Labeling of fusion proteins with synthetic fluorophores in live cells (original) (raw)
Abstract
A general approach for the sequential labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase (AGT) with different fluorophores in mammalian cells is presented. AGT fusion proteins with different localizations in the cell can be labeled specifically with different fluorophores, and the fluorescence labeling can be used for applications such as multicolor analysis of dynamic processes and fluorescence resonance energy transfer measurements. The facile access to a variety of different AGT substrates as well as the specificity of the labeling reaction should make the approach an important tool to study protein function in live cells.
Advances in fluorescence microscopy and the isolation and engineering of different autofluorescent proteins have revolutionized the way protein function is studied in the living cell (1–3). Beyond localization studies, fusion proteins of autofluorescent proteins can be used to study dynamic processes, follow conformational changes, and detect protein–protein interactions (1, 2, 4). Despite this enormous range of applications for autofluorescent fusion proteins, there are two reoccurring limitations of the approach. The first limitation is the restriction to the fluorophores of the existing autofluorescent proteins. Properties such as emission and excitation wavelength, extinction coefficient, and photostability are characteristic properties of each autofluorescent protein that do not necessarily match the requirements of the envisioned application. The second limitation is caused by the fusion of the autofluorescent protein to the protein of interest. In general, autofluorescent proteins possess a molecular mass of >25 kDa, and certain autofluorescent proteins furthermore form oligomers, raising the question of to what extent the creation of a fusion protein affects the function of the protein of interest (1, 3).
Alternative approaches to make proteins amenable to fluorescence studies in live cells include introduction of chemically labeled proteins through invasive techniques such as microinjection, site-specific incorporation of unnatural fluorescent amino acids, and the selective labeling of fusion proteins (5–10). An example of the latter approach is the so-called tetracysteine tag, which reacts with biarsenical fluorophores to form stable and highly fluorescent complexes (7, 8). We recently developed an approach that allows for the labeling of fusion proteins of human O6-alkylguanine-DNA alkyltransferase (AGT) with synthetic molecules (9). The labeling is based on the irreversible and specific reaction of AGT with O6-benzylguanine (BG) derivatives, leading to the transfer of the synthetic probe to a reactive cysteine residue (Fig. 1). Wild-type human AGT is a monomeric protein of 207 aa, the 30 C-terminal residues of which can be deleted without affecting the activity against BG, making it slightly smaller than autofluorescent proteins (11). Importantly, the rate of the reaction of AGT fusion proteins with BG derivatives is independent of the nature of the label, opening up the possibility to label a single AGT fusion protein with a variety of different probes (9, 12–14). Specific labeling of AGT fusion proteins in mammalian cells can be achieved by using AGT-deficient cell lines, and we have shown previously that nuclear-localized AGT can be labeled specifically with fluorescein in such cells (9). Here we demonstrate that synthetic BG derivatives can be used for the sequential labeling of AGT fusion proteins with a variety of different fluorophores in live mammalian cells and how the approach can be used for multicolor analysis of cellular processes and fluorescence resonance energy transfer (FRET) measurements.
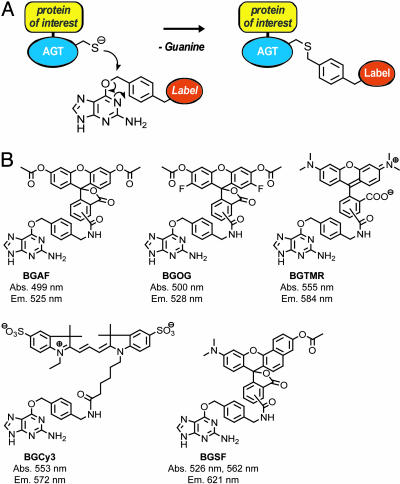
Fig. 1.
General mechanism of (A) and substrates used for (B) fluorescence labeling of AGT fusion proteins. (A) Covalent labeling of AGT fusion proteins with BG derivatives. (B) Structures of BG derivatives used for fluorescence labeling of AGT fusion proteins. The maxima in the absorbance (Abs.) and fluorescence emission spectra (Em.) of the substrates after reaction with AGT are listed; the data for BGAF, BGOG, and BGSF are those of the deacetylated molecules.
