hybridomas Yielded Three Unique Sequence-Independent and One Sequence-Dependent mAb
Hybridomas generated from combined splenocytes from rabbits 7303 and 7304 were screened by ELISA using the 3-pTza peptide library (data not shown). 30 ELISA-positive multiclonal cell lines were selected for secondary screening using 3-pHis-specific assays including pPGAM as described above. The four best 3-pHis mAb cell lines (MC39, MC44, MC56 and MC60) were subcloned resulting in up to 12 ELISA-positive subclones from each parental multiclone. E. coli transformed with a pGEX-PGAM plasmid were induced and crude lysates were supplemented with 2,3-DPG, spiked with purified, untagged PGAM and loaded on preparative minigels. High throughput, slot blotting was performed as described for 1-pHis mAb hybridomas (Figure S2E). A small-scale screen was performed in parallel using identical E. coli lysates treated with and without heat (Figure S2F). All of the detected bands were heat-sensitive, indicating the mAbs are 3-pHis-specific. Strong signals for the positive control proteins, GST-PGAM and PGAM (untagged) as well as many other heat-sensitive bands were detected by SC39s, SC56s and SC60s, suggesting that these mAbs lack strong sequence specificity, but do not produce an identical pattern of bands. In contrast, SC44s primarily detected a strong band corresponding to bacterial SCS (Figure S2E–F). SCS also uses a 3-pHis phospho-enzyme intermediate (Fraser et al., 2000), and SC44s detected both bacterial SCS and mammalian SCS and ACLY (Figure S2G), which share the sequence motif; G-H-A-G-A (Figure S2H). Cell lysates prepared from a stably transfected HEK293 cell line expressing FLAG-NME1 were blotted with 3-pHis mAb SC39-4 and 3-pHis on endogenous pPGAM was detected, but not 1-pHis on FLAG-NME1, indicating the 3-pHis mAbs do not cross-react with 1-pHis generated in vivo (Figure S2I). Sequence analysis of 3-pHis mAb VH and VL domains allowed us to identify SC39-4, SC56-2 and SC60-2 as unique, sequence-independent 3-pHis mAbs, as well as a distinct sequence-dependent mAb SC44-8 that has bias towards the A/G motif present in SCS and ACLY.
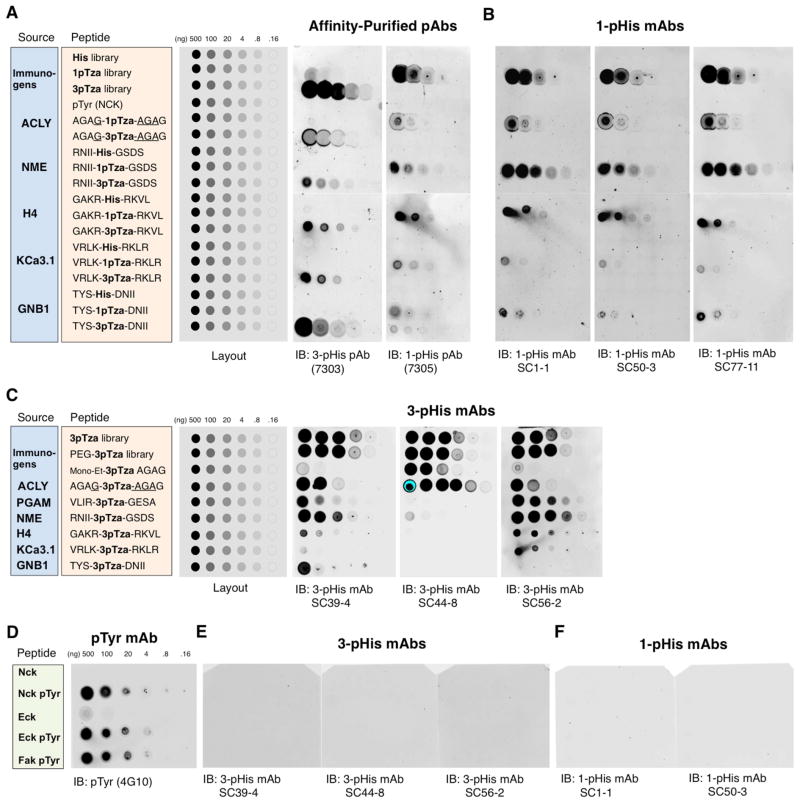
1-pHis and 3-pHis Antibodies Detect Various pTza Peptides of Defined Sequence
Synthetic 1-pTza and 3-pTza peptide dot blot arrays were used to confirm the pHis isoform specificity of the pHis mAbs and determine if they have any local amino acid sequence specificity. We synthesized peptides of defined sequence based on the best-characterized mammalian pHis proteins; ACLY, NME1, PGAM, histone H4, KCa3.1 and GNB1. Peptides corresponding to the pHis sites in these proteins were synthesized with either His, 1-pTza or 3-pTza flanked by 4 amino acids on either side (Figure S3 and Table S2). Serial dilutions of each peptide and the immunizing pTza and control His peptide libraries were spotted onto nitrocellulose and blotted with affinity-purified, polyclonal 1-pHis or 3-pHis antibodies. The 3-pHis antibodies bound only the 3-pTza peptides and the 1-pHis antibodies bound only the 1-pTza peptides, regardless of sequence (Figure 4A). Identical membranes were probed with 1-pHis mAbs as part of our screening process to select mAbs with the broadest sequence recognition (Figure 4B). The 1-pHis mAbs displayed similar binding profiles and detected as little as 1 ng of the NME1/2 H118 peptide. Each of the 1-pTza peptides tested was detected, suggesting these mAbs will be useful for detecting 1-pHis in a broad range of sequence contexts. Since 3-pHis mAbs did not cross-react with either 1-pTza or His peptides (Figure 4A), we probed peptide arrays consisting of just 3-pTza peptides, including a PGAM peptide, to determine their sequence specificity (Figure 4C). In contrast to the 1-pHis mAbs, the 3-His mAbs displayed some variation in binding profiles. 3-pHis mAb SC39-4 was able to detect all 3-pTza peptides down to 800 pg; however, binding to the KCa3.1 peptide was relatively poor (100 ng). 3-pHis mAb SC56-2 showed similar binding characteristics; however, it was better at detecting the KCa3.1 peptide (4 ng) while worse at binding the GNB1 peptide. SC44 detected the A/G motif peptide (based on ACLY) and the immunizing peptide library down to 160 pg and 800 pg respectively, confirming its sequence bias.
