Comprehensive Genetic Landscape of Uveal Melanoma by Whole-Genome Sequencing (original) (raw)
Abstract
Uveal melanoma (UM) is a rare intraocular tumor that, similar to cutaneous melanoma, originates from melanocytes. To gain insights into its genetics, we performed whole-genome sequencing at very deep coverage of tumor-control pairs in 33 samples (24 primary and 9 metastases). Genome-wide, the number of coding mutations was rather low (only 17 variants per tumor on average; range 7–28), thus radically different from cutaneous melanoma, where hundreds of exonic DNA insults are usually detected. Furthermore, no UV light-induced mutational signature was identified. Recurrent coding mutations were found in the known UM drivers GNAQ, GNA11, BAP1, EIF1AX, and SF3B1. Other genes, i.e., TP53BP1, CSMD1, TTC28, DLK2, and KTN1, were also found to harbor somatic mutations in more than one individual, possibly indicating a previously undescribed association with UM pathogenesis. De novo assembly of unmatched reads from non-coding DNA revealed peculiar copy-number variations defining specific UM subtypes, which in turn could be associated with metastatic transformation. Mutational-driven comparison with other tumor types showed that UM is very similar to pediatric tumors, characterized by very few somatic insults and, possibly, important epigenetic changes. Through the analysis of whole-genome sequencing data, our findings shed new light on the molecular genetics of uveal melanoma, delineating it as an atypical tumor of the adult for which somatic events other than mutations in exonic DNA shape its genetic landscape and define its metastatic potential.
Main Text
Despite having the very rare incidence of 5–8 new cases per million per year,1, 2 uveal melanoma (UM [MIM: 155720]) is the most common primary intraocular tumor of the adult. It develops from melanocytes in the choroid, the ciliary body, or the iris (collectively called the “uvea,” one of the inner layers of the eye) and usually metastasizes through the blood stream to the liver.3, 4 Symptoms include variable and painless visual disturbances, often presenting when the tumor has already reached a considerable mass. Survival and potential therapeutic options depend, among other things, on the presence of specific genetic alterations.5 Although population studies suggest ethnic predisposition,6, 7 environmental factors that are directly involved in the transformation process have not been clearly delineated. For instance, a possible association with UV light exposure has been suggested,8, 9, 10, 11 but questioned recently by molecular data.12 Research on UM molecular genetics has been performed mostly by investigating coding mutations or copy-number variations detectable by direct sequencing of target genes, karyotype, array-CGH, MLPA, or SNP-array analyses.13, 14, 15, 16, 17 As a result, mutations at codon 209 of the paralogous oncogenes GNAQ (MIM: 600998) and GNA11 (MIM: 139313)18, 19 and in the tumor suppressor BAP1 (MIM: 603089)20 have been identified in the majority of UMs, whereas insults in EIF1AX (MIM: 300186) and SF3B1 (MIM: 605590) or other genes seem to be less frequent, accounting for at most 20% of cases.12, 21, 22, 23, 24, 25, 26 Moreover, copy gains and losses are common events in this tumor, typically involving chromosome 3 monosomy, 6p gain, and 8q gain.14, 17 After whole-genome sequencing of a series of tumor-control pairs, we present here an analytically unbiased and comprehensive assessment of the genetic landscape of UM.
We screened 33 UM samples (24 primary tumors and 9 unrelated metastases; Table S1) and corresponding normal tissue pairs by deep-coverage whole-genome sequencing (WGS), using the processing platform by Complete Genomics.27 Written informed consent was obtained from all individuals enrolled in this study, and approval for human subject research was obtained from the Institutional Review Boards of all participating Institutions. Sequence reads were mapped to the human reference genome, assembly GRch37, and somatic variants in tumors were called by comparison with the matched normal genomes, as previously described.28 All samples were surgically collected from eye enucleations or resected from liver metastases, allowing very clear post-surgery macroscopic isolation of tumor tissue from the surrounding environment. None of the 33 affected individuals received any treatment prior to primary tumor removal. Average coverage was 112× (range 102–118) for both tumor and control samples (>96% of the genome was covered 40× or more times), with minimal inter-individual variations (not shown). Mutation calls were performed genome-wide and included single-nucleotide variants (SNVs), copy-number variations (CNVs), as well as structural variations (SVs) such as chromosomal rearrangements. CNVs and SVs were assessed by comparing sequence coverage and especially de novo assembly of reads defining novel genetic junctions.28 Data were extracted from MasterVar files and other relevant matrices by ad hoc Perl, bash, and R scripts, available upon request. Overall, we detected 37,321 SNVs (average per sample 1,166; range 576–2,131), 1,584 SVs (average per sample 50; range 13–182), and a number of CNVs corresponding to an average of 13.6% of the genome (range 0.03%–33.9%) (Table S2).