Materials and Methods
Syntheses of AGT Substrates. All substrates for the labeling of AGT fusion proteins were prepared by reacting a previously described BG derivative with commercially available _N_-hydroxysuccinimide esters of the corresponding fluorophores (9). SNARF-1 (SF), fluorescein, Oregon green (OG), and tetramethylrhodamine (TMR) were used as mixtures of isomers. Reactions were performed at ambient temperature in _N,N_-dimethylformamide in the presence of 1 eq of triethylamine. AGT substrates were purified by reversed-phase HPLC on a C18-column by using linear gradients from 0.1% trifluoroacetic acid to acetonitrile. After solvent evaporation substrates were dissolved in dimethyl sulfoxide and stored at -20°C. Each AGT substrate was analyzed by high-resolution mass spectrometry and purity-checked by HPLC. Electrospray ionization (ESI)-MS (m/z) calculated for BGTMR [C38H35N8O5]+: 683.2730; found: 683.2747. ESI-MS (m/z) calculated for BGOG [C38H27F2N6O9]]+: 749.1807; found: 749.1807. ESI-MS (m/z) calculated for BGSF [C42H34N7O7]+: 748.2519; found: 748.2524. ESI-MS (m/z) calculated for BGCy3 [C44H50N8O8S2]+: 882.3193; found: 882.3203. For the determination of the fluorescence spectra of the fluorophores bound to AGT, purified glutathione _S_-transferase (GST)-AGT fusion protein (10 μM) was incubated with an excess of the corresponding AGT substrate (20 μM) in 50 mM Hepes/1 mM DTT, pH 7.2. After 1 h, excess AGT substrate was removed through gel filtration by using a NAP-10 column (Amersham Pharmacia Biosciences), and fluorescence spectra were recorded on a Spectramax Gemini plate reader (Molecular Devices). Absorption and fluorescence spectra for BG- and AGT-bound fluorophores as well as their relative quantum yields are shown in the supporting information, which is published on the PNAS web site.
Construction of AGT Fusion Proteins. The AGT mutant used throughout this study is based on human AGT, which was modified by the following amino acid changes: Lys-125 → Ala, Ala-127 → Thr, Arg-128 → Ala, Asn-157 → Gln, and Ser-159 → Glu (14). The mutations at positions 157 and 159 have been shown to increase the activity of AGT against BG, and the mutations at positions 125, 127, and 128 were introduced to disrupt the interaction of the protein with DNA (14, 15). For targeting AGT to different subcellular localizations, the following constructs were made. For expression of farnesylated AGT (AGT-CaaX), the peptide RSKLNPPDESGPGCMSCKCVLS containing a CaaX motif was fused to the C terminus of AGT (16). The corresponding gene was inserted into the _Nhe_I and _Eco_RI sites of the vector pEGFP-F (Clontech). For expression of AGT-β-galactosidase (β-Gal), AGT (residues 1–177) was fused via an RSGSG linker to the N terminus of β-Gal. The corresponding gene was inserted into the _Nhe_I and _Xba_I sites of the vector pECFP-Nuc (Clontech). For expression of the thermosensitive glycoprotein of vesicular stomatitis virus (tsVSVG)-AGT, AGT was fused to the C terminus of tsVSVG via an SGLRS linker (17). The corresponding gene was inserted into the _Nhe_I and _Bam_HI sites of the vector pECFP-Nuc (Clontech). For expression of AGT-α-tubulin, AGT was fused to the N terminus of human α-tubulin via an RSR linker by ligating the AGT gene into the vector pEGFP-Tub (Clontech) by using the _Nhe_I and _Bgl_II restriction sites. For the expression of AGT-neurokinin-1 (NK1), the signal sequence of the 5-HT3 receptor (Sig5HT3) was fused to the N terminus of AGT via a DYV linker, and NK1 was fused to the C terminus of AGT via a TS linker (18, 19). In the resulting construct, a FLAG tag and a 6×-His tag were also attached to the C terminus of NK1. The corresponding gene of the fusion protein was inserted into the _Nhe_I and _Bam_HI sites of the vector pCEP4 (Invitrogen). For the expression of AGT-EGFP-NLS3, AGT was fused to the N terminus of enhanced GFP (EGFP) via a GSG linker, and three consecutive simian virus 40 nuclear localization sequences (NLS3) were attached to the C terminus of EGFP (20). The corresponding gene was inserted into the _Nhe_I and _Bam_HI sites of the vector pECFP-Nuc (Clontech).