Figure 4. 1-pHis and 3-pHis mAbs Detect Isomer-Specific pTza Peptides but not pTyr.
Synthetic peptide dot blot arrays consisting of the His, 1-pTza or 3pTza libraries (Figure 1B), a pTyr (NCK) peptide and peptides with either His, 1-pTza or 3pTza incorporated into defined sequences (based on the pHis protein substrates; ACLY, NME1/2, histone H4, KCa3.1 and GNB1) were spotted on nitrocellulose and probed with: (A) affinity-purified polyclonal 3-pHis (7303) or 1-pHis (7305) antibodies or (B) 1-pHis mAbs SC1-1, SC50-3 and SC77-11. Peptide layouts, sequences and their sources are shown in the grey, orange and blue boxes respectively. (C) 3-pTza peptide dot blot characterization of 3-pHis mAbs. Peptide layouts, sequences and their sources are shown in the grey, orange and blue boxes respectively. A partially-deprotected, mono-ethyl ester version of the ACLY-based peptide (AGAG-mono-Et-3-pTza-AGAG) was also included. (D–F) Synthetic pTyr peptide dot blots. Peptides based on Nck, Eck and FAK were spotted on nitrocellulose and probed with: (D) pTyr mAb 4G10, (E) 3-pHis mAbs SC39-4, SC44-8 and SC56-2 or (F) 1-pHis mAbs SC1-1 and SC50-3. See also Figure S3 and Extended Experimental Procedures.
pHis mAbs Do Not Cross-React with pTyr Peptides
Since some of the first described pTyr mAbs cross-reacted with pHis (Frackelton et al., 1983) and recently reported polyclonal pHis antibodies displayed only a 10-fold higher selectivity for pHis over pTyr (Kee et al., 2013), we tested our pHis mAbs for cross-reactivity using synthetic pTyr peptides. Serial dilutions of pTyr peptides (Nck, and the Eck/EphA2 and FAK tyrosine kinases) were spotted on nitrocellulose along with their unphosphorylated counterparts. The pTyr mAb 4G10 detected only the pTyr peptides (Figure 4D), whereas none of the peptides were detected by 3-pHis (Figure 4E) or 1-pHis mAbs (Figure 4F).
pHis mAbs Do Not Cross-React with pTyr Proteins in Cell Lysates
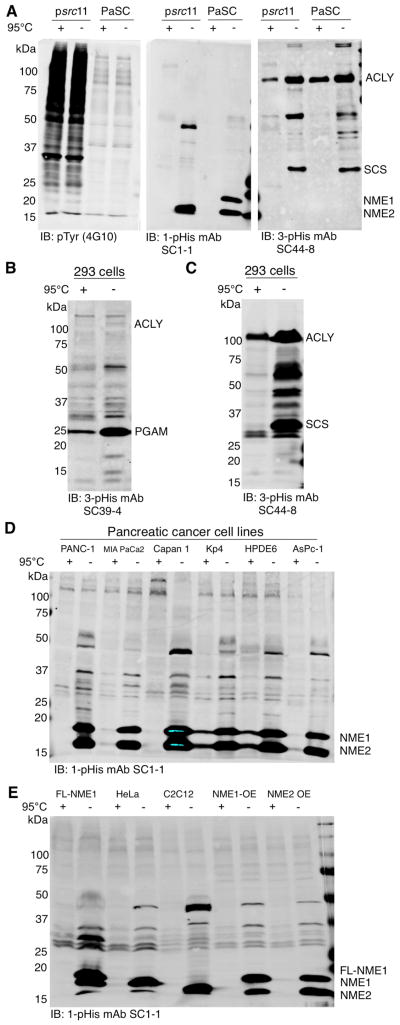
To test for pTyr cross-reactivity of our pHis mAbs on cell lysates, cultures of v-Src-transformed NIH/3T3 fibroblasts (p_src_11(Johnson et al., 1985)) were pre-incubated with 1 mM orthovanadate for 30 min to enhance pTyr signals. Non-transformed fibroblasts (pancreatic stellate cells PaSC) were tested in parallel as a negative control. To preserve pHis in cell lysates for analysis by immunoblotting, we adopted a modified SDS-PAGE method to maintain the sample pH above 8 to stabilize pHis (Extended Experimental Procedures). As expected, pTyr mAb 4G10 detected an elevated signal in the p_src_11 cells but not in the PaSC negative control cells, but neither the 1-pHis nor 3-pHis mAbs detected the elevated pTyr signal in p_src_11 cells (Figure 5A).