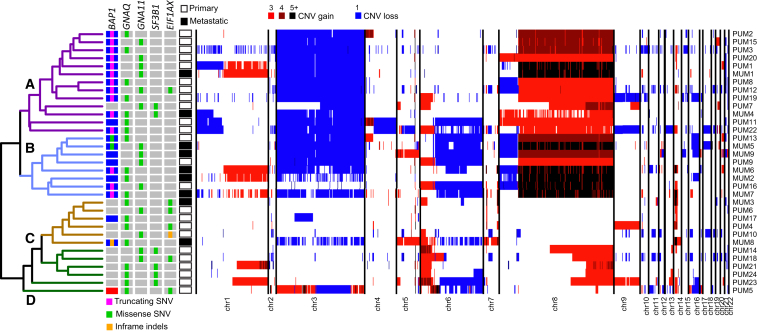
Unsupervised hierarchical clustering of our samples on CNVs revealed four major subgroups associated with mutational and metastatic status, branched two by two (Figure 1). Classes A and B (first branch) involved samples carrying chromosome 3 monosomy (by Fisher test, p value = 4.4 × 10−6), chr 8q gain (p value = 2.8 × 10−9), and in some instances chr 8p loss (p value = 3.0 × 10−2). In addition, class B tumors also had loss of chr 6q (p value = 1.0 × 10−3). Conversely, classes C and D represented more distinct subtypes with relatively few chromosomal rearrangements; class C tumor had no major aneuploidies, whereas class D reported gains of the distal part of chr 8q (p value = 2.0 × 10−3). Seven samples presenting chr 1q gain were scattered across all classes, whereas loss of chr 1p was typical of class A tumors (p value = 5.0 × 10−3).
Figure 1.
Unsupervised Hierarchical Clustering and Global Genetic Landscape of All Tumors Analyzed in This Study
Sample IDs are indicated on the right. CNV events are depicted in blue (copy losses) or in shades of red (copy gains) and ploidy is indicated in the legend provided at the top. SNVs in genes found to carry mutations in six or more individuals are shown on the left, with mutation classes provided within gray or blue boxes. Clustering identifies four classes—A, B, C, and D (see text)—indicated within the dendrogram.
Samples with monosomy of chr 3 were also associated (77% of cases) with somatic mutations in the tumor suppressor BAP1, lying on chr 3p21.1, in accordance with Knudson’s two-hit model of tumorigenesis.29 Indeed, BAP1 SNVs included all kinds of somatic events, but mostly mutations leading to premature stop codons and therefore to functional protein knockout (Figure S1, Table S3). Hallmark driver mutations in the GNAQ and GNA11 paralogs, encoding the components of the alpha subunit of the Gq protein heterotrimer, were present in 100% of the samples examined. They occurred in a perfectly mutually exclusive pattern and involved only four specific missenses—c.626A>T (p.Gln209Leu) (GenBank: NM_002072.3; 11 samples) and c.626A>C (p.Gln209Pro) (GenBank: NM_002072.3; 8 samples) in GNAQ, and c.626A>T (p.Gln209Leu) (GenBank: NM_002067.2; 13 samples) and c.626A>C (p.Gln209Pro) (GenBank: NM_002067.2; 1 sample) in _GNA11_—affecting the same functional amino acid residue and conferring oncogenic potential to this G protein.18, 19 Six tumors (18%) had missense mutations in SF3B1 (Splicing Factor 3B, subunit 1), affecting codon 625 (5 cases) and codon 626 (1 case) (Figure S1, Table S3), a previously described hotspot region.21 Finally, 7 other tumors had mutations impacting the first codons of EIF1AX (Eukaryotic Translation Initiation Factor 1A, X-Linked) (Figure S1, Table S3).22 Mutations in SF3B1 and EIF1AX seemed to occur in a mutually exclusive pattern and to be enriched with classes C and D (p value = 1.6 × 10−4), with SF3B1 preferentially mutated in class D. Except for one sample, BAP1 mutations were never observed in case subjects carrying mutations in SF3B1 or EIF1AX (p value = 1.4 × 10−5), in agreement with findings from previous literature.23, 30 Also, consistent with the fact that all of the tumors analyzed harbored variants affecting either GNAQ or GNA11 Gln209, no somatic SNVs were observed in PLCB4 or CYSLTR2, two genes that have been found to be mutated in a mutually exclusive pattern with respect to GNAQ or GNA11 variants.25, 26
Five genes (TP53BP1 [MIM: 605230], CSMD1 [MIM: 608397], TTC28 [MIM: 615098], DLK2, and KTN1 [MIM: 600381]) harbored somatic missense or truncating mutations in at least two samples, across all tumor classes (Table S3). TP53BP1 is a partner of the tumor suppressor protein p53, known to play a crucial role in maintaining genomic integrity as a mediator and effector of homologous recombination in response to double-strand breaks. This protein acts as a molecular scaffold that recruits responsive proteins, in order to repair damaged chromatin,31 and its depletion has been associated with increased cell proliferation.32 CSMD1 (Cub