Fluorescence Labeling of AGT Fusion Proteins. Chinese hamster ovary (CHO)-9-neo-C5, deficient of AGT, and HEK293 were grown in DMEM/F12 supplemented with 2.2% FCS (GIBCO/BRL) (21). Transient transfection was performed as described (9). For the labeling experiments, all AGT substrates were added to a final concentration of 5 μM in PBS. BG linked to diacetylated fluorescein (BGAF), BGOG, and BGCy3 were added at room temperature for 5 min before washing the cells three times with PBS. Cells were imaged in PBS 30 min after washing using appropriate filter sets for each fluorophore. The labeling with BGSF was followed online for 1–2 h at room temperature. If necessary, excess BGSF was removed through washing steps as described above. For labeling with BGTMR, cells were incubated for at least 1.5 h before excess fluorophore was removed through washing steps.
Laser-scanning confocal micrographs were recorded by using a 488-nm Ar/Kr laser line or a 543- or 633-nm HeNe laser line on a Zeiss LSM 510 microscope (Zeiss) with an ×63 water (1.2 numerical aperture) objective. Scanning speed and laser intensity were adjusted to avoid photobleaching of the fluorescent probes and damage or morphological changes of the cells. The microscope was equipped with a microcultivation system (Incubator S, CTI controller 3700 digital, Zeiss) to control temperature, humidity, and CO2 for maintaining physiological conditions during long-term labeling experiments. For the determination of the stability of AGT fusion proteins, transient transfected CHO cells expressing AGT-CaaX or AGT-NLS3 were labeled with BGAF as described and then incubated in medium containing 10 μM BG. For AGT-CaaX, the fluorescence intensity of at least 12 randomly picked cells was determined 70, 180, and 400 min after labeling (supporting information). For AGT-NLS3, the fluorescence intensity of the nucleus of at least 20 randomly picked cells was determined 80, 160, 285, 430, and 510 min after labeling (supporting information).
Results and Discussion
Fluorophores for the Labeling of AGT Fusion Proteins. AGT fusion proteins offer the possibility to label individual fusion proteins with a variety of different fluorophores. Although we have shown previously that AGT fusion proteins can be labeled specifically with fluorescein, fluorophores exhibiting higher photostability and higher emission wavelengths would be advantageous for functional studies in living cells (9). We therefore synthesized AGT substrates in which BG is linked to the fluorophores OG, TMR, SF, and Cy3 (Fig. 1). All AGT substrates were prepared in one step from a common precursor. The OG and SF derivatives BGOG and BGSF were synthesized as the corresponding acetates to increase cell permeability. Inside the cell, the nonfluorescent acetates are hydrolyzed by esterases, liberating the fluorophore. Except for BGCy3, all of the fluorescent AGT substrates proved to be cell-permeable, allowing for the labeling of nuclear-localized AGT (AGT-NLS3) in AGT-deficient CHO cells (Fig. 2 A_–_C). In general, labeling comprises incubation of the cell with the AGT substrate for a period characteristic for each substrate and subsequent washing steps to remove excess fluorophores. An exception is BGSF, for which fluorescence labeling of AGT-NLS3 with SF can be followed online without the need to remove excess dye through washing steps (Fig. 2_C_). This can be rationalized assuming that intracellular BGSF is scavenged by AGT-NLS3 before it is hydrolyzed to liberate the fluorophore. A subsequent washing step can then be used to further decrease background fluorescence.
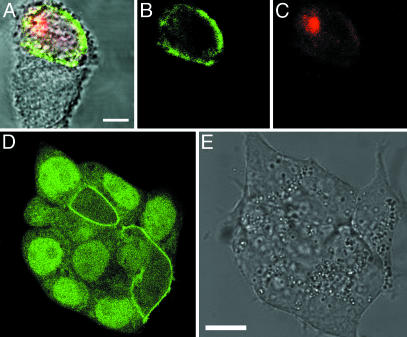
Fig. 2.