Figure 5. pHis Proteins, but not pTyr, are Detected in Mammalian Cells.
(A) Src-transformed and non-transformed fibroblast cell lines (psrc11) and pancreatic stellate cells (PaSCs) were analyzed by immunoblotting with pTyr, 1-pHis or 3-pHis mAbs. Cells were pre-treated with 1 mM sodium orthovanadate for 30 min prior to lysis. (B–C) HEK 293 cell lysates were immunoblotted with 3-pHis mAbs SC39-4 and SC44-8. (D–E) Pancreatic cancer cell lysates, FLAG-NME1 293, HeLa, C2C12 and NME1 or NME2 over-expressing (OE) melanoma cells were immunoblotted with 1-pHis mAb SC1-1. All lysates (A–E) were prepared by scraping cells into 2x pH 8.8 sample buffer and treated with or without heating for 10 min. See also Figure S4.
pHis mAbs Detect pHis Proteins in Mammalian Cell Lysates
A number of heat-sensitive bands were detected in the p_src_11 and PaSC lysates by 1- and 3-pHis mAbs. We next immunoblotted lysates from a variety of other mammalian cell lines to characterize the levels of pHis in different cell types. 3-pHis mAbs SC39-4 (Figure 5B) and SC44-8 (Figure 5C) were used to blot lysates from 293 cells, and 1-pHis mAb SC1-1 was used to blot lysates from several pancreatic cancer (PC) cell lines and HPDE6, a normal pancreatic epithelial cell line (Figure 5D). Common patterns of heat-sensitive bands were observed indicating many proteins in these cancer cell lines are similarly regulated by 1-pHis modification. Lysates from FLAG-NME1 293, HeLa, C2C12, NME1 and NME2 over-expressing (OE) melanoma cell lines (Hamby et al., 2000) were also blotted with 1-pHis mAb SC1-1 (Figure 5E). 1-pHis was detected on NME family members in mouse, human and bacterial cells including; NME1, NME2, NME4, NME5 (Figure S4A–C) and NME7 (data not shown), despite differing sequences flanking the pHis residue (Figure S4A), as well as the E. coli NME1 homolog NDK (Figure S4C). While the major pHis proteins detected appear to be known enzymes (i.e. NME1/2 [1-pHis], PGAM, SCS and ACLY [3-pHis], the detection of many unidentified, heat-sensitive bands, particularly by the 3-pHis mAbs, suggests they will be useful for identification of many pHis substrates.
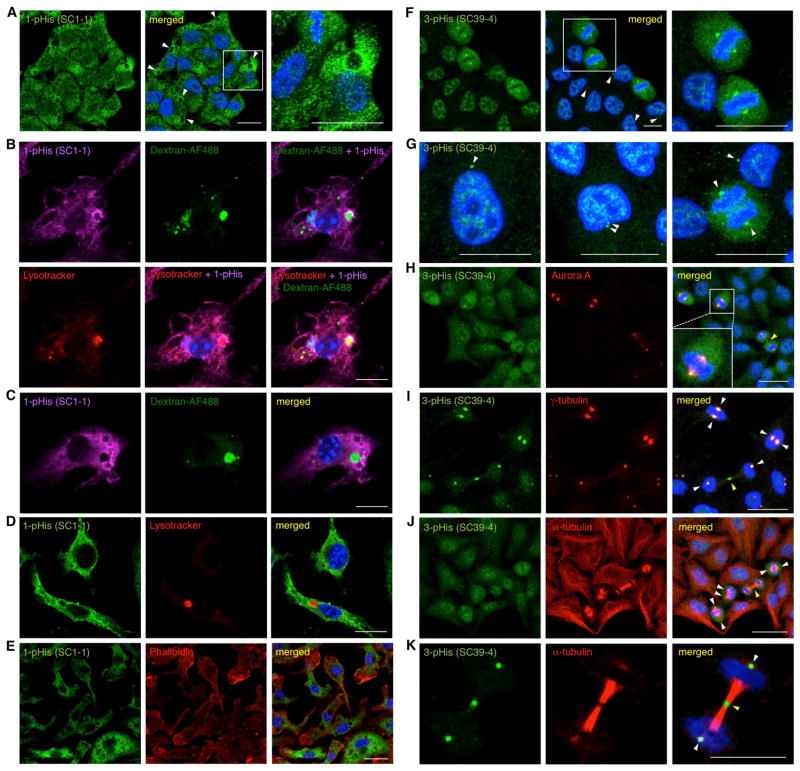
Immunofluorescence Staining Reveals Association of 1-pHis with Outer Membrane of Phagosomes
To test the ability of these mAbs to detect pHis proteins by immunofluorescence staining, HeLa cells were stained with the 1-pHis mAb SC1-1. We observed a distinct staining pattern in which most cells had a large (1–2 μm) compartment that stained strongly in the surrounding region but lacked interior pHis staining (Figure 6A). We surmised these might be acidic compartments such as phagosomes or autophagosomes, and proceeded to test this hypothesis by using primary murine macrophages to look for specific staining of phagosomes. Macrophages isolated from bone marrow were incubated with fluorescently-labeled dextrans to track internalization into phagosomes. Cells were also incubated with LysoTracker prior to fixation to label acidic compartments. 1-pHis staining was absent in nuclei as well as the interior of phagosomes in macrophages co-labeled with the internalized dextrans and LysoTracker, but staining was pronounced in the regions surrounding these compartments (Figure 6B–D). Remodeling of the actin cytoskeleton supports the extension of pseudopodia at sites of particle engulfment and F-actin assembles around nascent phagosomes. Co-staining with mAb SC1-1 and phalloidin-TRITC revealed a lack of co-localization of 1-pHis with actin filaments (Figure 6E).