and Sushi Multiple Domains-1) is a candidate tumor suppressor gene, the hyper-expression of which increases survival in mice with xenografted tumors.33 Loss of CSMD1 was detected in a large set of cancers, including head and neck, lung, breast, and skin primary tumors,34 and associated with high tumor grade in invasive ductal breast carcinoma.35 TTC28 (Tetratricopeptide Repeat Domain 28) is a ubiquitous protein, associated with diverse biological functions. Of note, TCC28 plays a critical role in the progress of mitosis and cytokinesis during mammalian cell cycle and its dysfunction was described as a potential component of tumorigenesis and tumor progression.36, 37 DLK2 (Delta-Like 2 homolog) is a transmembrane epidermal growth factor-like protein. It is highly homologous to DLK1, a protein that was found to be present at high levels in gliomas and involved in cell proliferation.38 Similar to DLK1, DLK2 can bind to NOTCH1,39 modulating the oncogenic potential of cultured melanoma cells.40 Finally, KTN1 (Kinectin 1) is a protein of the endoplasmic reticulum membrane that interacts with kinesins.41, 42 Its role in cancer may be linked to dysregulation of cytoskeletal activity and mitosis. Two of these five genes were previously found to be mutated in UM: a missense in TTC28 was detected in 1 out 35 samples from a WES screen,22 and the cBioPortal repository reports a missense mutation in KTN1 in one out of 80 tumors profiled by TCGA.43, 44
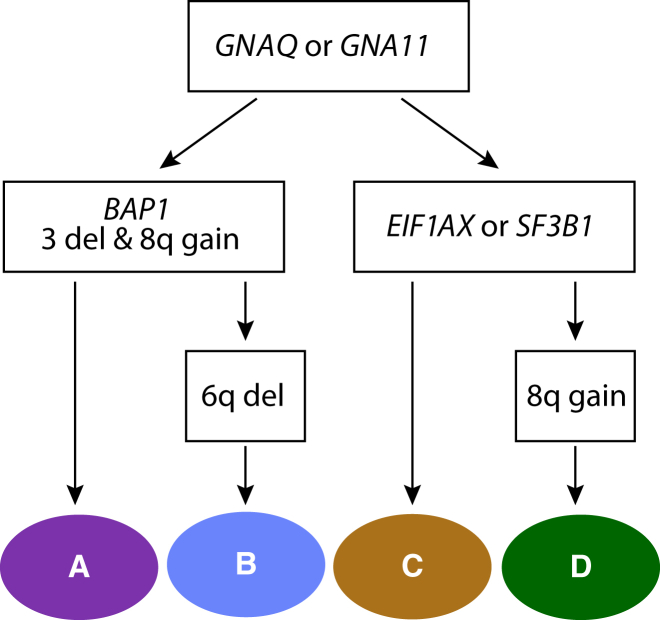
Taken together, our clustering analysis indicates that initial events involve GNAQ or GNA11 mutations, followed by a major branching determined by the functional loss of BAP1 and copy gains of chr 8q versus cases with a relatively normal chromosomal ploidy. These latter samples have conversely mutations in EIF1AX or in SF3B1 (classes A and B versus C and D, respectively). In tumors with BAP1 mutations, the long arm of chr 6 could eventually be lost, differentiating class B from class A. In tumors that are negative for BAP1 mutations, part of chr 8q could undergo amplification, differentiating class D from class C (Figure 2).
Figure 2.
Inferred Somatic Events Defining Tumor Classes, as Identified by Clustering
Colors are the same as those shown in Figure 1. All steps determining branching are statistically significant.
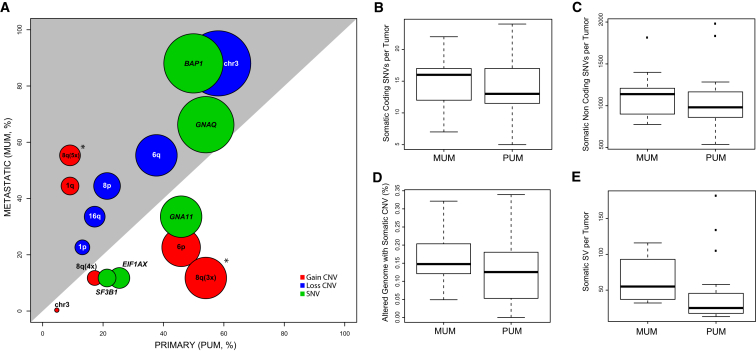
Aggregate analyses on genetic data showed no significant differences between primary UM (PUM) and metastatic UM (MUM) samples, in terms of number of somatic coding SNVs, non-coding SNVs, CNVs, and SVs, indicating that the extent of genomic instability was here not associated with metastatic potential (Figures 3B–3E). Although singularly none of the main somatic drivers (chr 3, 6q, and 8q aneuploidies, as well as SNVs in BAP1, GNAQ, GNA11, SF3B1, EIF1AX) were computed as being statistically different, enrichment in PUMs versus MUMs showed very clear association trends (Figure 3A).
Figure 3.
Genetic Features in Primary and Metastatic Tumors
(A) Overview of all major somatic events with respect to PUMs and MUMs. Each circle indicates a specific genetic event; its center corresponds to the percentage of samples carrying this feature in PUMs versus MUMs, whereas its diameter indicates the total number of such samples. Asterisks indicate statistical significance. The gray area depicts the surface of the plot for which there is an enrichment in MUMs.
(B–E) Boxplots of different types of genetic alterations, at the genome-wide scale.
Remarkably, when considering specific levels of 8q amplification detectable by algorithms querying non-coding WGS data for CNVs and aneuploidies, we found a very clear association between metastatic potential and 8q ploidy of five copies or more (p value = 8.6 × 10−4, Figure 3A). In addition, single-copy amplification of 8q (ploidy = 3) was indeed associated with primary tumors (p value = 4.7 × 10−3, Figure 3A). Similarly, when mutational sets defining tumor sub-classes were considered, a significant association between sub-class B and metastases was identified (Figure 1, p value = 2.0 × 10−2).