Transiently transfected CHO cells expressing various AGT fusion proteins labeled with fluorescent BG derivatives. (A) AGT-NLS3 labeled with OG using BGOG. (B) AGT-NLS3 labeled with TMR using BGTMR. (C) AGT-NLS3 labeled with SF using BGSF. (D) AGT-β-Gal labeled with fluorescein using BGAF. (E) AGT labeled with fluorescein. (F) EGFP. (G) AGT-CaaX labeled with TMR. (H) AGT-α-tubulin labeled with fluorescein. (I) tsVSVG-AGT labeled with fluorescein. (Scale bars, 10 μm.)
Fluorescence Labeling of Different AGT Fusion Proteins. To test the scope of our labeling approach for live-cell applications, we first investigated the fluorescence labeling of AGT fusion proteins at different subcellular localizations. The AGT fusion proteins investigated included β-Gal, α-tubulin, a farnesylation motif, the tsVSVG, and the G protein-coupled receptor NK1.
When β-Gal was fused to the C terminus of AGT, fluorescein labeling of the corresponding AGT-β-Gal fusion in CHO cells revealed a diffuse cytosolic localization of the fusion protein (Fig. 2_D_). These data indicate that fusion of AGT to cytosolic proteins does not lead to active transport of these fusions into the nucleus of a cell, an important prerequisite for functional studies of cytosolic proteins. It should be noted that the predominant nuclear localization of wild-type human AGT is due to its affinity for DNA and that the AGT mutant used throughout this study possesses mutations that interfere with its interaction with DNA and enhance its activity against BG (12, 14). Both EGFP and fluorescein-labeled AGT display an almost even distribution between cytosol and nucleus when expressed without any fusion partners (Fig. 2 E and F).
As an example for a cytoskeletal protein, a fusion between AGT and α-tubulin was transiently expressed in CHO cells. Labeling with fluorescein revealed that AGT-α-tubulin is incorporated into microtubules, demonstrating that both AGT and α-tubulin are functional in the fusion protein (Fig. 2_H_). The image furthermore indicates that labeled AGT-α-tubulin is also present to some extent in the cytosol. It has been shown previously that, in CHO cells, ≈40% of total tubulin is present in the cytosol (22). In addition, analyzing CHO cells transiently expressing the corresponding EGFP-α-tubulin fusion protein revealed a similar distribution of EGFP-α-tubulin (supporting information). To investigate whether AGT fusion proteins can be localized at the cytoplasmic side of the plasma membrane, we fused a peptide possessing a CaaX farnesylation motif to the C terminus of AGT. The farnesylation of AGT leads to its functional insertion into the plasma membrane, as visualized by fluorescence staining with TMR (Fig. 2_G_). AGT was also fused to the cytoplasmic C terminus of the temperature-sensitive glycoprotein of vesicular stomatitis virus (tsVSVG-AGT). tsVSVG is a membrane protein that is transported via the secretory pathway to the plasma membrane at permissive temperatures, whereas at the nonpermissive temperature of 40°C, the protein reversibly misfolds and is retained in the endoplasmic reticulum (23). Transiently expressing tsVSVG-AGT in CHO cells at the permissive temperature of 34°C leads to efficient transport of the fusion protein to the plasma membrane, as demonstrated by labeling with fluorescein (Fig. 2_I_). We then examined whether AGT fusion proteins, despite possessing a reactive cysteine, can also be functionally expressed on the extracellular side of the plasma membrane. Toward this end, AGT was fused to the exoplasmic N terminus of the G protein-coupled NK1 receptor (AGT-NK1) (19). HEK293 cells transiently expressing AGT-NK1 could be labeled with fluorescein as well as with the nonpermeable dye Cy3, indicating a preserved activity of extracellular AGT toward its substrate (Fig. 3 A and B). The use of a nonpermeable AGT substrate for extracellular applications has the advantage that both endogenous AGT as well as not properly translocated or internalized AGT-NK1 will not be labeled. This differentiation between different receptor populations is not possible when using protein fusions with autofluorescent proteins. To investigate whether AGT-NK1 is functional, the binding of the TMR-labeled, NK1-specific ligand substance P (SP-rho) to cells expressing AGT-NK1 was studied. Fluorescence signals resulting from labeling of AGT-NK1 with fluorescein and binding of its cognate ligand SP-rho overlapped (Fig. 3 A and C). In addition, fluorescence labeling resulting from binding of SP-rho could be readily reversed by adding excess of unlabeled SP, indicating preserved functionality of the AGT-NK1 receptor in terms of ligand binding (Fig. 3_D_). Ligand-induced activation of the AGT-NK1 fusion protein also couples to the intracellular Ca2+ signaling pathway. Binding of SP (1 μM) to HEK293 cells coexpressing the AGT-NK1 receptor and the promiscuous Gαq protein elicited a transient Ca2+ response, with a rapid increase in intracellular Ca2+ concentration and slower decay (10–20 s) back to baseline as detected by confocal microscopy using a calcium-sensitive fluorescence indicator (unpublished data) (24). These experiments demonstrate that the localization of the investigated AGT fusion proteins was determined exclusively by either specific targeting sequences or the natural subcellular localization of the fusion partner to which the AGT was genetically attached.