Figure 6. 1-pHis mAbs Negatively Stain Macrophage Phagosomes and 3-pHis mAbs Stain Centrosomes and Spindle Poles in HeLa Cells.
(A) HeLa cells were fixed with PFA and stained with 1-pHis mAb SC1-1. White arrows indicate acidic compartments. (B) Macrophages were fed Dextran-AF488 and labeled with LysoTracker for 60 min prior to fixation with PFA and staining with 1-pHis mAb SC1-1 was detected by Cy5 conjugated secondary antibodies. Bar, 10 μm. (C) Macrophages were incubated with Dextran-AF488 for 60 min and staining with mAb SC1-1 was detected by Cy5-conjugated secondary antibodies. (C) Macrophages were labeled with LysoTracker for 60 min prior to fixation and mAb SC1-1 staining was detected by AF-488 conjugated secondary antibodies. (E) Co-staining of macrophages with mAb SC1-1 and Phalloidin-TRITC. (F–K) HeLa cells were fixed with; PFA (F–G), or pre-permeabilized with 0.5% Triton X-100 and fixed with PFA (I and K) or methanol (H and J) and stained with 3-pHis mAb SC39-4 alone (F–G) or co-stained with Aurora A (H), γ-tubulin (I) or α-tubulin (J–K) antibodies. (F) Metaphase cells are shown in an expanded view in the right panel. (G) From left to right, interphase, an early prophase and anaphase cells. (H–K) Cells in metaphase, prometaphase and telophase are shown. White arrows indicate centrosomes and spindle poles and yellow arrows indicate midbodies in telophase cells. Nuclei were visualized with DAPI. Size Bar, 20 μm. See also Figure S5.
3-pHis mAb Immunofluorescence Reveals Staining of Centrosomes, Spindle Poles and Midbodies
Macrophages stained with 3-pHis mAbs displayed a pattern distinct from 1-pHis staining. Punctate structures were observed throughout the cytosol; however, no co-localization was observed when antibodies specific for organelle markers (e.g. ATP Synthase, LC3, Rab5, α-tubulin and LAMP1, (Figure S5A–B and data not shown) were tested for co-staining. In contrast to macrophages, staining of HeLa cells with 3-pHis mAbs was primarily nuclear (though curiously absent from nucleoli) and distinctive cell cycle-dependent patterns were observed. Cells in prometaphase through telophase displayed remarkable 3-pHis staining of spindle poles (Figure 6F–K). Interphase cells displayed staining of centrosomes and cells in prophase were observed with duplicated centrosomes (Figure 6G). An apparent burst of 3-pHis signals was observed in dividing cells and this seemed to last from prometaphase through anaphase. To confirm this observation, HeLa cells were co-stained with 3-pHis mAbs and spindle pole markers Aurora-A and γ-tubulin (Figure 6H–I). To demonstrate that 3-pHis mAbs stained primarily spindle poles and not spindles, cells were co-stained with α-tubulin (Figure 6J). 3-pHis mAbs also stained structures devoid of Aurora-A, γ-tubulin and α-tubulin in both HeLa and U2Os cells and these appeared to be the midbody of cells in late telophase. (Figure 6H–K and S5C–E). A series of negative controls using the immunizing pTza peptide libraries were performed. Only the 1-pTza peptides could block 1-pHis staining (Figure S5F–I, P–Q) while only the 3-pTza peptides could block 3-pHis staining (Figure S5K–N, R–S). Additionally, boiling slides for 10 min in citrate buffer reduced both 1-pHis and 3-pHis staining (Figure S5J and O).
Enrichment and Identification of Proteins by pHis mAb Immunoaffinity Purification and SILAC LC-MS/MS
Traditional immunoprecipitation methods are not amenable to pHis preservation and detection. A method for immunoaffinity purification of pHis substrates using immobilized pHis mAbs was developed. Reusable pHis mAb resins were packed in chromatography columns and used to enrich pHis phosphoproteins from cell lysates prior to analysis by LC-MS/MS (Extended Experimental Procedures). pNME1 and pPGAM were used to test the pHis isomer selectivity of the high density mAb columns. NME1 and PGAM were phosphorylated in vitro, denatured (6 M urea, pH 10), mixed together and incubated with either a 1-pHis or 3-pHis mAb column. Purification fractions were immunoblotted with 1- and 3-pHis mAbs as well as NME1 and PGAM antibodies and quantification demonstrates that pNME1 was enriched in elution fractions from the 1-pHis mAb column while pPGAM was enriched in elutions from the 3-pHis mAb column (Figure S6A–B).