With respect to metastatic samples, the aneuploidies identified correlated well with those of 66 liver metastases from UM investigated previously, detecting chr 3 monosomy (73%), 8q gain (89%), 6q loss (64%), 1p loss (47%), 8p loss (45%), 1q gain (35%), and 16q loss (32%).14 Similarly, the identified SNVs matched those on another study on five liver metastases.45 Finally, the identification of a SF3B1 mutation in one metastatic sample from our series is also in line with late-onset metastasis occurring in individuals with SF3B1 mutations.23
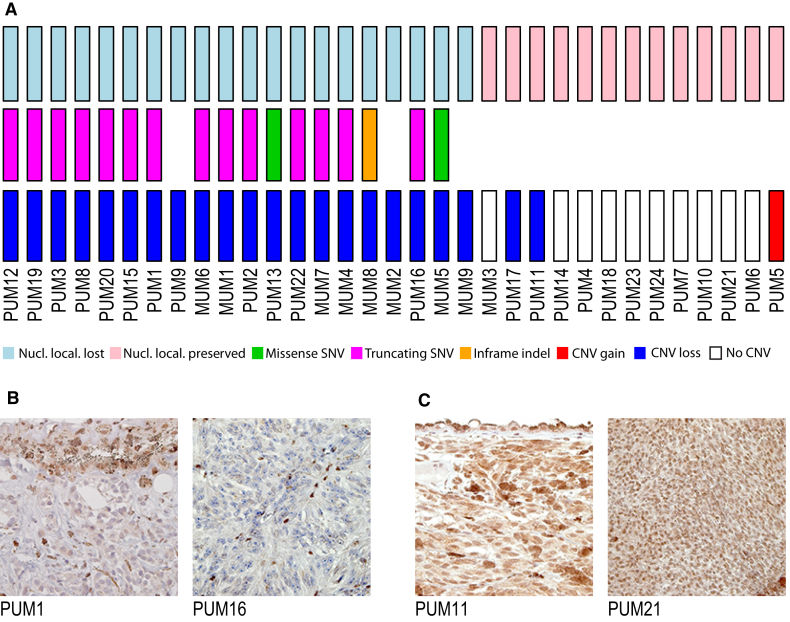
Mutations targeting BAP1 are one of the genetic landmarks of UM20 and were found here to be associated with classes A and B (p value = 1.2 × 10−4), classes that are in fact defined mostly by the presence of chr 3 monosomy. To test for functional inactivation of the BAP1 protein, we assessed its nuclear staining in histological sections of all tumors (Figure 4). Of the 33 samples (60%), 20 displayed loss of nuclear localization (Figure 4). Of these, 17 (85%) presented chr 3 monosomy and a coding SNV (a truncating SNV in 14 cases and a missense or an in-frame deletion in 3 cases), accounting for loss of heterozygosity and protein delocalization.
Figure 4.
Landscape of Genetic Alterations Involving BAP1 and Immunohistochemistry of BAP1 Protein
(A) Samples are ordered with respect to absence/presence of BAP1 nuclear localization, indicating loss or preservation of protein function, respectively. In general, loss of BAP1 function correlates with presence of a somatic SNV and loss of heterozygosity.
(B and C) Representative micrographs of paraffin-embedded UM samples showing absence (B) and presence (C) of BAP1 protein. Magnification: 252×.
The number of somatic SNVs involving coding and non-coding regions was strikingly low (Figure S2). Globally, the average load of coding mutations was 0.24/Mb (range 0.08–0.42/Mb, Table S4), one of the lowest detected so far in tumors. Comparison with other cancer types revealed that UM mutational load for coding regions was closer to that of pediatric tumors such as rhabdoid tumors, medulloblastoma (MIM: 155255), neuroblastoma, etc.,46 rather than that of adult cancers (Figure S3). Pediatric tumors typically develop over a shorter period than most adult malignancies, frequently harbor few driver mutations, and may thereby have fewer sources of heterogeneity, facilitating the assessment of both the genetic and epigenetic determinants underlying their pathogenesis. Our data seem to suggest that, similarly to pediatric tumors, UM development may rely more on epigenetic drivers of transformation and tumor progression, rather than the classical accumulation of genetic events observed in the vast majority of adult malignancies.
The number of non-coding SNVs was also relatively low (736 per tumor on average, range 371–1,347) and mostly proportional to the number of coding SNVs (17 per tumor on average, range 7–28) (Figure S2), confirming that, compared to both cutaneous and conjunctival melanomas, which also originate from melanocytes, UM follows a different oncogenic pathway, characterized by significantly fewer mutations.47, 48 In addition, we failed to identify any statistically relevant non-coding SNVs for tumor-specific sites that were present in four samples or more, suggesting the absence of common regulatory variants in the landscape of these tumors, at least in our cohort.
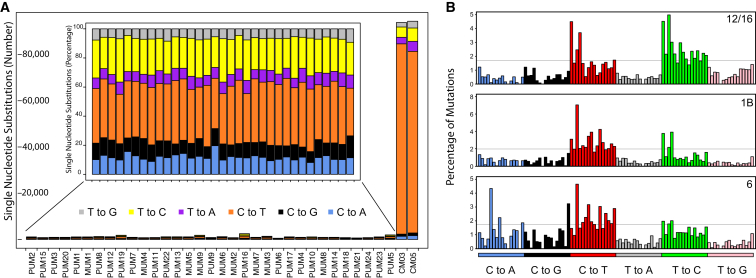
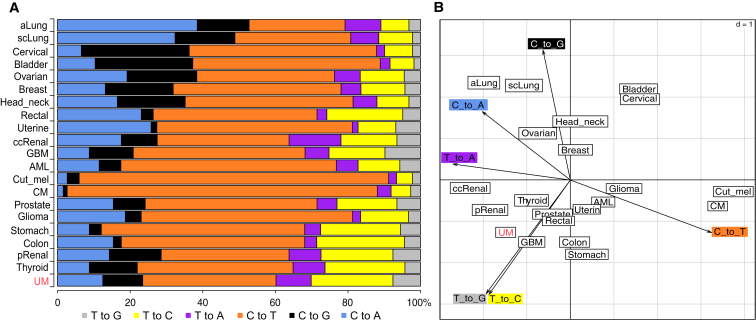
Another difference between UM and cutaneous and conjunctival melanomas involved its mutational spectrum (Figure 5). Analysis of all coding and non-coding somatic single-nucleotide substitutions (SNSs) from our series showed the clear absence of an UV-induced mutation signature. This particular spectrum results from sunlight-driven formation of pyrimidine dimers on the DNA49 and is found in both cutaneous and conjunctival melanomas (Figure 6A).47, 48 Direct analysis of genes known to be involved in cutaneous melanoma, such as BRAF (MIM: 164757), NRAS (MIM: 164790), and NF1 (MIM: 162200) revealed no somatic mutations in UM, supporting again the notion that uveal and cutaneous melanomas have a different molecular etiology.50, 51 Conversely, the UM mutational spectrum was remarkably similar across all PUMs and MUMs and resembled that of apparently unrelated tumors, such as clear cell renal carcinoma, thyroid tumor, and glioblastoma (Figure 6). Notably, despite a different cellular origin, UM shares with these tumors recurrent genetic modifications; BAP1 mutations and chr 3 monosomy are frequently seen in clear cell renal carcinoma,52 while hotspot mutations in the first codons of EIF1AX are recurrent in papillary thyroid carcinoma.53
Figure 5.
