Real-Time Visualization of ZBP1 Association with β-Actin mRNA during Transcription and Localization (original) (raw)
. Author manuscript; available in PMC: 2016 Feb 24.
Abstract
Background
mRNA localization in somatic cells is an important mechanism for gene expression regulation. In fibroblasts, the protein ZBP1 associates with the sequence that localizes β-actin mRNA to the leading edge of fibroblasts, augmenting motility. β-actin mRNA localizes in a cytoskeleton-dependent manner, depending on intact actin and myosin ATP-hydrolysis, and is largely bound to the actin cytoskeleton. The ZBP1 protein contains four KH RNA binding domains and a classic RBD RNA binding domain. It also contains a putative nuclear import and export sequence, suggesting a nuclear phase in this protein's function.
Results
Using high-speed imaging, we show here the targeting of this RNA binding protein to β-actin pre-mRNA transcripts in the nuclei of living cells and measure the residence time of the RNA-protein complex before it leaves the transcription site. Then, the RNA-protein particle is exported to the cytoplasm, where it localizes at velocities of 0.6 µm/s by using actin filaments and/or microtubules. This RNA-ZBP1 complex is required for cytoplasmic localization in fibroblasts; mislocalizing the protein also mislocalizes the RNA, and expressing the protein in a ZBP1-deficient cell line induces β-actin mRNA localization.
Conclusions
This work demonstrates that the RNA-protein association, essential for cytoplasmic localization, begins as soon as the RNA is transcribed. The ZBP1 then forms a ribonucleoprotein particle and moves in a myosin-dependent fashion by using the cytoskeleton for directional transport.
Introduction
RNA localization is an important and efficient mechanism for redistribution and spatial restriction of protein expression in various types of somatic and developmental cells. In Drosophila melanogaster oogenesis, gurken, bicoid, and oskar RNAs are redistributed to determine dorsal, anterior, and posterior compartments of a future embryo [1]. In Saccharomyces cerevisiae, a repressor of transcription, Ash1p, localizes exclusively to the nucleus of a budding daughter cell to repress mating type switching [2, 3]. In motile fibroblasts, the mRNA encoding β-actin is localized just proximal to the leading edge, where the mRNA restricts the spatial distribution of actin protein synthesis and augments cell asymmetry and motility [4, 5]. The “zipcode” sequence in the 3′ UTR of β-actin mRNA is required for its localization in motile fibroblasts [6, 7]. We isolated and cloned a protein binding to this sequence, the zipcode binding protein (ZBP1) [8], that is the founding member of an ever increasing family of proteins involved in RNA regulation [9]. ZBP1 is an RNA binding protein containing KH and RRM domains [10, 11, 12] and putative Rev-like nuclear export and nuclear localization sequences [13, 14]. This raised the possibility that there may be an important nuclear component to mRNA localization in the cytoplasm [8]. Using both antibodies raised to ZBP1 and fusions of ZBP1 to GFP, we followed the association of the protein with β-actin mRNA and its subsequent transport. In this study, we demonstrate that ZBP1 associates with β-actin mRNA cotranscriptionally and measure its residence time at the transcription site. We show that ZBP1-β-actin mRNA interaction occurs during transcription and is essential for localization in the cytoplasm; this localization is also dependent on intact actin cytoskeleton and actomyosin interactions. At the leading edge, ZBP1 particles are associated with actin filaments and microtubules. The particles redistribute at high speeds as the leading edge of a live cell is changing and new protrusions are formed.
Results
Recombinant ZBP1 Directly and Specifically Bound the β-actin mRNA Zipcode ZBP1
A recombinant ZBP1-GST fusion bound the 54-nucleotide β-actin mRNA zipcode by gel mobility shift assay. The mutant zipcode that was inactive in the in vivo localization assay was deficient in binding to endogenous ZBP1 in chicken embryo fibroblast (CEF) extracts [8]. The recombinant ZBP1-GST fusion therefore possessed the same nucleic acid specificity as the native protein, distinguishing between closely related RNA molecules (data not shown). This demonstrated that the cloned recombinant ZBP1 directly and specifically bound to the β-actin mRNA zipcode and could do so in the absence of additional factors. The bound complex was weaker and of different mobility than the endogenous complex, suggesting that additional factors augmenting RNA binding were involved in in vivo complex formation.
ZBP1 Colocalized to the Leading Edge with β-actin mRNA and Bound to the Cytoskeleton
To determine if the intracellular distribution of endogenous ZBP1 was consistent with its binding to β-actin mRNA intracellularly, an antiserum developed to a specific peptide in ZBP1 was used. The antiserum stained CEFs brightly, producing a granular staining pattern that resembled the RNA distribution in that it was predominantly at the leading lamella of polarized fibroblasts as well as in the perinuclear region (Figure 1A). Despite the presence of nuclear localization and export sequences in ZBP1, there was almost no detectable protein in the nuclei of all cells. In order to determine whether cytoskeletal structural components were retaining the protein in the cytoplasm, the cells were extracted with Triton before fixation. ZBP1 staining remained at the leading edge but was mostly removed from the perinuclear regions, indicating that ZBP1 in the lamella was cytoskeleton associated (Figure 1B, see below), while, in the perinuclear region, it was soluble or membrane associated. To verify that ZBP1 was bound to its target mRNA while localized to the leading edge of the fibroblasts, we detected β-actin mRNA by using fluorescent probes and demonstrated that ZBP1 colocalized with it in the same cells by immunofluorescence or by fusion to GFP (Figure 1C). Like the endogenous protein, the fusion protein was also significantly absent from the nucleus, in contrast to the diffuse nuclear and cytoplasmic distribution of GFP alone. A deconvolved section through the cell confirmed that the nucleus had undetectable levels of β-actin mRNA and ZBP1-GFP (Figure 1C, right). The highest concentrations of protein and the RNA were colocalized near the lamellipod. The majority of localized β-actin mRNA and ZBP1, or ZBP1-GFP, overlapped in regions of protrusion (yellow color in Figure 1C). Both components were seen distributed as granules throughout the cytoplasm. Many granules contained β-actin mRNA and also stained for ZBP1, but some appeared to contain only one or the other.