In order to determine which proteins bind the columns in a pHis-dependent manner and which are likely false positives that bind non-specifically, we used stable isotope labeling by amino acids in cell culture (SILAC) to metabolically label FLAG-NME1 293 cells. Initially, both the light and heavy labeled (Arg 13C6/15N4 and Lys 13C6) cells were lysed using identical denaturing and alkaline conditions (6 M urea, pH 10) to preserve pHis. The heavy lysates were then acidified (pH 6) and heated at 65°C for 30 min to reduce pHis. A dramatic decrease of pHis in the heavy lysates was confirmed by immunoblotting (data not shown). Both lysates were neutralized, diluted (1:5 to 1 M urea, pH 8), mixed together and one half was passed over the 1-pHis mAb column while the other half was passed over the 3-pHis mAb column. LC-MS/MS analysis was performed on tryptic peptides derived from proteins that eluted off the columns at pH 11. A SILAC ratio (light/heavy) was calculated for each peptide to determine which proteins were enriched in the untreated, light vs. heavy lysates that had been treated to reduce pHis. We observed significant enrichment of NME1/2 (55-fold) by the 1-pHis mAb column as well as enrichment of PGAM (4-fold) and other known 3-pHis proteins including; histone H4 (22-fold) and ACLY (11-fold). Proteins corresponding to recently identified pHis phosphopeptides (Lapek et al., 2015) including; TUBB, TCP1/CCT1, YWHAB, LDHA, RPS3A and GAPDH were also enriched from 5 to 11-fold (Tables 1 and S1). In all, 630 proteins (58% of proteins quantified in 1-pHis column elutions E1–E3) and 506 proteins (54% of 3-pHis E1–E3) were enriched by at least 2-fold (Table S1) for a total of 786 different proteins. 280 of these were unique to the 1-pHis column and 156 were unique to the 3-pHis column. Gene Ontology analysis by biological process (DAVID v6.7(Huang da et al., 2009)) revealed 97 of the 786 genes are involved in cell cycle processes including; PP1, CDK1, cyclin B1, CUL1 and multiple proteasome subunits. Network analysis (STRING v9.1(Franceschini et al., 2013)) was performed to visualize the protein-protein interaction network of these cell cycle related genes (Figure S6C) with known pHis proteins (Table 1).
Table 1.
SILAC Ratios Determined by LC-MS/MS Analysis Indicate Enrichment of Known pHis Proteins by pHis mAb Immunoaffinity Purification
| Protein | Sequence | Site | 1-pH E1 | 1-pH E2 | 3-pH E1 | 3-pH E2 |
|---|---|---|---|---|---|---|
| NME1 b | QVGRNIIHGSDSVES | H118 | 15.05 | 51.86 | 3.18 | 2.14 |
| NME2 b | QVGRNIIHGSDSVKS | H118 | 15.59 | 55.96 | 3.46 | - |
| NME4 | HISRNVIHASDSVEG | H151 | - | - | - | - |
| NME5 a | DDLRNALHGSNDFAA | H127 | - | - | - | - |
| NME6 a | TDTRNTTHGSDSVVS | H129 | - | - | - | - |
| NME7 | DGIRNAAHGPDSFAS | H206 | - | - | - | - |
| Histone H4 b | GKGGAKRHRKVLRDN | H18 | 19.14 | 9.8 | 22.82 | 17.9 |
| KCa3.1 a | FRQVRLKHRKLREQV | H358 | - | - | - | - |
| TRPV5 a | LRQNTLGHLNLGLNL | H711 | - | - | - | - |
| GNB1 b | QELMTYSHDNIICGI | H226 | - | - | - | 1.21 |
| PGAM | YKLVLIRHGESAWNL | H11 | - | 4.25 | 4.01 | 2.44 |
| ACLY | SSEVQFGHAGACANQ | H760 | 5.29 | 7.2 | 11.62 | 2.99 |
| SCS | PPGRRMGHAGAIIAG | H299 | - | 1.68 | - | - |
| P-Selectin a | GKCPLNPHSHLGTYG | H771 | - | - | - | - |
| TUBB b | GNNWAKGHYTEGAEL | H105 c | 3.55 | 4.49 | 4.33 | 2.09 |
| TCP1 b | EETERSLHDAIMIVR | H346 c | 6.10 | 5.99 | 5.98 | 3.20 |
| YWHAB b | KTALCFRHLMKQLLN | H202 c | - | 2.04 | 7.43 | 1.63 |
| LDHA b | GEMMDLQHGSLFLRT | H67 c | 4.18 | 6.50 | 6.60 | 3.08 |
| RPS3A | LGKLMELHGEGSSSG | H232 c | 7.21 | 9.10 | 10.41 | 4.98 |
| GAPDH | TMEKAGAHLQGGAKR | H111c | 3.40 | 4.45 | 4.76 | 3.32 |
Discussion
Our goal was to develop tools for the study of pHis in mammalian cells and to begin to determine its importance as a regulatory process in normal cells and in diseases, such as cancer. Our immunoblotting and proteomic data (Tables 1 and S1) suggest that these mAbs can recognize pHis independent of sequence context. However, little is known regarding the prevalence and relative levels of pHis proteins in mammalian cells, making it unclear whether all pHis substrates are recognized. For example, NME1/2 were the major cellular 1-pHis proteins detected by the 1-pHis mAbs, but this could reflect either a sequence bias of our 1-pHis mAbs (perhaps influenced by our screening approach) or simply that abundant expression levels of NME1/2 combined with their ability to auto-phosphorylate results in stronger detection by immunoblotting.