Mutational Signature of Our Samples, for SNSs, Genome-wide
(A) Main graph: comparison of mutational load of the UM samples studied with respect to two melanomas of the conjunctiva (CM), sequenced and processed according to the same methods.48 The number of mutations is radically different in UM versus CM. Inset: mutational spectrum of each UM sample, in percentage, showing a relatively homogeneous pattern.
(B) Results of the analysis of mutational events according to the methods and the classification proposed by Alexandrov et al.54, 55 Three main signatures are detected in our samples, evocative of Alexandrov’s signatures 12/16, 1B, and 6. The different peaks indicate specific genetic contexts of the altered nucleotide and are ordered according to the original article.54
Figure 6.
Analysis of the Mutational Spectrum in Our Samples versus Other Cancer Types, in Coding Regions Only
(A) The spectrum from UM is dissimilar from those from cutaneous and conjunctival melanomas, which are dominated by UV light-induced events (C to T transitions) and is conversely closer to that of thyroid and renal papillary cancer.
(B) Principal-component analysis (PCA) of the same data, showing the relatedness of UM with a few cancers but, again, not with other melanomas. Dimensions of the PCA are indicated by the arrowed axes. Primary data other than UM are from previously published sources.48, 62 Cancer types are indicated by the following abbreviations. aLung: lung, adenocarcinoma; scLung: lung, squamous cell; pRenal: renal, papillary; ccRenal: renal, clear cell; GBM: glioblastoma multiforme; AML: acute myeloid leukemia; Cut_mel: cutaneous melanoma; CM: conjunctival melanoma.
Analysis of all specific SNS types along with composition of the flanking bases allowed us to determine specific mutational signatures for UM, according to the classification of Alexandrov et al.54 Our samples appeared to be enriched for signatures 12 or 16 (55% of the score), signature 1B (25%), and signature 6 (20%) (Figure 5B). Signature 1B corresponds to a rather ubiquitous pattern in cancer, resulting from the spontaneous deamination of 5-methyl-cytosine, which in turn is thought to correlate with the process of aging.54, 55 Conversely, signatures 12/16 and 6 are associated with defects in nucleotide excision repair and DNA mismatch repair, respectively.
A more global approach, considering the intersection between the SNVs detected in our series and the most frequently mutated genes in cancer (TCGA PANCAN list),56 also revealed a minimal overlap, limited to BAP1 and SF3B1 (Figure S4).
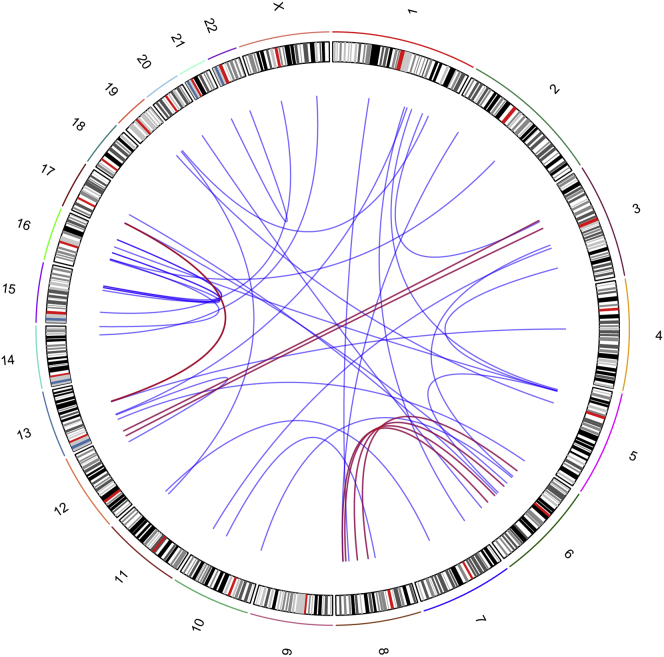
A non-negligible number of structural variants (SVs) such as deletions, duplications, inversions, or inter- and intra-chromosomal rearrangements were also observed (Figure S5). Only a few of these events were recurrent, indicating the absence of major common drivers constituted by genetic events involving large parts of the genome. Among these, however, there were three inter-chromosomal events that were present in at least two individuals (Figure 7). Three samples (PUM20, PUM18, and PUM5) had a translocation involving chr 6 and chr 8 disrupting UBE2W (MIM: 614277) and MYO6 (MIM: 600970) for the two first samples, respectively. The third event occurred in intergenic regions. Translocations between chr 13 and chr 17 (no genes involved) were also present in MUM9 and PUM5 and between chr 3 and chr 12 in PUM17 (no genes involved) and in PUM5 (disrupting KDM2B [MIM: 609078]) (Table S5). Although these translocations impacted roughly the same genomic areas, highlighting possible hotspot regions in UM genome, they neither targeted the same genes nor defined a specific tumor sub-category. Of note, one individual (PUM5) appeared to harbor a higher number of interchromosomal events and SVs than the average value of the other cases (Figure S5). Notably, this individual was also an outlier of our clustering analysis (Figure 1). However, neither the medical history nor tumor pathology revealed any uncommon feature, compared to the rest of the cohort.
