AIM2 inflammasome activation and regulation: A structural perspective (original) (raw)
. Author manuscript; available in PMC: 2018 Dec 1.
Published in final edited form as: J Struct Biol. 2017 Aug 13;200(3):279–282. doi: 10.1016/j.jsb.2017.08.001
Abstract
Absent in melanoma 2 (AIM2) inflammasome is a multi-protein platform that recognizes aberrant cytoplasmic dsDNA and induces cytokine maturation, release and pyroptosis. It is composed of AIM2, apoptosis-associated speck-like protein containing a CARD (ASC), and caspase-1. Recent X-ray crystallographic and high resolution cryo-electron microscopic (cryo-EM) studies have revealed a series of structures in AIM2 inflammasome activation and regulation. One prominent feature common in multiple steps is the assembly of high-order structures, especially helical filaments nucleated by upstream molecules, rather than stoichiometric complexes. In this review, we track the AIM2 inflammasome activation process step by step, using high-resolution structures to illustrate the overall architecture of AIM2 inflammasome and its assembly and regulatory mechanisms.
Keywords: AIM2, inflammasome, helical filament, high-order assembly, polymerization, ASC, caspase-1, PYD, CARD, p202, INCA
Inflammasomes are cytoplasmic multi-protein platforms that mediate cytokine maturation, secretion, and pyroptosis. There are three tiers of molecules in a typical inflammasome: sensor, adaptor, and effector (Figure 1A). Sensor proteins detect the presence of pathogen-derived molecular features or perturbation in cellular environment, adaptor proteins relay the signal to effectors through protein-protein interactions, and the effector inflammatory caspases process pro-inflammatory cytokines or gasdermin D into their mature forms, leading to cytokine secretion and pyroptosis (Broz and Dixit, 2016).
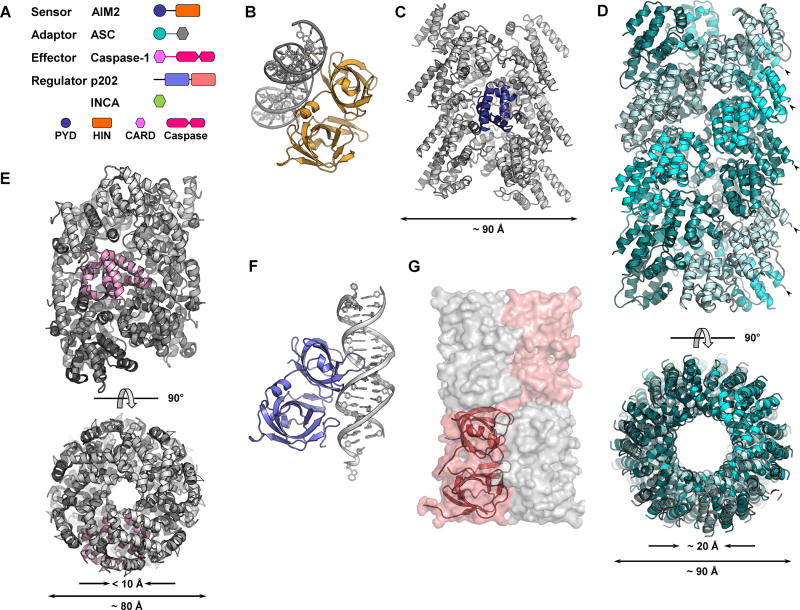
Figure 1.
(A) Domain organization of AIM2, ASC, caspase-1, p202, and INCA. Spheres, rectangles, and hexagons represent PYD, HIN, and CARD domains, respectively. (B) Crystal structure of AIM2HIN binds to a 19-bp dsDNA (PDB ID: 3RN5). AIM2HIN is colored in orange and DNA is in gray. (C) AIM2PYD filament structure reconstructed at ~ 5.0 Å. One protomer is colored in blue while the rest are in gray. (D) ASCPYD filament structure reconstructed at ~ 3.8 Å (PDB ID: 3J63). Two orthogonal views are shown. ASCPYD protomers are colored in three different shades of cyan. Arrowheads indicate ASCPYD C-termini. (E) Two orthogonal views of Casp1CARD filament reconstructed at ~ 4.8 Å (PDB ID: 5FNA). The filament is colored in a gray gradient with one protomer colored in pink. (F) Crystal structure of p202HIN1 in complex with a 20-bp dsDNA (PDB ID: 4L5R) with p202HIN1 colored in light blue and DNA in gray. The orientation of p202HIN1 is the same as AIM2HIN in (B). (G) Surface representation of p202HIN2 tetramer. Two protomers are colored in shades of salmon and the other two in gray. One protomer is also shown as a ribbon diagram.