As is the case with pTyr blotting, activation of specific signaling pathways or enrichment of certain proteins by fractionation or immunoprecipitation prior to immunoblotting may be necessary to visualize low abundance, organelle-specific or protein substrates with low stoichiometry of phosphorylation. The greater thermodynamic instability of 1-pHis vs. 3-pHis may also contribute to the difficulty in detection of some 1-pHis proteins. While detection of pNME1/2 by 1-pHis mAb immunoblotting is quite heat sensitive, brief heat treatment may not be harsh enough to reduce pHis in all proteins. There is a prominent, heat-resistant doublet detected in most of these cell lines (Figure 5D–E) that we suspect is NME4, the mitochondrial NME family member (Figure S4B).
Striking differences were observed between 1-pHis and 3-pHis mAb immunofluorescence staining. SC1-1 will be useful for studying acidic compartments (i.e. phagosomes, autophagosomes and lysosomes) since staining is absent from the interior of these acidic compartments and often very bright in the surrounding regions. Analysis of the “phagosome proteome” identified NME2 as an outer membrane associated phagosomal protein (Garin et al., 2001). Dynamin-2 is required for macrophage phagocytosis (Gold et al., 1999), and NME1/2 were recently shown to interact with dynamin and dynamin-induced tubules and be required for efficient dynamin-mediated endocytosis by providing a localized source of GTP (Boissan et al., 2014). The strong 1-pHis signals we observed in regions surrounding phagosomes (Figure 6A–E) could be the local, enzymatic activity of NME1/2 working to replenish GTP. Alternatively, the 1-pHis signals could reflect the presence of non-enzymatic, pHis protein substrates that are phosphorylated by NME1/2 or an unknown His kinase.
The 786 enriched proteins (Table S1) represent a starting point for identification and analysis of the mammalian pHis proteome. We preserved existing pHis on proteins in the lysates as fully as possible (i.e. denaturing and alkaline conditions to prevent protein phosphatase driven- or acid-catalyzed hydrolysis). However, we do not yet know what conditions or stimuli induce or increase pHis (analogous to EGF or orthovanadate treatment for pTyr) to increase the yield of pHis proteins. As it stands, the most abundant proteins with the highest stoichiometry of His phosphorylation and stability are most likely to have been observed and quantified. Therefore, the complete list of pHis substrates is likely to be significantly longer than what we have detected. The enrichment of cell cycle proteins by pHis mAbs is of great interest, since we also observe specific staining of mitotic structures by 3-pHis mAbs (i.e. spindle poles, centrosomes and midbodies (Figure 6F–K and S5C–E) that indicates pHis signals might be critical for regulating multiple cell cycle events. The 97 cell cycle related proteins analyzed by STRING v9.1 have multiple interaction nodes that fall into 7 main categories; proteasome, cytokinesis, ribosome, cell cycle regulation, protein phosphatase PP1, chromosome condensation and chromatin organization and spindle organization (Figure S6C). In addition, gene ontology analysis (DAVID v6.7) by biological process revealed that 239 of the enriched proteins correspond to genes involved in nucleic acid related processes including (in order of P-value); translation (116), translational elongation (65), RNA processing (89), ribonucleoprotein complex biogenesis (46), RNA splicing (56) and ribosome biogenesis (35) suggesting these may also be dependent on pHis signaling. In this regard, His phosphorylation (unlike pSer, pThr and pTyr) results in a positive to negative charge switch that may help regulate binding of proteins to nucleic acids.
While these pHis mAbs are isoform specific when used for immunoblotting and immunofluorescence, there is significant overlap of proteins (350 or 45%) that bound to the 1-pHis and 3-pHis mAb columns (Tables 1 and S1). We suspect that base-catalyzed isomerization of pHis may occur under the high pH conditions in which the proteomic experiments were performed (Gonzalez-Sanchez et al., 2013). We have made the initial steps towards using these mAbs to study the mammalian pHis proteome; however, there are many other cell lines and tissues to investigate and further optimization of our pHis mAb immunoaffinity purification and LC-MS/MS methods is necessary to allow the purification of pHis phosphopeptides and direct identification of specific sites of His phosphorylation by MS.
If the mature field of phosphoester phosphorylation is any guide, the nascent and slow-growing field of His phosphorylation will benefit from the development of these immunological reagents and methods, but still missing is the development of pHis phosphatase inhibitors and a full accounting of cellular pHis phosphatases, kinases and substrates. These mAbs will accelerate progress in all of these areas and help determine the importance of pHis in human health and disease.
Experimental Procedures
Synthesis of 1-pTza and 3-pTza Peptides and Degenerate Peptide Libraries
Detailed methods for chemical synthesis and analysis of the immunizing pTza peptide libraries can be found in Extended Experimental Procedures. In general, pTza analogues were synthesized by solid phase peptide synthesis according to according to published procedures (McAllister and Webb, 2012).
NME and PGAM in vitro Phosphorylation Assays
In vitro autophosphorylation of purified NME1 and NME2 (10–30 ng/μl) was performed by incubating in TMD buffer (20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM DTT) at RT for 10 min. Reactions were stopped by addition of 5X pH 8.8 sample buffer and analyzed immediately by modified SDS-PAGE (Extended Experimental Procedures). Autophosphorylation of PGAM was performed as described for NME1 except 2,3-DPG was used as the phosphate donor instead of ATP and incubations were carried out at 30°C. Reactions lacking ATP or 2,3-DPG or treated briefly with heat or acid served as negative controls. Heat treatment was performed after addition of 5X pH 8.8 sample buffer for 10–15 min at 95°C. Acid treatment was performed by adding 25 μl 1 N HCl to a 100 μl reaction and incubating at 37°C for 15 min. Reactions were neutralized with 25 μl 1 N NaOH.