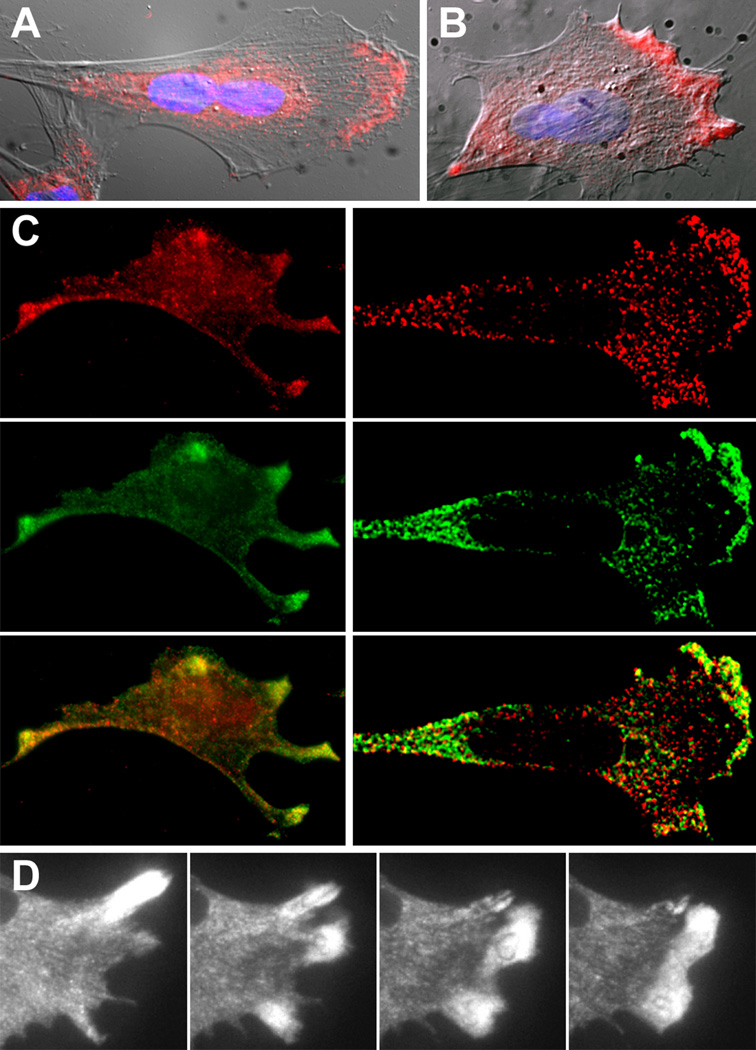
Figure 1. Cytoplasmic Distribution and Relocalization of ZBP1.
(A) Cells were fixed and stained with anti-ZBP1 antiserum (red), which was digitally superimposed on the Nomarski image and nuclear staining (DAPI, blue).
(B) Cells extracted with Triton X-100 for 30 s before treatment described in (A).
(C) Colocalization of endogenous (left) and GFP-fused (right) ZBP1 (green) and β-actin mRNA (red) in chicken embryo fibroblasts. Left: cells were stained with anti-ZBP1 serum and were subsequently hybridized with β-actin mRNA fluorescent oligonucleotide probes. Right: cells were transfected with the ZBP1-GFP fusion, and green cells were visualized by deconvolution microscopy after fixation and in situ hybridization. The yellow staining in the protrusions demonstrates RNA and protein colocalization. The scale bar represents 10 µm.
(D) ZBP1-GFP particles moving in living cells and localizing to a dynamic, moving lamella. Panels are shown at approximately 100-s intervals. The scale bar represents 10 µm. See Movies 1–11 in the Supplementary Material.
ZBP1 Visualized in Real Time Moved to the Motile Leading Edge
The movement of the ZBP1-GFP fusion in living cells was observed in real time by using a rapid-acquisition CCD camera. This allowed us to determine the speed and directionality of this transport. Particles were seen moving between the nucleus and the lamellipod (Figure 1D and Movies 1–11 in the Supplementary Material available with this article online). The particles within the lamella could occasionally be seen to be aligned and moving bidirectionally toward and away from the nucleus. As the fibroblast responded to serum stimulation, ZBP1-GFP that was accumulated in an older protrusion rapidly (within a minute) redistributed to a newly forming protrusion. ZBP1-GFP particles were often seen “streaming” at speeds at speeds of 0.6 µm/s (see Table S1 in the Supplementary Material). This suggested that the movement of ZBP1-GFP particles was an active motor-driven process. The movement was concurrent with β-actin mRNA localization and consequently the motility of the cell [4, 5]; therefore, some of these ZBP1-GFP particles were presumably transporting β-actin mRNA. The dynamic nature of this movement emphasizes how rapidly the moving cell can reposition sites of β-actin protein synthesis in response to changes in cell morphology.
ZBP1 Localization Depended on Intact Actin Cytoskeleton and Actomyosin Interactions
The rapid movements of the GFP-ZBP1 and their detergent resistance suggested an association with cytoskeletal elements. Both microtubules and actin filaments were colocalized with anti-ZBP1 antibody by light microscopy (Figures 2A and 2B) and by immunogold staining (Figure 2F). We found that ZBP1 particles in the cytoplasm were associated predominately with actin filament bundles but occasionally with microtubules. These findings suggested a role for both systems in localization and anchoring. The identities of the structural elements that were functionally important in localization were determined by using cytochalasin or colchicine to affect the microfilaments or microtubules, respectively. β-actin mRNA localization has been shown to be sensitive to cytochalasin compared to colchicine [15]. Like the mRNA, the distribution of ZBP1 was significantly affected in the CEFs treated with cytochalasin (Figure 2C); very few cells (<5%) contained ZBP1 localized to the lamella, and most cells accumulated in the perinuclear area, but not within the nucleus. Also like the mRNA, fibroblasts treated with colchicine sometimes showed a gradient of ZBP1 from the periphery to the perinuclear area but maintained polarity and localization (Figure 2D). These results were consistent with a model in which the actin cytoskeleton played the major role in the transport of ZBP1-β-actin mRNA-containing particles in CEFs while microtubules played a minor role in either transporting or anchoring the particles at the leading edge. In order to test whether myosin was involved in actin-based transport, we used an inhibitor of myosin ATPase, 2,3-butanedione-2-monoxime (BDM) [16], and evaluated ZBP1 localization. It has recently been shown that β-actin mRNA localization can be disrupted by BDM and that the localization is dependent on myosin IIB [17]. ZBP1 rapidly delocalized in the presence of this inhibitor; almost all of the cells (95%) had evenly distributed ZBP1 throughout the cytoplasm (Figure 2E; see Movie 12 in the Supplementary Material), but it was still excluded from the nucleus. The movement of the ZBP1-GFP in living cells did not show any directionality compared to the cell in Figure 1D (see Movie 1 in the Supplementary Material). This result suggested that the cytoplasmic localization of the ZBP1-β-actin mRNA complex in fibroblasts was dependent on actin-myosin interactions.