Figure 7.
Circos Plot of All Somatic Interchromosomal Events, in All UM Samples
Red lines indicate events involving the same chromosomal regions in more than two individuals.
Amplification of chr 8q is a well-known and important feature of UM.17, 57, 58 Levels of chr 8q amplification seem to define prognostic status and metastatic potential in UM and differentiate class C from D (Figures 1 and 2). However, the molecular bases for this phenomenon are not known. One possible explanation is that the amplification is driven by the MYC oncogene (MIM: 190080), which lies in this region.59 By comparing the pattern of chr 8q amplification in our samples, we determined the minimal region of overlap, involving a 2.3 Mb fragment toward its telomeric site (chr 8: 126,404,000–128,682,000).
Surprisingly, this region was very close to MYC (chr 8: 128,748,314–128,753,680) but did not include it. Conversely, it harbored six other genes (POU5F1B [MIM:615739], FAM84B [MIM: 609483], TRIB1 [MIM: 609461], LOC100130231/LINC00861, LOC100507056/CCAT1, and LOC727677/CASC8). The most interesting of them was POU5F1B (POU Class 5 Homeobox 1B), a pseudogene of the POU5F1/OCT4 family, recently shown to be involved in prostatic and gastric cancer.60, 61 Real-time quantitative PCR experiments showed that only POU5F1B, TRIB1, LINC00861, and CCAT1 were expressed in our UM samples, but no statistically significant correlation between their expression levels and 8q amplification or tumor class could be detected. The same held true for the MYC transcript, suggesting that none of these genes play a key role in UM pathogenesis (Figures S6 and S7).
By using a WGS-based, untargeted approach to investigate the genetic components of UM, we had the unique opportunity of assessing its genomic landscape as a whole, from single-nucleotide variants to interchromosomal rearrangements, providing the basis for future functional studies that go beyond the scope of our analysis. The global picture emerging from our work indicates that, genetically, UM is a relatively atypical tumor, mostly in virtue of the paucity of somatic events that characterize it. Driver mutations are very few and are confined to a relatively low number of genes, such as BAP1, GNAQ, and GNA11. Other genes, including those that were identified in this study, may have a role in tumorigenesis, but they are nonetheless present in a small fraction of the samples studied. Conversely, larger events such as extended copy-number and structural variations seem to shape UM’s genome in a much more relevant way, possibly determining tumor progression and fate. Taken together, our results point to a critical role for non-canonical mechanisms of cellular transformation in UM development, where chromosomal rearrangements and non-coding SNVs potentially affecting distal regulatory elements may collaborate in the establishment of a permissive oncogenic landscape.
Acknowledgments
This work was supported by the “Fond'action contre le cancer” foundation, Switzerland, the Swiss National Science Foundation (Div III grant# 156260 to C.R. and SNF Professorship grant# PP00P3-157468/1 to N.R.), and the MEDIC Foundation. We wish to thank Complete Genomics (Mountain View, CA) for providing the whole-genome sequencing and Patricia Martin for her invaluable help. We thank as well the Lausanne Institutional Biobank and Danielle Minaidis for help in collecting biological material from affected individuals.
Published: October 13, 2016
Footnotes
Web Resources
- GenBank, http://www.ncbi.nlm.nih.gov/genbank/
- OMIM, http://www.omim.org/
- Primary dataset, https://drive.switch.ch/index.php/s/7GjD2zQEoPOyaj9
- RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq
Supplemental Data
Document S1. Figures S1–S7 and Tables S1–S3 and S5
Table S4. Somatic Coding SNVs Detected
Document S2. Article plus Supplemental Data
References
- 1.Singh A.D., Turell M.E., Topham A.K. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 2.Bergman L., Seregard S., Nilsson B., Ringborg U., Lundell G., Ragnarsson-Olding B. Incidence of uveal melanoma in Sweden from 1960 to 1998. Invest. Ophthalmol. Vis. Sci. 2002;43:2579–2583. [PubMed] [Google Scholar]
- 3.Luke J.J., Triozzi P.L., McKenna K.C., Van Meir E.G., Gershenwald J.E., Bastian B.C., Gutkind J.S., Bowcock A.M., Streicher H.Z., Patel P.M. Biology of advanced uveal melanoma and next steps for clinical therapeutics. Pigment Cell Melanoma Res. 2015;28:135–147. doi: 10.1111/pcmr.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodman S.E. Metastatic uveal melanoma: biology and emerging treatments. Cancer J. 2012;18:148–152. doi: 10.1097/PPO.0b013e31824bd256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shields J.A., Shields C.L. Management of posterior uveal melanoma: past, present, and future: the 2014 Charles L. Schepens lecture. Ophthalmology. 2015;122:414–428. doi: 10.1016/j.ophtha.2014.08.046. [DOI] [PubMed] [Google Scholar]
- 6.Park S.J., Oh C.M., Kim B.W., Woo S.J., Cho H., Park K.H. Nationwide incidence of ocular melanoma in South Korea by using the national cancer registry database (1999-2011) Invest. Ophthalmol. Vis. Sci. 2015;56:4719–4724. doi: 10.1167/iovs.15-16532. [DOI] [PubMed] [Google Scholar]