Peptide Dot Blots
Peptide dot blots were used to screen rabbit antisera titer and characterize mAb specificity. The 1-pTza and 3-pTza peptide libraries, His control library and a pTyr peptide (Nck pY105) were dissolved in water at 1 mg/ml. 1:5 serial dilutions (500, 100, 20, 5, 1 and 0.2 ng/μl) were prepared for each peptide and 1 μl was spotted on nitrocellulose and allowed to dry for 1–2 hr at RT. Membranes were blocked 1 hr at RT in Casein Blocking Buffer (0.1% casein, 0.2x PBS −/−) and incubated with rabbit antisera (diluted 1:1,000 in Blocking Buffer with 0.1% Tween-20) for 1 hr at RT or overnight at 4°C. pTza peptide dot blots were also used to characterize pHis mAbs. 1-pTza, 3-pTza or His was incorporated into synthetic peptides of defined sequences (Extended Experimental Procedures) from mammalian proteins with mapped pHis sites (Table 1). Peptides were dissolved in water at 1 mg/ml. 1:5 serial dilutions were prepared and spotted as described above.
Immunofluorescence
Primary murine macrophages were differentiated from bone marrow progenitors (Zhang et al., 2008) plated on cover slips and incubated O/N in fresh medium. Cells were incubated with 10 μg/ml Oregon Green-Dextran®488 and/or LysoTracker (50 nM) for 1–2 hr prior to fixation with 4% PFA for 10 min. Negative controls were performed by boiling slides for 5–10 min in 0.01 M citrate buffer or by pre-incubation of pHis mAbs with pTza blocking peptides [5 μg/ml]. Cells were permeabilized in blocking buffer (PBS, 5% serum (2nd Ab species), 2% BSA, 0.1% Tween) with 0.1% Triton-X100 for 1 hr at 4°C. Primary antibodies were diluted to 1 μg/ml in blocking buffer and incubated with slides for 2 hr at 4°C. Slides were washed 5X with cold PBS + 0.1% Tween and incubated with 2nd Ab diluted 1:400 in blocking buffer for 1 hr at 4°C. Slides were mounted on cover slips after washing 5X with cold PBS + 0.1% Tween. See also Extended Experimental Procedures for immunostaining of HeLa cells.
Supplementary Material
1
2
3
4
5
6
7
8
Acknowledgments
Acknowledgement of funding sources: S.R.F. NIH training grant (2 T32 CA009370) and a Salk Institute Innovation Grant. This work was supported by USPHS R01 grants to T.H. (CA80100, CA82683, CA194584) and made use of the Peptide Synthesis and Biophotonics Cores supported by P30CA14195. The Helmsley Center for Genomic Medicine also provided support. T.H. is a Frank and Else Schilling American Cancer Society Professor, and holds the Renato Dulbecco Chair in Cancer Research. A.Z. Human Frontiers Science Program. G.L. NIH R01 NS085296. J.R.Y. NCRR grant (5P41RR011823-17) and NIGMS grant (8P41GM103533-17). We thank Joseph Kim and Ronghua Li at Sanofi for peptide synthesis.
Footnotes
Author Contributions
S.R.F. conceived of and performed experiments, developed methods and pHis mAb screening assays, characterized mAbs, cultured hybridomas, performed pAb and mAb purification and wrote the paper. J.M. performed peptide synthesis, assisted with rabbit immunization strategy and discussed results. A.A. performed pHis mass spectrometry analysis in **J.R.Y.**’s lab at TSRI. T.H. conceived of degenerate peptide strategy and experiments and edited manuscript. L.M. HeLa and U2OS cell IF. A.Z. performed pHis mAb IF experiments on macrophages in **G.L.**’s lab at Salk. Our collaborators at Sanofi designed and synthesized the pTza peptide libraries; J.M. and A.B. designed pTza peptide library synthesis, J.M., M.S. and F.A. synthesized pTza analogues and peptides. All authors reviewed and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attwood PV, Piggott MJ, Zu XL, Besant PG. Focus on phosphohistidine. Amino acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- Besant PG, Attwood PV. Histone H4 histidine phosphorylation: kinases, phosphatases, liver regeneration and cancer. Biochem Soc Trans. 2012;40:290–293. doi: 10.1042/BST20110605. [DOI] [PubMed] [Google Scholar]
- Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M, Sauvonnet N, Lagache T, Lascu I, Raposo G, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–1515. doi: 10.1126/science.1253768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PD. Identification of Phosphohistidine in Digests from a Probable Intermediate of Oxidative Phosphorylation. J Biol Chem. 1962:3306–3308. [PubMed] [Google Scholar]
- Cai X, Srivastava S, Surindran S, Li Z, Skolnik EY. Regulation of the epithelial Ca2+ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014;25:1244–1250. doi: 10.1091/mbc.E13-04-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conery AR, Sever S, Harlow E. Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal stability by activating the GTPase dynamin during cytokinesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15461–15466. doi: 10.1073/pnas.1010633107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frackelton AR, Ross AH, Eisen HN. Characterization and use of monoclonal antibodies for isolation of phosphotyrosyl proteins from retrovirus-transformed cells and growth factor-stimulated cells. Mol Cell Biol. 1983;3:1343–1352. doi: 10.1128/mcb.3.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser ME, James MNG, Bridger WA, Wolodko WT. Phosphorylated and Dephosphorylated Structures of Pig Heart, GTP-specific Succinyl-CoA Synthetase. J Mol Biol. 2000;299:1325–1339. doi: 10.1006/jmbi.2000.3807. [DOI] [PubMed] [Google Scholar]
- Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. The Journal of cell biology. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. Dynamin 2 is required for phagocytosis in macrophages. The Journal of experimental medicine. 1999;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sanchez MB, Lanucara F, Helm M, Eyers CE. Attempting to rewrite History: challenges with the analysis of histidine-phosphorylated peptides. Biochemical Society transactions. 2013;41:1089–1095. doi: 10.1042/BST20130072. [DOI] [PubMed] [Google Scholar]
- Hamby CV, Abbi R, Prasad N, Stauffer C, Thomson J, Mendola CE, Sidorov V, Backer JM. Expression of a catalytically inactive H118Y mutant of nm23-H2 suppresses the metastatic potential of line IV Cl 1 human melanoma cells. International journal of cancer Journal international du cancer. 2000;88:547–553. doi: 10.1002/1097-0215(20001115)88:4<547::aid-ijc5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PJ, Coussens PM, Danko AV, Shalloway D. Overexpressed pp60c-src can induce focus formation without complete transformation of NIH 3T3 cells. Molecular and cellular biology. 1985;5:1073–1083. doi: 10.1128/mcb.5.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee JM, Muir TW. Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS chemical biology. 2012;7:44–51. doi: 10.1021/cb200445w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee JM, Oslund RC, Couvillon AD, Muir TW. A second-generation phosphohistidine analog for production of phosphohistidine antibodies. Organic letters. 2015;17:187–189. doi: 10.1021/ol503320p. [DOI] [PubMed] [Google Scholar]
- Kee JM, Oslund RC, Perlman DH, Muir TW. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol. 2013;9:416–421. doi: 10.1038/nchembio.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee JM, Villani B, Carpenter LR, Muir TW. Development of Stable Phosphohistidine Analogues. J Am Chem Soc. 2010;132:14327–14329. doi: 10.1021/ja104393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapek JD, Jr, Tombline G, Kellersberger KA, Friedman MR, Friedman AE. Evidence of histidine and aspartic acid phosphorylation in human prostate cancer cells. Naunyn-Schmiedeberg’s archives of pharmacology. 2015;388:161–173. doi: 10.1007/s00210-014-1063-4. [DOI] [PubMed] [Google Scholar]
- Matthews H. Protein Kinases and phosphatases that act on Histidine lysine or arginine residues in eukaryotic proteins a possible regulator of the MAPK cascade. Pharmac Ther. 1995;67:232–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]
- McAllister TE, Nixa MG, Webb ME. Fmoc-chemistry of a stable phosphohistidine analogue. Chem Commun. 2011;47:1297–1299. doi: 10.1039/c0cc04238b. [DOI] [PubMed] [Google Scholar]
- McAllister TE, Webb ME. Triazole phosphohistidine analogues compatible with the Fmoc-strategy. Org Biomol Chem. 2012;10:4043–4049. doi: 10.1039/c2ob25517k. [DOI] [PubMed] [Google Scholar]
- Moréra S, LeBras G, Lascu I, Lacornbe ML, Véron M, Janin J. Refined X-ray Structure of Dictyostelium discoideum NDPK at 1.8 Å resolution. J Mol Biol. 1994;243:873–890. doi: 10.1006/jmbi.1994.1689. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pesis KH, Wei Y, Lewis M, Matthews HR. Phosphohistidine is found in basic nuclear proteins of Physarum polycephalum. FEBS letters. 1988;239:151–154. doi: 10.1016/0014-5793(88)80563-5. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Li Z, Ko K, Choudhury P, Albaqumi M, Johnson AK, Yan Y, Backer JM, Unutmaz D, Coetzee WA, et al. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol Cell. 2006;24:665–675. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a Novel Gene Associated With Low Tumor Metastatic Potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- Thakur RK, Yadav VK, Kumar P, Chowdhury S. Mechanisms of non-metastatic 2 (NME2)-mediated control of metastasis across tumor types. Naunyn-Schmiedeberg’s archives of pharmacology. 2011;384:397–406. doi: 10.1007/s00210-011-0631-0. [DOI] [PubMed] [Google Scholar]
- Tso PH, Wang Y, Yung LY, Tong Y, Lee MM, Wong YH. RGS19 inhibits Ras signaling through Nm23H1/2-mediated phosphorylation of the kinase suppressor of Ras. Cellular signalling. 2013;25:1064–1074. doi: 10.1016/j.cellsig.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner PD. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. The Journal of biological chemistry. 1995 doi: 10.1074/jbc.270.37.21758. [DOI] [PubMed] [Google Scholar]
- Wieland T, Hippe HJ, Ludwig K, Zhou XB, Korth M, Klumpp S. Reversible Histidine Phosphorylation in Mammalian Cells. Methods in Enzymology. 2010;471:379–402. doi: 10.1016/S0076-6879(10)71020-X. [DOI] [PubMed] [Google Scholar]
- Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 14. Chapter 14. 2008. p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8