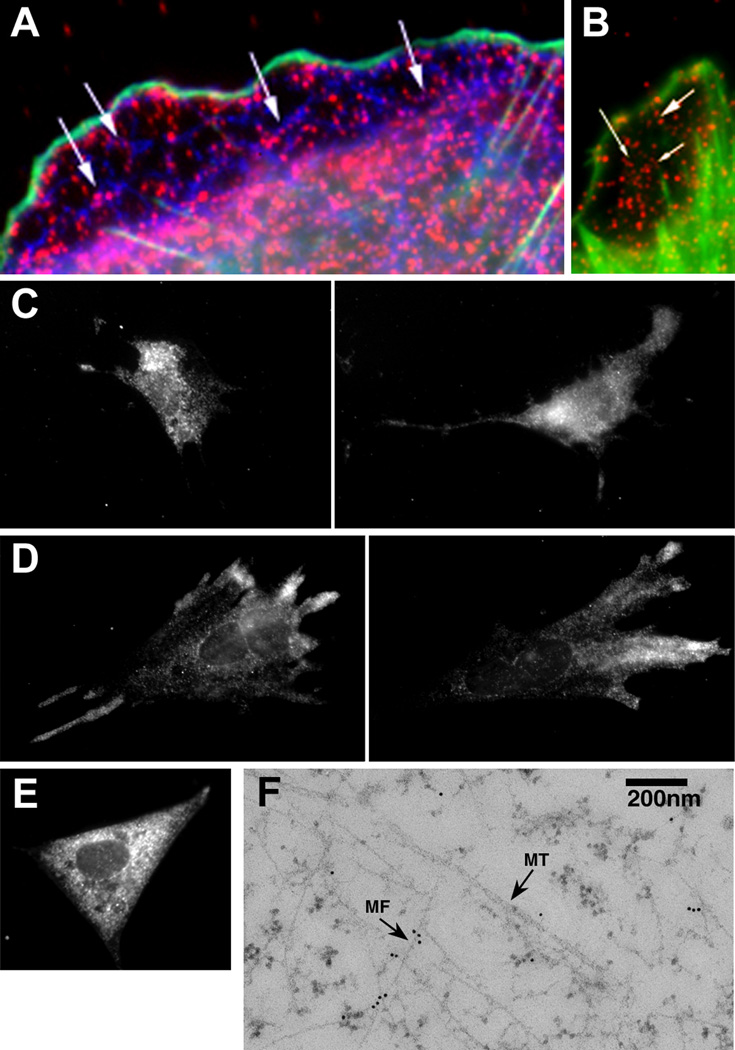
Figure 2. ZBP1 Associates with the Cytoskeleton.
(A) Immunofluorescence staining of ZBP1 (red), actin (green), and tubulin (blue). Arrows indicate localization of ZBP1 along microtubules.
(B) Immunofluorescence staining of ZBP1 (red) and actin (green). Arrows indicate ZBP1 along filament bundles.
(C) ZBP1 distribution in cytochalasin-treated cells.
(D) ZBP1 distribution in colchicine-treated cells.
(E) ZBP1 distribution in BDM-treated cells. See Movie 12 in the Supplementary Material.
(F) Electron microscopy of ZBP1 in CEFs. Thin-sectioned cells were stained with anti-ZBP1 antibody and secondary gold-conjugated antibody. MT, microtubule; MF, actin.
The scale bar in (B) represents 2 µm in (A) and (B), the scale bar in (C) represents 10 µm in (C)–(E), and the scale bar in (F) represents 200 nm.
Formation of the ZBP1-β-actin mRNA Complex Was Essential for Localization
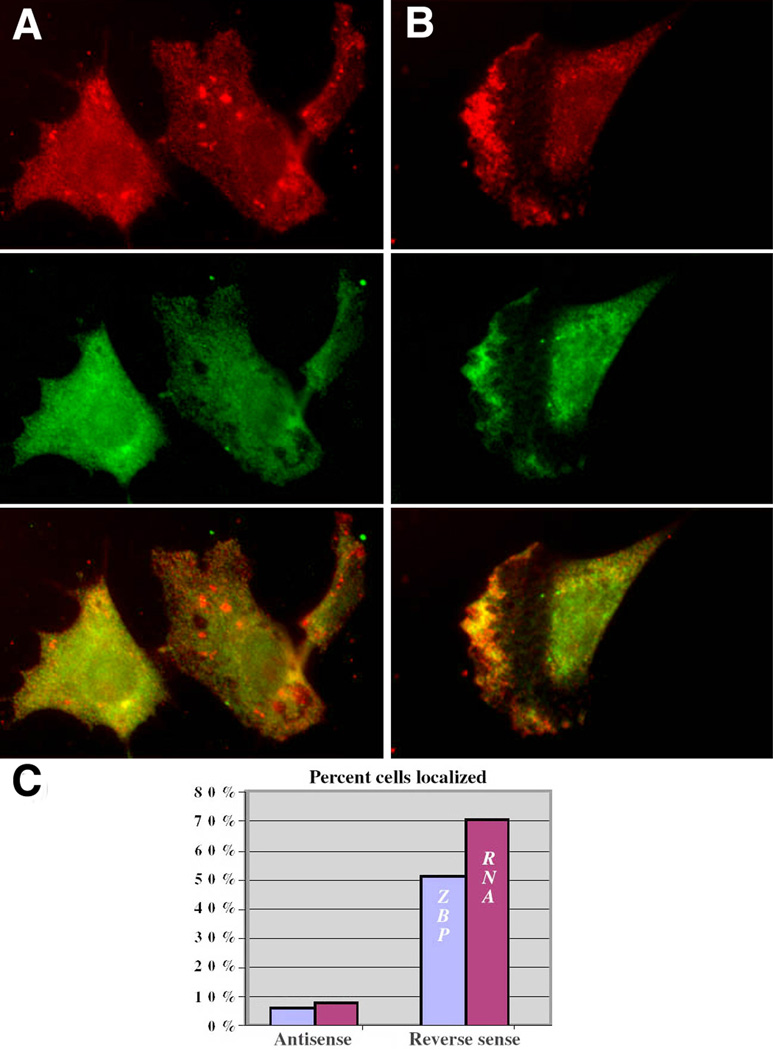
The protein-RNA complex formation was likely essential for the localization of both β-actin mRNA and ZBP1 [7]. Alternatively, ZBP1 or β-actin mRNA could localize independently. In order to test these possibilities, the association of the RNA and the protein was disrupted by using zipcode-antisense oligodeoxynucleotides (ODN). The application of phosphorothioate oligonucleotides was capable of eliminating the localization of β-actin mRNA in CEFs [7], presumably because of the disruption of the complex by the antisense. We confirmed this in gel shift assays: the ODNs blocked complex formation between the zipcode and ZBP1 in fibroblast (data not shown) and neuronal [18] cell extracts. A surprising result was obtained when β-actin mRNA and ZBP1 distribution were analyzed after a 12-hr treatment of CEFs with zipcode antisense ODNs. Delocalization of both β-actin mRNA and ZBP1 was observed (Figure 3A). In contrast, the presence of a reverse sequence control ODN that did not bind the zipcode left the complex intact, and ZBP1 remained colocalized with β-actin mRNA in the lamella of polarized CEFs (Figures 3B and 3C).
Figure 3. ZBP1 and β-actin mRNA Association Are Required for Localization.
(A and B) Antisense to the zipcode inhibits localization of ZBP1 and β-actin mRNA. CEFs were incubated with (A) antisense oligonucleotides to the zipcode or (B) reverse control ODN for 12 hr, fixed, and stained for ZBP1 (green) and β-actin mRNA (red). The scale bar represents 10 µm.
(C) Number of cells localizing mRNA and ZBP1 after antisense or control treatment (n = 115 in each group).