- 7.Hu D.N., Yu G.P., McCormick S.A., Schneider S., Finger P.T. Population-based incidence of uveal melanoma in various races and ethnic groups. Am. J. Ophthalmol. 2005;140:612–617. doi: 10.1016/j.ajo.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 8.Seddon J.M., Gragoudas E.S., Glynn R.J., Egan K.M., Albert D.M., Blitzer P.H. Host factors, UV radiation, and risk of uveal melanoma. A case-control study. Arch. Ophthalmol. 1990;108:1274–1280. doi: 10.1001/archopht.1990.01070110090031. [DOI] [PubMed] [Google Scholar]
- 9.Holly E.A., Aston D.A., Ahn D.K., Smith A.H. Intraocular melanoma linked to occupations and chemical exposures. Epidemiology. 1996;7:55–61. doi: 10.1097/00001648-199601000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Vajdic C.M., Kricker A., Giblin M., McKenzie J., Aitken J., Giles G.G., Armstrong B.K. Sun exposure predicts risk of ocular melanoma in Australia. Int. J. Cancer. 2002;101:175–182. doi: 10.1002/ijc.10579. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt-Pokrzywniak A., Jöckel K.H., Bornfeld N., Sauerwein W., Stang A. Positive interaction between light iris color and ultraviolet radiation in relation to the risk of uveal melanoma: a case-control study. Ophthalmology. 2009;116:340–348. doi: 10.1016/j.ophtha.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 12.Furney S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de la Grange P., Roman-Roman S. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prescher G., Bornfeld N., Hirche H., Horsthemke B., Jöckel K.H., Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 14.Trolet J., Hupé P., Huon I., Lebigot I., Decraene C., Delattre O., Sastre-Garau X., Saule S., Thiéry J.P., Plancher C. Genomic profiling and identification of high-risk uveal melanoma by array CGH analysis of primary tumors and liver metastases. Invest. Ophthalmol. Vis. Sci. 2009;50:2572–2580. doi: 10.1167/iovs.08-2296. [DOI] [PubMed] [Google Scholar]
- 15.Cassoux N., Rodrigues M.J., Plancher C., Asselain B., Levy-Gabriel C., Lumbroso-Le Rouic L., Piperno-Neumann S., Dendale R., Sastre X., Desjardins L., Couturier J. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br. J. Ophthalmol. 2014;98:769–774. doi: 10.1136/bjophthalmol-2013-303867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Essen T.H., van Pelt S.I., Versluis M., Bronkhorst I.H., van Duinen S.G., Marinkovic M., Kroes W.G., Ruivenkamp C.A., Shukla S., de Klein A. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br. J. Ophthalmol. 2014;98:1738–1743. doi: 10.1136/bjophthalmol-2014-305047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damato B., Dopierala J.A., Coupland S.E. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin. Cancer Res. 2010;16:6083–6092. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 18.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O’Brien J.M., Simpson E.M., Barsh G.S., Bastian B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., Obenauf A.C., Wackernagel W., Green G., Bouvier N. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour J.W., Onken M.D., Roberson E.D., Duan S., Cao L., Worley L.A., Council M.L., Matatall K.A., Helms C., Bowcock A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harbour J.W., Roberson E.D., Anbunathan H., Onken M.D., Worley L.A., Bowcock A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M., Maßhöfer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yavuzyigitoglu S., Koopmans A.E., Verdijk R.M., Vaarwater J., Eussen B., van Bodegom A., Paridaens D., Kiliç E., de Klein A., Rotterdam Ocular Melanoma Study Group Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 24.Cimino P.J., Kung Y., Warrick J.I., Chang S.H., Keene C.D. Mutational status of IDH1 in uveal melanoma. Exp. Mol. Pathol. 2016;100:476–481. doi: 10.1016/j.yexmp.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Johansson P., Aoude L.G., Wadt K., Glasson W.J., Warrier S.K., Hewitt A.W., Kiilgaard J.F., Heegaard S., Isaacs T., Franchina M. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore A.R., Ceraudo E., Sher J.J., Guan Y., Shoushtari A.N., Chang M.T., Zhang J.Q., Walczak E.G., Kazmi M.A., Taylor B.S. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 28.Carnevali P., Baccash J., Halpern A.L., Nazarenko I., Nilsen G.B., Pant K.P., Ebert J.C., Brownley A., Morenzoni M., Karpinchyk V. Computational techniques for human genome resequencing using mated gapped reads. J. Comput. Biol. 2012;19:279–292. doi: 10.1089/cmb.2011.0201. [DOI] [PubMed] [Google Scholar]
- 29.Knudson A.G., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decatur C.L., Ong E., Garg N., Anbunathan H., Bowcock A.M., Field M.G., Harbour J.W. Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016;134:728–733. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 32.Bi J., Huang A., Liu T., Zhang T., Ma H. Expression of DNA damage checkpoint 53BP1 is correlated with prognosis, cell proliferation and apoptosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015;8:6070–6082. [PMC free article] [PubMed] [Google Scholar]
- 33.Tang M.R., Wang Y.X., Guo S., Han S.Y., Wang D. CSMD1 exhibits antitumor activity in A375 melanoma cells through activation of the Smad pathway. Apoptosis. 2012;17:927–937. doi: 10.1007/s10495-012-0727-0. [DOI] [PubMed] [Google Scholar]