Fig. 3.
Extracellular (A_–_D) and FRET (E_–_I) applications of AGT fusion proteins. (A_–_D) Transiently transfected HEK293 cells expressing AGT-NK1. (A) AGT-NK1 labeled with fluorescein. (B) AGT-NK1 labeled with Cy3 using BGCy3. (C) Binding of SP-rho (50 nM) to the same cell as in A. (D) Time-resolved binding of SP-rho (50 nM) to an HEK293 cell displaying AGT-NK1 and its replacement through the addition of unlabeled ligand (2.5 μM) at the indicated time (black arrow). (E_–_I) Transiently transfected CHO cells expressing AGT-EGFP-NLS3 labeled with SF for FRET detection. (E) Overlay of transmission and fluorescence channel for EGFP before labeling with SF (excitation 488 nm/emission 505–530 nm). (F) Fluorescence channel for SF before labeling with SF (excitation 488 nm/emission > 650 nm). (G) Fluorescence channel for EGFP 60 min after labeling with SF (excitation 488 nm/emission 505–530 nm). (H) Overlay of transmission and fluorescence channel for SF after labeling with SF (excitation 488 nm/emission > 650 nm). (I) Relative fluorescence intensities (RFI) of the EGFP channel before and after labeling with SF. Columns E and G correspond to the fluorescence intensities (RFI) of the EGFP channel of the cell shown in E and G. Columns J and K correspond to the fluorescence intensities (RFI) of the EGFP channel of a cell before and after incubation with SF but previously incubated with BG (20 μM). (Scale bars 10 μm.)
A characteristic property of AGT is that it is degraded after transfer of the alkyl group from O6-alkylated guanine-DNA to the reactive cysteine. It is believed that a conformational change of the alkylated protein triggers its degradation, presumably through ubiquitinylation of the alkylated protein and subsequent proteolysis (25). To investigate both the rate of degradation of AGT fusion proteins and to what extend the degradation after labeling interferes with fluorescence measurements, the decrease in fluorescence intensity for two AGT fusions proteins (AGT-NLS3 and AGT-CaaX) was determined over a period of 7 h. During these measurements, the fluorescence intensity for both proteins decreased, but not more than 12% per hour (supporting information). After 7 h of incubation, the intensity of the fluorescence signal for AGT-NLS3 and AGT-CaaX was ≈30% and 50%, respectively, of the value measured directly after the labeling. The observed rate of degradation is comparable with that measured in earlier experiments in which the fate of alkylated wild-type hAGT was followed by Western blotting using anti-hAGT antibodies (26). These data demonstrate that labeled AGT fusion proteins can be followed inside living cells for several hours after the labeling.