ZBP1 Localized β-actin mRNA in Cells Lacking ZBP1 and Mislocalized β-actin mRNA When Misdirected to Membranes
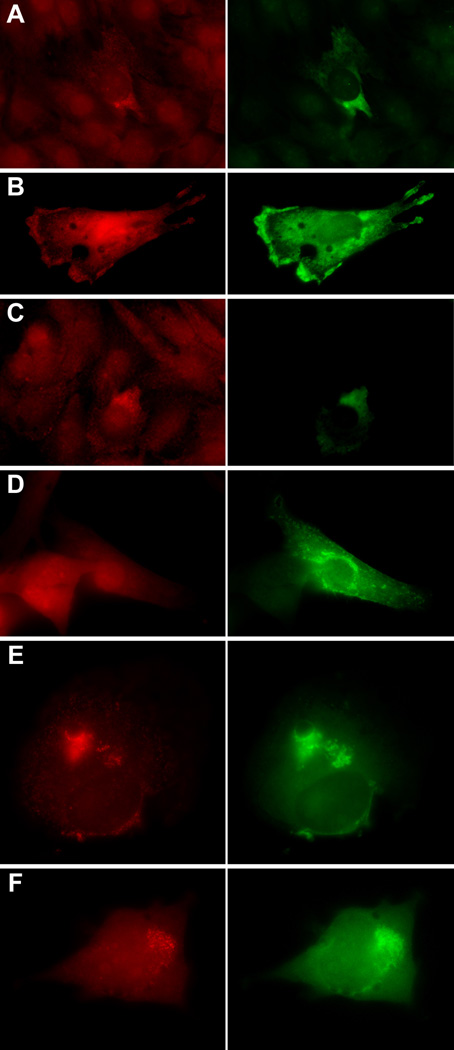
In order to demonstrate that ZBP1 was necessary for β-actin mRNA localization, ZBP1 was expressed in a C2C12 cell line that contained β-actin mRNA but no detectable ZBP1, as assessed by immunofluorescence or Western blot (data not shown). In these cells, β-actin mRNA was not localized and was diffuse, consistent with the absence of ZBP1. When ZBP1-GFP was expressed, the β-actin mRNA became punctate and showed increased polarity (Figures 4A–4C). When ZBP1 was mislocalized to membranous structures by fusion to CFP with a neuromodulin membrane-targeting signal [19], it accumulated at cellular membranes and mislocalized the majority of β-actin mRNA particles (Figures 4E and 4F). Membrane-targeted CFP had no effect on RNA localization (Figure 4D). This directly demonstrated that ZBP1 was responsible for determining β-actin mRNA localization, because it alone was able to mistarget it to a different location.
Figure 4. Distribution of ZBP1 Determines Distribution of β-actin mRNA.
(A–C) Expression of ZBP1-GFP in C2C12 cells. β-actin mRNA (red); ZBP1-GFP (green). C2C12 cells were transfected with ZBP1-GFP DNA, fixed, and β-actin mRNA-stained via in situ hybridization. The expression of ZBP1-GFP in cells without endogenous ZBP1 causes the formation and polarized location of particles of β-actin mRNA. Nontransfected C2C12 cells in the same field exhibit a diffuse RNA pattern. The scale bar represents 10 µm.
(D) Control, just CFP fused to the membrane signal.
(E and F) Expression of ZBP1 fused to the neuromodulin membrane-targeting signal (green) accumulates at cellular membranes and mislocalizes β-actin mRNA (red). The scale bar represents 5 µm.
ZBP1 Shuttles between Nucleus and Cytoplasm
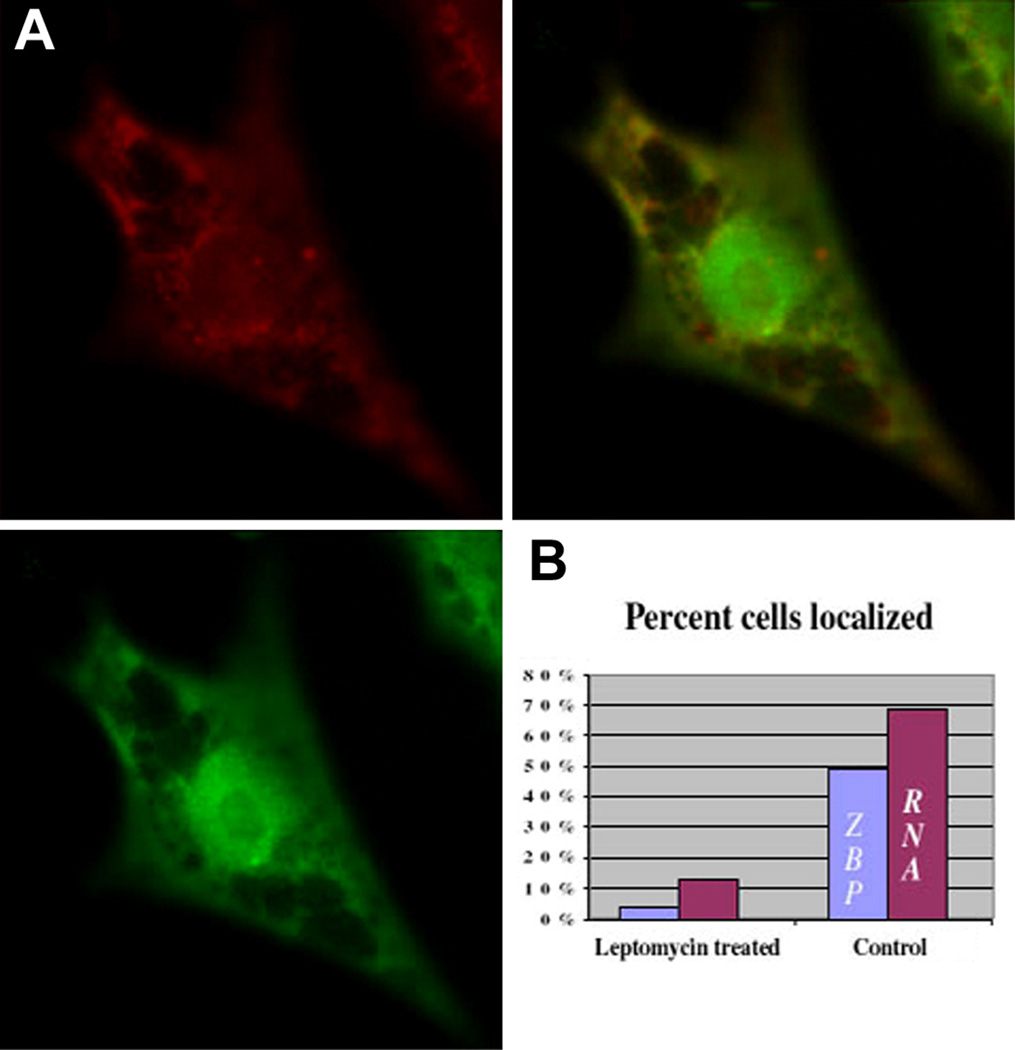
We postulated that the association of ZBP1 with actin filaments kept the vast majority of the protein in the cytoplasm. Since ZBP1 contains a putative Rev-like, leucine-rich nuclear export signal and a nuclear localization sequence closely related to SV40 T-antigen [8], it was likely that the protein shuttled, spending a short time in the nucleus. In order to test this hypothesis, Leptomycin B (LMB), an inhibitor of Rev-like, Crm1-mediated nuclear export [20], was used in order to determine whether the distribution of the protein changed. In the presence of LMB, the distribution of ZBP1 was markedly different from normal (Figure 5A). ZBP1 was not localized to the lamellipodia, and occasionally fibroblasts were observed with a significant fraction of the protein present in the nucleus, excluding nucleoli. In contrast, actinomycin D treatment, known to block nuclear transport of some hnRNPs [21], had no effect on ZBP1 export (not shown). Interestingly, β-actin mRNA localization was also affected. The fraction of cells with localized ZBP1 and β-actin mRNA decreased significantly (Figure 5B). These results indicated that ZBP1 shuttled between the nucleus and cytoplasm and that its export was at least in part mediated by the LMB-sensitive exportin-1 (Crm1). This also demonstrated that Crm1-dependent shuttling was involved in both ZBP1 and β-actin mRNA localization in the cytoplasm.