- 34.Ma C., Quesnelle K.M., Sparano A., Rao S., Park M.S., Cohen M.A., Wang Y., Samanta M., Kumar M.S., Aziz M.U. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol. Ther. 2009;8:907–916. doi: 10.4161/cbt.8.10.8132. [DOI] [PubMed] [Google Scholar]
- 35.Kamal M., Shaaban A.M., Zhang L., Walker C., Gray S., Thakker N., Toomes C., Speirs V., Bell S.M. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res. Treat. 2010;121:555–563. doi: 10.1007/s10549-009-0500-4. [DOI] [PubMed] [Google Scholar]
- 36.Izumiyama T., Minoshima S., Yoshida T., Shimizu N. A novel big protein TPRBK possessing 25 units of TPR motif is essential for the progress of mitosis and cytokinesis. Gene. 2012;511:202–217. doi: 10.1016/j.gene.2012.09.061. [DOI] [PubMed] [Google Scholar]
- 37.Pitkänen E., Cajuso T., Katainen R., Kaasinen E., Välimäki N., Palin K., Taipale J., Aaltonen L.A., Kilpivaara O. Frequent L1 retrotranspositions originating from TTC28 in colorectal cancer. Oncotarget. 2014;5:853–859. doi: 10.18632/oncotarget.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin D., Xie D., Sakajiri S., Miller C.W., Zhu H., Popoviciu M.L., Said J.W., Black K.L., Koeffler H.P. DLK1: increased expression in gliomas and associated with oncogenic activities. Oncogene. 2006;25:1852–1861. doi: 10.1038/sj.onc.1209219. [DOI] [PubMed] [Google Scholar]
- 39.Sánchez-Solana B., Nueda M.L., Ruvira M.D., Ruiz-Hidalgo M.J., Monsalve E.M., Rivero S., García-Ramírez J.J., Díaz-Guerra M.J., Baladrón V., Laborda J. The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other’s activities. Biochim. Biophys. Acta. 2011;1813:1153–1164. doi: 10.1016/j.bbamcr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Nueda M.L., Naranjo A.I., Baladrón V., Laborda J. The proteins DLK1 and DLK2 modulate NOTCH1-dependent proliferation and oncogenic potential of human SK-MEL-2 melanoma cells. Biochim. Biophys. Acta. 2014;1843:2674–2684. doi: 10.1016/j.bbamcr.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 41.Fütterer A., Kruppa G., Krämer B., Lemke H., Krönke M. Molecular cloning and characterization of human kinectin. Mol. Biol. Cell. 1995;6:161–170. doi: 10.1091/mbc.6.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machleidt T., Geller P., Schwandner R., Scherer G., Krönke M. Caspase 7-induced cleavage of kinectin in apoptotic cells. FEBS Lett. 1998;436:51–54. doi: 10.1016/s0014-5793(98)01095-3. [DOI] [PubMed] [Google Scholar]
- 43.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luscan A., Just P.A., Briand A., Burin des Roziers C., Goussard P., Nitschké P., Vidaud M., Avril M.F., Terris B., Pasmant E. Uveal melanoma hepatic metastases mutation spectrum analysis using targeted next-generation sequencing of 400 cancer genes. Br. J. Ophthalmol. 2015;99:437–439. doi: 10.1136/bjophthalmol-2014-305371. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas N., Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivolta C., Royer-Bertrand B., Rimoldi D., Schalenbourg A., Zografos L., Leyvraz S., Moulin A. UV light signature in conjunctival melanoma; not only skin should be protected from solar radiation. J. Hum. Genet. 2016;61:361–362. doi: 10.1038/jhg.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikehata H., Ono T. The mechanisms of UV mutagenesis. J. Radiat. Res. (Tokyo) 2011;52:115–125. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 50.Foster W.J., Fuller C.E., Perry A., Harbour J.W. Status of the NF1 tumor suppressor locus in uveal melanoma. Arch. Ophthalmol. 2003;121:1311–1315. doi: 10.1001/archopht.121.9.1311. [DOI] [PubMed] [Google Scholar]
- 51.Cruz F., 3rd, Rubin B.P., Wilson D., Town A., Schroeder A., Haley A., Bainbridge T., Heinrich M.C., Corless C.L. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63:5761–5766. [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy S.R., Salk J.J., Schmitt M.W., Loeb L.A. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9:e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamborero D., Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Kandoth C., Reimand J., Lawrence M.S., Getz G., Bader G.D., Ding L., Lopez-Bigas N. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehlers J.P., Worley L., Onken M.D., Harbour J.W. Integrative genomic analysis of aneuploidy in uveal melanoma. Clin. Cancer Res. 2008;14:115–122. doi: 10.1158/1078-0432.CCR-07-1825. [DOI] [PubMed] [Google Scholar]
- 58.Sisley K., Rennie I.G., Parsons M.A., Jacques R., Hammond D.W., Bell S.M., Potter A.M., Rees R.C. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19:22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 59.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi H., Arao T., Togashi Y., Kato H., Fujita Y., De Velasco M.A., Kimura H., Matsumoto K., Tanaka K., Okamoto I. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34:199–208. doi: 10.1038/onc.2013.547. [DOI] [PubMed] [Google Scholar]
- 61.Breyer J.P., Dorset D.C., Clark T.A., Bradley K.M., Wahlfors T.A., McReynolds K.M., Maynard W.H., Chang S.S., Cookson M.S., Smith J.A. An expressed retrogene of the master embryonic stem cell gene POU5F1 is associated with prostate cancer susceptibility. Am. J. Hum. Genet. 2014;94:395–404. doi: 10.1016/j.ajhg.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burns M.B., Temiz N.A., Harris R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S7 and Tables S1–S3 and S5
Table S4. Somatic Coding SNVs Detected
Document S2. Article plus Supplemental Data