FRET and Multicolor Analysis. FRET measurements between pairs of (auto)fluorescent proteins are an important tool to investigate the spatial distance between proteins in a time-resolved manner (27). Furthermore, various reporter systems to monitor cellular parameters are based on intramolecular FRET between two autofluorescent proteins fused in a single polypeptide chain (1). To demonstrate the use of AGT fusion proteins in FRET applications, we constructed a fusion protein between EGFP and AGT-NLS3 (AGT-EGFP-NLS3). Here, EGFP would be the donor (488 nm) and SF-labeled AGT would be the acceptor for intramolecular FRET. The broad absorbance of SF-labeled AGT in the region from 500 to 580 nm at pH 7.4 makes it an ideal acceptor in such experiments (supporting information). Labeling of AGT-EGFP-NLS3 with 5 μM BGSF for 1 h led to a 98% decrease in EGFP emission at 505–530 nm, indicating both efficient FRET and an efficient labeling of AGT-EGFP-NLS3 with SF (Fig. 3 E_–_I). Because SF can also be excited to some extent at 488 nm, we assume that the observed emission >650 nm is caused by both direct laser excitation and FRET (Fig. 3_H_). Preincubation of cells with BG (20 μM) completely prevented the quenching of the EGFP fluorescence of AGT-EGFP-NLS3 after incubation with BGSF (Fig. 3_I_). Furthermore, simultaneous expression of AGT-NLS3 and EGFP-NLS3 as two separate proteins and subsequent labeling with SF did not lead to any significant quenching of the fluorescence of EGFP-NLS3 (unpublished data). Potential applications of intramolecular FRET between an autofluorescent protein and a fluorescence-labeled AGT are as reporter systems for measuring biochemical events in live cells (1). Here, such a FRET pair might prove superior to the currently used donor–acceptor pair of enhanced cyan fluorescent protein and yellow fluorescent protein (1). In addition, these experiments demonstrate that AGT fusion proteins in general are well suited for FRET applications.
Sequential labeling of proteins with different fluorophores offers the possibility to distinguish older and newer copies of the same protein within one cell through multicolor analysis. Multicolor analysis using the tetracysteine tag and biarsenical dyes has been applied to study the dynamics of gap-junction channels (28). To demonstrate the feasibility of multicolor analysis of AGT fusion proteins, we attempted to differentiate tsVSVG-AGT populations expressed before and after a temperature shift through sequential labeling with different fluorophores (Fig. 4). In these experiments, CHO cells transiently expressing tsVSVG-AGT were incubated for 20 h at 34°C and subsequently labeled with fluorescein. To quench residual AGT fusion protein not labeled with fluorescein, cells were incubated for 15 min with the AGT inhibitor O6-(4-bromothenyl)-guanine (29). The temperature of the medium then was shifted to 40°C, which should lead to misfolding of newly synthesized tsVSVG-AGT and its retention in the endoplasmic reticulum. After incubation of the cells for 75 min at 40°C, tsVSVG-AGT synthesized at this temperature was labeled with SF. Subsequent fluorescence imaging of cells demonstrated that fluorescein-labeled tsVSVG-AGT was located predominantly in the plasma membrane (77%), whereas SF-labeled tsVSVG-AGT was predominantly located in internal membrane structures (71%), most likely perinuclear endoplasmic reticulum or Golgi (Fig. 4 A_–_C) (17, 30). The exact localization of retained tsVSVG-AGT was not characterized further. The labeling and washing at 40°C unavoidably lead to temperature fluctuations, and because tsVSVG rapidly folds at permissive temperatures and remains transport-competent once it exits the endoplasmic reticulum, one would expect a heterogeneous distribution of retained tsVSVG-AGT (31, 32). Despite the ambiguities with respect to the localization of retained tsVSVG-AGT, the data clearly demonstrate that within live cells older and newer copies of AGT fusion proteins can be discriminated by sequential labeling with different fluorophores. In addition to sequential labeling, AGT fusion proteins can also be labeled simultaneously with two different fluorophores. If fluorophores with different photostability would be used, such double-labeling experiments could be exploited to study dynamic processes by using fluorescence localization after photobleaching (33). The experiments suggest that multicolor analysis of AGT fusion proteins has the potential to become an important tool to study dynamic processes in live cells.
Fig. 4.
Multicolor analysis of tsVSVG-AGT and labeling of AGT-CaaX in cells possessing endogenous AGT. (A_–_C) Sequential labeling of tsVSVG-AGT: Labeling with fluorescein at permissive temperature (34°C) and with SF at nonpermissive temperature (40°C). (A) Overlay of transmission and fluorescence micrographs. (B) Fluorescence channel for fluorescein-labeled tsVSVG-AGT (excitation 488 nm/emission 505–530 nm). (C) Fluorescence channel for SF-labeled tsVSVG-AGT (excitation 543 nm/emission > 650 nm). (D and E) Labeling of AGT-CaaX in HEK293 cells possessing endogenous AGT. (D) Fluorescence channel after labeling with fluorescein (excitation 488 nm/emission 505–530 nm). (E) Transmission channel of same cells as in G. (Scale bar, 10 μm.)