Figure 5. Nuclear Distribution of ZBP1 in LMB-Treated Cells.
(A) ZBP1 (green) and β-actin mRNA distribution (red) in chicken embryo fibroblasts treated with LMB for 6 hr. The mid-nuclear focal plane is displayed. Retention of ZBP1 in the nuclei resulted in significant delocalization of β-actin mRNA (overlap). The scale bar represents 10 µm.
(B) Percentage of β-actin mRNA and ZBP1 localized after treatment (n = 113).
ZBP1 Associates with Newly Transcribed β-actin mRNA
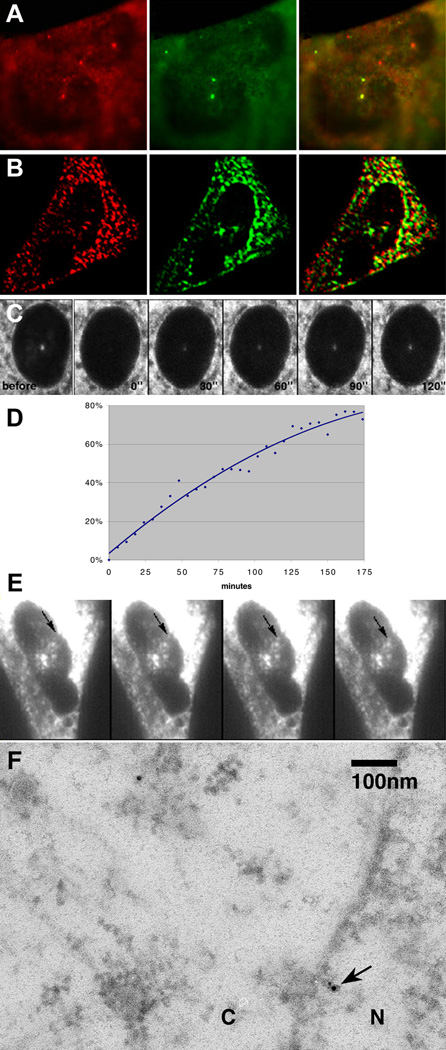
Why would ZBP1, a protein important for localization of β-actin mRNA in the cytoplasm, shuttle into and out of the nucleus? One possibility was that ZBP1 identified the β-actin mRNA transcripts while in the nucleus. In order to test this possibility, β-actin mRNA transcription was activated by serum, resulting in the rapid (within 3 min) appearance of transcription sites of nascent β-actin RNA [22]. The intranuclear distribution of ZBP1 was then followed over a period of 30 min. At 5 min after induction, a significant number (around 30%) of fibroblasts contained bright, dot-like nuclear foci of ZBP1 (Figure 6A) or ZBP1-GFP (Figure 6B) that were identified as transcription sites of β-actin RNA by in situ hybridization. Cell populations stimulated for either shorter (2 min) or longer (30 min) periods of time rarely showed ZBP1 foci in the nucleus; this finding is consistent with the time necessary for the first polymerases to transcribe the 3′ UTR and the observed decrease in transcription rate after 15 min [22]. These data suggested that ZBP1 associated with β-actin mRNA cotranscriptionally. Either ZBP1 entered the nucleus, or undetectable levels of ZBP1 existed in the nucleoplasm. Using high-speed imaging of a ZBP1-GFP fusion after 10 min of serum stimulation, we were able to capture an intranuclear particle moving between the nuclear envelope and a transcription site (Figure 6E and Movie 14, available in the Supplementary Material). The particle traveled directly from the edge of the nucleus to the transcription site at high speed. This same cell was later subjected to in situ hybridization and immunofluorescent staining to confirm transcription site position, as shown in Figure 6B (rotated to the vertical).
Figure 6. ZBP1 Nuclear Transport and Accumulation at β-actin Transcription Sites.
(A) Accumulation of endogenous ZBP1 at β-actin transcription sites in CEF nuclei. Serum was applied to chick embryo fibroblasts for 5 min after synchronization by serum starvation. The cells were immediately fixed and stained for β-actin mRNA (red) by in situ hybridizaton and ZBP1 (green) by immunofluorescence. ZBP1 and β-actin mRNA colocalize (yellow) at the transcription sites.
(B) Accumulation of ZBP1-GFP fusion at β-actin transcription sites. CEFs were transfected with ZBP1-GFP fusion DNA and were imaged live after treatment as in (A). After identification and imaging of transcription sites in live cells (see [E]), they were fixed and stained by in situ hybridization for the β-actin mRNA (red). The image was subjected to deconvolution to yield the optical section shown.
(C) ZBP1-GFP fusion proteins move into the nucleus and bind to nascent chains of β-actin at the transcription site shortly after serum induction. The transcription site in a ZBP1-GFP-transfected CEF was photobleached, and fluorescence recovery was monitored by confocal microscopy. Half-recovery occurred within 90 s. The panels shown are at 30-s intervals. “Before,” before photobleaching; “0,” after photobleaching for 20 s. See Movie 13 in the Supplementary Material.
(D) Time plot of ZBP1-GFP fluorescence recovery at a transcription site. Relative intensity was calculated from raw imaging data and was plotted as a fraction of intensity prior to bleaching. The time in seconds after bleaching was complete is indicated on the horizontal axis.
(E) The same cell as in (B) before fixation and in situ hybridization. ZBP1 particle movement between the nuclear envelope and β-actin transcription site can be seen in live cells. The time between the frames shown is 800 ms. See Movie 14 in the Supplementary Material.
(F) Electron microscopy of ZBP1 at the nuclear pore (arrow). Sections of cells treated in (A) were stained with anti-ZBP1 antibody and secondary gold-conjugated antibody. N, nucleus; C, cytoplasm.
The scale bars in (A)–(C) represent 5 µm, the scale bar in (E) represents 10 µm, and the scale bar in (F) represents 100 nm.
ZBP1 Turnover Rate at the Transcription Site
In order to study the rate of ZBP1-GFP turnover in nuclear foci, the transcription site was photobleached at high laser power and fluorescence recovery was measured. The protein was seen at the transcription site within seconds after photobleaching, and the site returned to half its original brightness within 90 s and 80% brightness at approximately 180 s (Figures 6C and 6D; see Movie 13 in the Supplementary Material). Therefore, the ZBP1-GFP fusion protein continuously arrived at the transcription site, associated with the RNA, and left, consistent with what is shown in Figure 6E. The residence of ZBP1-GFP at the site is consistent with measurements made to determine the time for nascent transcripts of β-actin mRNA to be released from the transcription site [21, 23]. Since the ZBP1 binding site is almost a kilobase from the end of the transcript, 2 min is the expected time that it takes to finish transcription and then terminate [22]. The lack of a significant amount of ZBP1 in the nucleus, except where β-actin mRNA transcripts were present, was consistent with a model wherein the protein is immediately exported (or, possibly, dissipated) unless its nascent RNA target is available for binding. Once bound, the protein transports with the RNA into the cytoplasm, localizes, and anchors with it on the cytoskeleton, thereby resulting in low nuclear ZBP1 content. To estimate the steady-state amount of nuclear and cytoskeletal ZBP1, ZBP1 particle distribution was analyzed by using electron microscopy (Figure 6F). Using immunogold staining and counting gold particles in nuclear and cytoplasmic sections, we determined that the concentration of ZBP1 in the nucleus was approximately 2.5 times lower than in the cytoplasm. Because the ratio of nuclear volume to cytoplasmic volume in these cells is 1:5 [24], cytoplasm contains 92% of the cellular ZBP1, while the nucleus contains approximately 8%.
Discussion
Our experimental data support the shuttling of ZBP1 and its essential role in β-actin mRNA transport and localization. FRAP experiments demonstrated that ZBP1 accumulates at β-actin RNA transcription sites in the nucleus, indicating the rapid association of the protein with the β-actin pre-mRNA. In real-time analysis, the protein resided at this site only long enough for the nascent chains to terminate, indicating that the RNA-protein complex moved away from the site of transcription rapidly with the RNA. This does not support a model wherein the RNA stays around the site of synthesis after transcriptional termination. The residence time in the cytoplasm is likely longer than in the nucleus, based on the relative concentrations in each compartment. A longer cytoplasmic residence could be due to its association in a complex with the RNA and with cytoskeletal filaments.
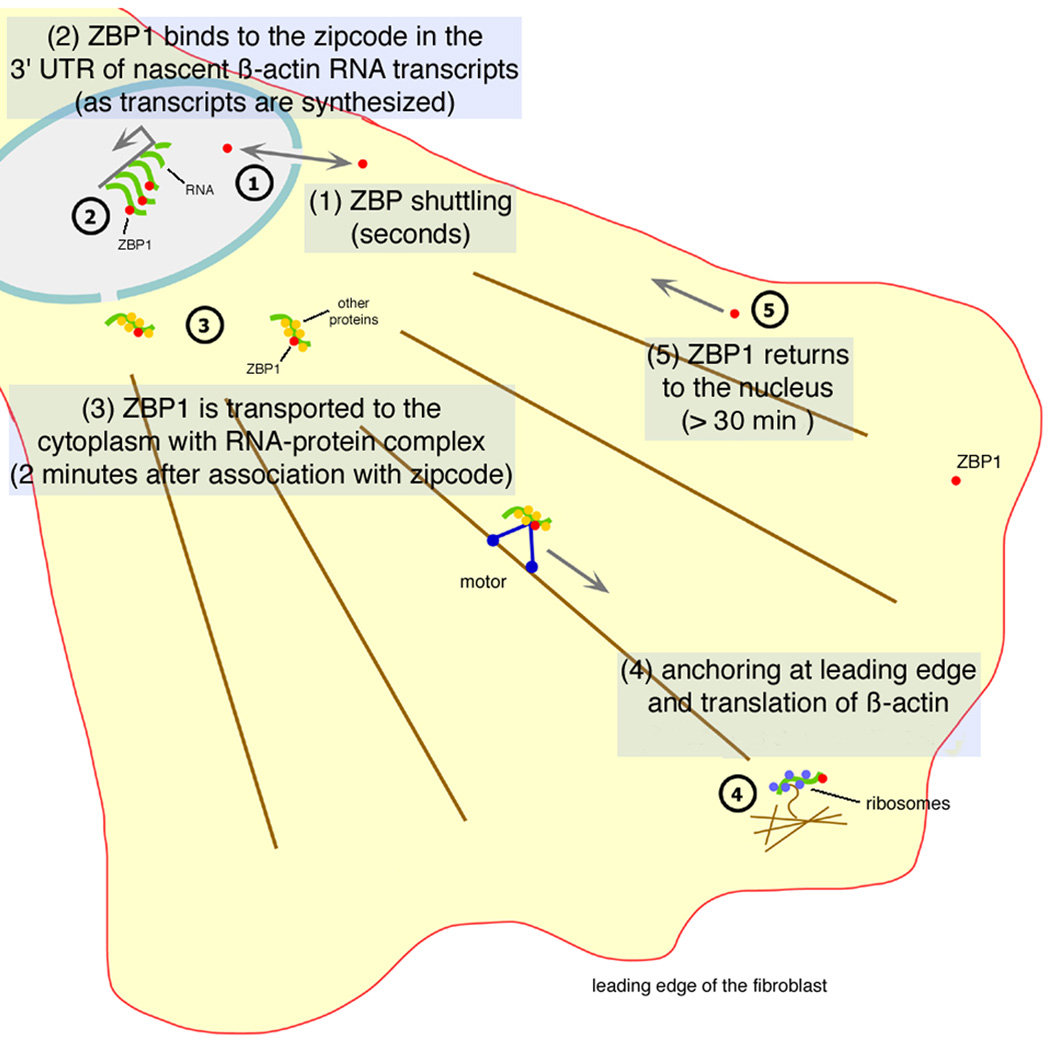
LMB treatment affected ZBP1 export from the nucleus and delocalized both β-actin mRNA and ZBP1 in the cytoplasm. This suggested that Crm1-dependent nuclear export of ZBP1 or some other factor was required for continued β-actin mRNA localization. In addition, the disruption of the localization of the ZBP1-mRNA complex to the zipcode by using antisense oligonucleotides demonstrated that neither component of the complex could localize independently of the other. β-actin mRNA could still be exported from the nucleus by an independent process, since nuclear levels didn't increase after exposure to the zipcode antisense [7] but its localization was abolished. We propose that ZBP1 chaperoned the β-actin mRNA toward a specific localization pathway by packaging the RNA with other proteins into a localization-competent form, the “locasome,” during synthesis (Figure 7). The splicing of β-actin RNA occurs cotranscriptionally [25]. ZBP2, which also bound the β-actin zipcode and is a homolog of the splicing factor KSRP [26], may also play a role. These proteins are likely to interact with β-actin RNA throughout the downstream processing and transport stages marking it for proper location. Similarly, Y14 and Mago proteins associate with mRNA exon-exon junctions upon splicing and are required for its cytoplasmic localization [27, 28, 29, 30]. The key role of ZBP1 in the localization process is supported by the finding that it is able to induce and enhance localization in C2-C12 cells that express β-actin mRNA but have no ZBP1. The ability of misdirected ZBP1 to mislocalize β-actin mRNA also indicates that it is the active localization component and that its location is causal for the mRNA distribution.