Labeling in Cell Lines Expressing Endogenous AGT. The scope of applications for the fluorescence labeling of AGT fusion proteins would be significantly broadened if AGT fusion proteins could be labeled specifically in cells expressing endogenous wild-type AGT. Because of the 20-fold higher activity of the here-used AGT mutants relative to wild-type AGT, a specific labeling of (overexpressed) AGT fusion proteins in cells possessing wild-type AGT is already possible in certain cases. An example is the fluorescein labeling of transiently expressed AGT-CaaX in HEK293 cells: Transfected cells can be identified by a fluorescent plasma membrane, whereas nontransfected cells display a fluorescent nucleus (Fig. 4 D and E). We assume that the specificity of the labeling observed in the transfected cells, i.e., the absence of a fluorescent nucleus, relies on the increased activity of AGT-CaaX relative to wild-type AGT and a substoichiometric labeling.
Conclusions
AGT fusion proteins can be labeled specifically in a sequential manner with synthetic fluorophores in live cells. The localization of the investigated fusion proteins was determined exclusively by either specific targeting sequences or the natural subcellular localization of the fusion partner to which the AGT was genetically attached. The stability of fluorescence-labeled AGT fusion proteins allows following their fate over periods of hours. Importantly, a single AGT fusion protein can be (sequentially) labeled with a variety of different fluorophores for applications such as multicolor analysis or FRET applications. These features should make AGT fusion proteins an important tool for fluorescence-based studies of protein function in live cells.
Supplementary Material
Supporting Information
Acknowledgments
We thank Jan Ellenberg for advice and for providing the gene of tsVSVG. This work was supported by Swiss National Science Foundation Grant 3152A0-100103 (to K.J.) and by the TopNano program (H.V.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AGT, O6-alkylguanine-DNA alkyltransferase; BG, O6-benzylguanine; FRET, fluorescence resonance energy transfer; OG, Oregon green; TMR, tetramethylrhodamine; SF, SNARF-1; β-Gal, β-galactosidase; tsVSVG, thermosensitive glycoprotein of vesicular stomatitis virus; NK1, neurokinin-1; EGFP, enhanced GFP; NLS3, three consecutive simian virus 40 nuclear localization sequences; CHO, Chinese hamster ovary; AF, diacetylated fluorescein; SP-rho, NK1-specific ligand substance P.
References
- 1.Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3**,** 906-918. [DOI] [PubMed] [Google Scholar]
- 2.Lippincott-Schwartz, J. & Patterson, G. H. (2003) Science 300**,** 87-91. [DOI] [PubMed] [Google Scholar]
- 3.Miyawaki, A., Sawano, A. & Kogure, T. (2003) Nat. Cell Biol. 5**,** Suppl., S1-S7. [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz, J., Altan-Bonnet, N. & Patterson, G. H. (2003) Nat. Cell Biol. 5**,** Suppl., S7-S14. [PubMed] [Google Scholar]
- 5.Cornish, V. W., Benson, D. R., Altenbach, C. A., Hideg, K., Hubbell, W. L. & Schultz, P. G. (1994) Proc. Natl. Acad. Sci. USA 91**,** 2910-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turcatti, G., Nemeth, K., Edgerton, M. D., Meseth, U., Talabot, F., Peitsch, M., Knowles, J., Vogel, H. & Chollet, A. (1996) J. Biol. Chem. 271**,** 19991-19998. [DOI] [PubMed] [Google Scholar]
- 7.Griffin, B. A., Adams, S. R. & Tsien, R. Y. (1998) Science 281**,** 269-272. [DOI] [PubMed] [Google Scholar]