Figure 7. Proposed Model of ZBP1-mRNA Localization.
The travels of ZBP1. ZBP1 shuttles into and out of the nucleus. If it encounters the nascent zipcode sequences, it binds there as the nascent RNA and finishes transcription and processing/polyadenylation before export to the cytoplasm. Once in the cytoplasm, it associates with cytoskeletal elements (actin filament bundles are shown here) and localizes to the leading edge.
The localization of mRNAs in other systems also appears to involve proteins that shuttle. Since ZBP1 was first described, other hnRNPs have been implicated in RNA localization. In Drosophila, Squid, an hnRNPA/B, is involved with localization of gurken and _ftz_mRNAs [31], and Y14 and Mago are required for oskar localization [27, 28, 29]. The hnRNPA/B is also possibly involved in the localization of myelin basic protein mRNA in oligodendrocytes [32, 33]. Recently, hnRNP I was shown to be associated with Vg1 localization in Xenopus_[34, 35, 36], as was the Xenopus ortholog of ZBP1 [37, 38]. In yeast, the localization of ASH1 mRNA is enhanced by an exclusively nuclear protein, Loc1p [39]. All of these cases support a model in which RNA localization proteins perform_both nuclear and cytoplasmic functions to effect proper RNA transport and localization [8]. Perhaps this occurs by coupling synthesis, processing, or export [40, 41] with the localization pathway.
Once in the cytoplasm, ZBP1 is associated with actin filament bundles or microtubules (Figure 7). The ability of a minor fraction of ZBP1 in CEFs to associate with microtubules is consistent with the observation that the Xenopus homolog, Vg1RBP, can bind to microtubules [42]. Furthermore, ZBP1 transports β-actin mRNA along microtubules and down neuronal processes, where it localizes to the growth cone [18]. These results show that the same ZBP1-RNA complex can associate with either microfilaments or microtubules, or that two complexes form, each capable of interacting with a different cytoskeletal system. It seems likely that the ZBP1-RNA complex could interact with both microtubule and microfilament motors, so that either system would ensure the eventual transport to, and anchoring at, the leading edge. ZBP1 particles were associated with microtubules and microfilaments at the leading edge, but their transport to the leading edge and localization was critically dependent on intact actin cytoskeleton and actin-myosin interactions. This suggests a role for actin and myosin in motor-driven translocation of “locasomes” and cytoskeleton-based anchoring. The distribution of ZBP1 particles at the leading edge was shown here for the first time to be very dynamic and congruent with lamellar protrusions of a motile cell. Relocation of particles was concurrent with actual protrusion, suggesting an important role for localized RNA associated with the ZBP1. All of these observations suggest that ZBP1 directs and regulates β-actin mRNA from its synthesis to its final location in the cytoplasm, and perhaps even its translation into protein.
Experimental Procedures
Plasmids
ZBP1-GFP fusion was created by subcloning the BamHI-EcoRI ZBP1 coding region fragment into BglII-EcoRI-digested pEGFP-C1 (Clontech). Membrane-targeted GAP43-CFP-ZBP1 was made by excising an AseI-BsrGI fragment containing CFP and the GAP-43 targeting signal from pECFP-Mem (Clontech) and inserting it into a ZBP1-GFP fusion plasmid.
Peptide Synthesis and Polyclonal Antibody Production
The peptide sequences used for antipeptide antibody synthesis were #629, NH2-KITTILAQVRRQQXK-COOH and #627, NH2-KVRMVVIGPPEAQFK-COOH. Peptides were synthesized in milligram quantities by the peptide synthesis facility at the University of Massachusetts Medical Center, Worcester. Approximately 10 mg each of peptides #627 and 629 were combined with adjuvant and were injected into rabbits (East Acres Biologics). Antisera were tested by Western blotting and immunoprecipitation.
Immunofluorescence and In Situ Hybridization
Cells were Triton extracted, if necessary, by treatment with a 0.5% solution at room temperature for 30 s. Cells were fixed in 4% paraformaldehyde in PBS-5 mM MgCl2 for 10 min and were washed three times in PBS-5 mM MgCl2. Following permeabilization with 0.5% Triton X-100, the cells were blocked in 1% BSA in PBS-5 mM MgCl2 for 30 min. A 1:200 dilution of ZBP1 antipeptide antibody in BSA was applied for an hour at 37°C, followed by three washes in PBS-5 mM MgCl2. Tubulin was stained with mouse anti-α,β-tubulin antibody. Fluorescently labeled secondary antibody or phalloidin solution was applied subsequently for 1 hr at 37°C, followed by three washes in PBS-5 mM MgCl2 and a rinse in DAPI-PBS-5 mM MgCl2 solution. If in situ hybridization were to follow, DAPI staining was omitted and the cells were fixed in 4% formaldehyde for 15 min, followed by three washes in PBS-5 mM MgCl2. In situ hybridization was performed with a mixture of six Cy3-labeled oligonucleotides complementary to β-actin mRNA 3′ UTR for 3 hr in 50% formamide [22]. The cells were washed with SSC and PBS-5 mM MgCl2 solutions and were mounted on slides.
Electron Microscopy and Density Quantitation
Embedding and Sectioning
CEF cells were grown on glass coverslips and were fixed with 4% paraformaldehyde, 0.1% glutaraldehyde in 0.1 M Cacodylate buffer (pH 7.4) for 20 min. They were then dehydrated to 95% in ethanol and embedded en face in LR White (London Resin Company). Sections (90 nm) were taken beginning at the basal cell surface and were collected on nickel grids.
Immunolabeling
Grids were quenched with 0.5 M glycine and were blocked with 2.1% cold-water fish skin gelatin, 9.75% BSA, and 4.75% goat serum before being incubated in anti-ZBP1 at 1:1000 or incubation buffer (negative control). After copious rinsing, grids were incubated for 1 hr in either 6 nm or 10 nm gold conjugated to goat anti-rabbit IgG (Electron Microscopy Sciences). Immunodecorated grids were then counterstained with uranyl acetate and lead citrate before being viewed on a JEOL 1200EX transmission electron microscope (JEOL).
Quantitation
Micrographs of the nucleus, cytoplasm, and processes were taken of each cell at 20KX before moving on to photograph the neighboring cell. A grid of 0.25 µm2 was drawn on an acetate sheet and was aligned with the print. Only boxes exclusively containing the compartment being analyzed (cytoplasm or nucleus) were counted for gold particles. The total number of gold particles counted was divided by the total number of boxes to calculate the number of particles per unit area. Background was derived by using this method on negative control grids, and the value was subtracted from anti-ZBP1 values.
Transfection and Live Cell Imaging
The pEGFP-ZBP-1 construct was transfected into chicken embryo fibroblasts or C2C12 cells with Effectine reagent (Qiagen) according to the manufacturer's instructions at the time of plating onto 40 mm coverslips. Fibroblasts were allowed to express for 12–24 hr in serum-free medium. The cells were transferred to the FCS2 closed chamber system (Bioptechs) that was mounted onto an Olympus microscope equipped with a CCD camera and TILL Photonics image acquisition system (TILL Photonics). The frame sequences were exported into movie files or NIH Image for analysis and were stored on CDROM media. For particle tracking, successive frames were taken for a particle consistently appearing in every frame, and the movement was measured per frame and per total period of analysis. For confocal microscopy and FRAP experiments, the transfected cells in the closed chamber system were mounted and imaged on a BioRad Radiance 2000 Laser Scanning Confocal Microscope (Biorad). A cell of interest was bleached at 100% laser power under visual control for approximately 15–25 s until the fluorescence was no longer visible. The imaging sequence was collected immediately after the bleaching was complete. Fluorescence was quantitated with NIH Image software. Deconvolution was done with EPR (Scanalytics) software as has been described in previous publications [18, 39]. Movies were subjected to a two-dimensional deconvolution routine by using Huygens software (SVI).
C2C12 cells were transfected with a plasmid expressing the ZBP1-GFP fusion for 24 hr, serum starved overnight, and stimulated with 20% FBS for 2 hr. For mislocalization, the GAP43-CFP-ZBP1 fusion was transfected into C12 cells and was expressed for 24 hr, and the cells were then serum starved overnight and stimulated with 20% FBS for 2 hr. The cells were fixed and stained for β-actin mRNA via in situ hybridization.
Drug and Antisense Treatments
LMB at 5 ng/ml and actinomycin D at 5 µg/ml final concentration were applied for 3 hr in MEM supplemented with 10% fetal calf serum. BDM (20 nM), actinomycin D (5 µg/ml), cytochalasin D (5 µg/ml), and colchicine (100 µM) were all were used for 30 min in MEM + FCS. Cells were subsequently fixed and stained.
The antisense treatment protocol is described in detail in [7]. Briefly, the antisense and reverse antisense (reverse orientation of the antisense) phosphorothioate oligonucleotides at 8 µM concentration were applied in MEM media to chicken embryo fibroblasts for 12 hr, and fresh oligonucleotides were added every 4 hr. The cells were subsequently fixed and stained.
Supplementary Material
Movie 1
Movie 5
Movie 6
Movie 7
Movie 8
Movie 9
Supplemental
Movie 10
Movie 11
Movie 12
Movie 13
Movie 14
Movie 2
Movie 3
Movie 4
Acknowledgments
This work was supported by National Institutes of Health grant AR41480. We thank Shailesh M. Shenoy for his essential expertise in imaging and processing the data. We also thank Carolyn Marks and the Analytical Imaging Facility at Albert Einstein College of Medicine for expert electron microscopy assistance.
Footnotes
Supplementary Material
Supplementary Material including the movies that correspond to Figure 1, Figure 2, Figure 6, a deconvolved version of the movie corresponding to Figure 1D, and movies and tables for the movements of nine particles from the movie in Figure 1D is available at http://images.cellpress.com/supmat/supmatin.htm.
References
- 1.St. Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 2.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast con-trolled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 3.Takizawa PA, Sil A, Swedlow JR, Herzkowitz I, Vale RD. Actin dependent localization of an RNA encoding a cell fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 4.Kislauskis EH, Zhu EH, Singer RH. β actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kislauskis EH, Zhu X-C, Singer RH. Sequences required for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zip-code-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oleynikov Y, Singer RH. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- 10.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 12.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 13.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 14.Adam SA, Lobl TJ, Mitchell MA, Gerace L. Identification of specific binding proteins for a nuclear location sequence. Nature. 1989;337:276–279. doi: 10.1038/337276a0. [DOI] [PubMed] [Google Scholar]
- 15.Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin mRNAs. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- 16.Mckillop DFA, Fortune NS, Ranatungga KW, Geeves MA. The influence of 2,3-butanedione 2-monoxime(BDM) on interaction between actin and myosin in solution and in skinned muscle fibres. J. Mus. Res. Cell Motil. 1994;15:309–318. doi: 10.1007/BF00123483. [DOI] [PubMed] [Google Scholar]
- 17.Latham VM, Yu EHS, Tullio AN, Adelstein RS, Singer RH. A Rho-dependent signaling pathway operating through myosin localizes β-actin mRNA in fibroblasts. Curr. Biol. 2001;11:1010–1016. doi: 10.1016/s0960-9822(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neuro-trophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 19.Skene JH, Virag I. Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J. Cell Biol. 1989;108:613–624. doi: 10.1083/jcb.108.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 21.Pinol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- 22.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 23.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 24.Taneja KL, Lifshitz LM, Fay FS, Singer RH. Poly(A) RNA codistribution with microfilaments: evaluation by in-situ hybridization and quantitative digital imaging micros-copy. J. Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 26.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J. Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 28.Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 31.Lall S, Francis-Lang H, Flament A, Norvell A, Schupbach T, Ish-Horowicz D. Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell. 1999;98:171–180. doi: 10.1016/s0092-8674(00)81012-0. [DOI] [PubMed] [Google Scholar]
- 32.Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 33.Shan J, Moran-Jones K, Munro TP, Kidd GJ, Winzor DJ, Hoek KS, Smith R. Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J. Biol. Chem. 2000;275:38286–38295. doi: 10.1074/jbc.M007642200. [DOI] [PubMed] [Google Scholar]
- 34.Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 35.Mowry KL, Cote CA. RNA sorting in Xenopus oocytes and embryos. FASEB J. 1999;13:435–445. doi: 10.1096/fasebj.13.3.435. [DOI] [PubMed] [Google Scholar]
- 36.Gautreau D, Cote CA, Mowry KL. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development. 1997;124:5013–5020. doi: 10.1242/dev.124.24.5013. [DOI] [PubMed] [Google Scholar]
- 37.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 39.Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 2001;153:307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 41.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 42.Elisha Z, Havin L, Ringel I, Yisraeli JK. Vg1 RNA binding protein mediates the association of Vg1 RNA with microtubules in Xenopus oocytes. EMBO J. 1995;14:5109–5114. doi: 10.1002/j.1460-2075.1995.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1
Movie 5
Movie 6
Movie 7
Movie 8
Movie 9
Supplemental
Movie 10
Movie 11
Movie 12
Movie 13
Movie 14
Movie 2
Movie 3
Movie 4