- 8.Adams, S. R., Campbell, R. E., Gross, L. A., Martin, B. R., Walkup, G. K., Yao, Y., Llopis, J. & Tsien, R. Y. (2002) J. Am. Chem. Soc. 124**,** 6063-6076. [DOI] [PubMed] [Google Scholar]
- 9.Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H. & Johnsson, K. (2003) Nat. Biotechnol. 21**,** 86-89. [DOI] [PubMed] [Google Scholar]
- 10.Guignet, E. G., Hovius, R. & Vogel, H. (2004) Nat. Biotechnol. 22**,** 440-444. [DOI] [PubMed] [Google Scholar]
- 11.Pegg, A. E. (2000) Mutat. Res. 462**,** 83-100. [DOI] [PubMed] [Google Scholar]
- 12.Juillerat, A., Gronemeyer, T., Keppler, A., Gendreizig, S., Pick, H., Vogel, H. & Johnsson, K. (2003) Chem. Biol. 10**,** 313-317. [DOI] [PubMed] [Google Scholar]
- 13.Kindermann, M., George, N., Johnsson, N. & Johnsson, K. (2003) J. Am. Chem. Soc. 125**,** 7810-7811. [DOI] [PubMed] [Google Scholar]
- 14.Gendreizig, S., Kindermann, M. & Johnsson, K. (2003) J. Am. Chem. Soc. 125**,** 14970-14971. [DOI] [PubMed] [Google Scholar]
- 15.Lim, A. & Li, B. F. (1996) EMBO J. 15**,** 4050-4060. [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, J. F., Cadwallader, K., Paterson, H. & Marshall, C. J. (1991) EMBO J. 10**,** 4033-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschberg, K., Miller, C. M., Ellenberg, J., Presley, J. F., Siggia, E. D., Phair, R. D. & Lippincott-Schwartz, J. (1998) J. Cell Biol. 143**,** 1485-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, M. B. & Yakel, J. L. (1995) Annu. Rev. Physiol. 57**,** 447-468. [DOI] [PubMed] [Google Scholar]
- 19.Maggi, C. A. (1995) Gen. Pharmacol. 26**,** 911-944. [DOI] [PubMed] [Google Scholar]
- 20.Kalderon, D., Roberts, B. L., Richardson, W. D. & Smith, A. E. (1984) Cell 39**,** 499-509. [DOI] [PubMed] [Google Scholar]
- 21.Fritz, G., Tano, K., Mitra, S. & Kaina, B. (1991) Mol. Cell. Biol. 11**,** 4660-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodionov, V., Nadezhdina, E. & Borisy, G. (1999) Proc. Natl. Acad. Sci. USA 96**,** 115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmann, J. E. & Singer, S. J. (1983) J. Cell Biol. 97**,** 1777-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strathmann, M. & Simon, M. I. (1990) Proc. Natl. Acad. Sci. USA 87**,** 9113-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivenugopal, K. S., Yuan, X. H., Friedman, H. S. & Ali-Osman, F. (1996) Biochemistry 35**,** 1328-1334. [DOI] [PubMed] [Google Scholar]
- 26.Xu-Welliver, M. & Pegg, A. E. (2002) Carcinogenesis 23**,** 823-830. [DOI] [PubMed] [Google Scholar]
- 27.Chen, Y., Mills, J. D. & Periasamy, A. (2003) Differentiation (Berlin) 71**,** 528-541. [DOI] [PubMed] [Google Scholar]
- 28.Gaietta, G., Deerinck, T. J., Adams, S. R., Bouwer, J., Tour, O., Laird, D. W., Sosinsky, G. E., Tsien, R. Y. & Ellisman, M. H. (2002) Science 296**,** 503-507. [DOI] [PubMed] [Google Scholar]
- 29.McElhinney, R. S., Donnelly, D. J., McCormick, J. E., Kelly, J., Watson, A. J., Rafferty, J. A., Elder, R. H., Middleton, M. R., Willington, M. A., McMurry, T. B., et al. (1998) J. Med. Chem. 41**,** 5265-5271. [DOI] [PubMed] [Google Scholar]
- 30.Scales, S. J., Pepperkok, R. & Kreis, T. E. (1997) Cell 90**,** 1137-1148. [DOI] [PubMed] [Google Scholar]
- 31.Balch, W. E. & Keller, D. S. (1986) J. Biol. Chem. 261**,** 14690-14696. [PubMed] [Google Scholar]
- 32.de Silva, A. M., Balch, W. E. & Helenius, A. (1990) J. Cell Biol. 111**,** 857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn, G. A., Dobbie, I. M., Monypenny, J., Holt, M. R. & Zicha, D. (2002) J. Microsc. (Oxford) 205**,** 109-112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information