Absent in melanoma 2 (AIM2) is a protein that senses the presence of double-stranded DNA (dsDNA) in the cytoplasm, be it from bacterial, viral or host cellular origin (Man et al., 2016). AIM2 is the founding member of the AIM2-like receptor (ALR) family that consists of four members in human genome and fourteen in mouse (Cridland et al., 2012; Schattgen and Fitzgerald, 2011). ALRs are usually bipartite proteins: an N-terminal PYD domain followed by one or two HIN domains with the exception of p202, which lacks the PYD domain (Figure 1A). The ~ 90 aa PYD domain, belonging to the death domain (DD) superfamily, mediates homotypic protein-protein interaction; while the HIN domain binds to DNA or other proteins using its two oligonucleotide/oligosaccharide-binding folds (OB-folds). Activated AIM2 recruits apoptosis-associated speck-like protein containing a CARD (ASC), which in turn recruits caspase-1 (Casp1) to form the complete AIM2 inflammasome (Figure 1A).
Since the identification of AIM2 as a cytoplasmic sensor of dsDNA in 2009 (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009), our understanding of the mechanisms governing AIM2 inflammasome activation and regulation has grown substantially, especially through biochemical and structural methods including X-ray crystallography, high-resolution cryo-electron microscopy (cryo-EM), and light microscopy. Here we will briefly summarize the structures and mechanisms in AIM2 inflammasome activation and regulation, with a focus on helical assemblies repeatedly observed in this process.
dsDNA recognition by AIM2
DNA binding is the first step in a series of events in AIM2 inflammasome formation. In X-ray crystallographic structures, AIM2HIN binds to both strands of B-form dsDNA (Jin et al., 2012) (Figure 1B), explaining its specificity to dsDNA over ssDNA. The interaction is mainly electrostatic with lysine and arginine residues coordinating with phosphates and sugar moieties on DNA backbone. Mutagenesis of certain positively charged residues confirmed their role in DNA binding. This is consistent with sequence-independent recognition of dsDNA by AIM2.
AIM2 entry into competency
AIM2PYD is a structural domain with a strong tendency to self-aggregate. The two AIM2PYD structures are obtained either by fusion to an N-terminal MBP solubility tag or through introduction of surface mutations (Jin et al., 2013; Lu et al., 2014a). The PYD domain adopts a structure of six-helix bundle characteristic of the death domain superfamily (Jin et al., 2013; Lu et al., 2014a). Both in vitro pull-down assay and fluorescence polarization (FP) assay results are consistent with intramolecular interactions between wild-type AIM2PYD and AIM2HIN. Presence of dsDNA or mutation of a patch of acidic residues on AIM2PYD disrupted such interactions (Jin et al., 2013; Jin et al., 2012). Xiao and colleagues proposed that in resting state, AIM2HIN sequesters AIM2PYD through intramolecular interactions. Binding of dsDNA displaces AIM2PYD from the clutch of AIM2HIN, ready to engage the downstream adaptor ASC. This hypothesis has been challenged by Sohn and colleagues (Morrone et al., 2015). In full-length AIM2, binding between dsDNA and the acid patch mutant was much looser, undermining the inhibitory role of AIM2PYD proposed before. Furthermore, AIM2 could self-associate without dsDNA when protein concentration reached a certain threshold (Morrone et al., 2015). Based on the new findings, Sohn and colleagues proposed that dsDNA works as a one-dimensional ruler for AIM2 to cluster upon, therefore increasing its local concentration.
AIM2 clustering on dsDNA
Although the footprint of one AIM2HIN on dsDNA is 8–9 bp, and isolated AIM2HIN binds to 20-bp dsDNA with an affinity of ~ 30 nM, full activation of AIM2 in cells required dsDNA of a minimum length of ~ 80 bp (Jin et al., 2012). Indeed, electron micrographs showed both AIM2FL and AIM2HIN bound to and coated the same piece of DNA molecule (Lu et al., 2015; Morrone et al., 2015), even when DNA was in excess (Morrone et al., 2015). Although AIM2PYD lacks affinity to DNA, it contributes to AIM2FL clustering on DNA through its intrinsic capability of self-association. Potential weak interactions between AIM2HIN protomers also affect clustering on dsDNA. Indeed, mutation of residues potentially involved in HIN: HIN interactions decreased cooperative DNA binding by both AIM2FL and AIM2HIN (Morrone et al., 2015).
AIM2PYD forming helical filaments
Multiple AIM2 binding to the same DNA molecule greatly elevate local concentration of AIM2PYD free to interact with each other. Even in the absence of DNA, AIMPYD readily assembled into long helical filaments at moderate concentration in vitro (Lu et al., 2015; Morrone et al., 2015). Reconstruction of GFP-AIM2PYD filaments revealed the helical AIM2PYD core filament as a right-handed one-start hollow filament with an outer diameter of ~ 90 Å and an inner one of ~ 20 Å (Lu et al., 2015) (Figure 1C). Comparison between AIM2PYD crystal structures with cryo-EM AIM2PYD filament structure revealed minimal conformation change (Jin et al., 2013; Lu et al., 2014a; Lu et al., 2015).
AIM2-nucleated ASCPYD polymerization into helical filaments
DNA sensor AIM2 engages adaptor protein ASC through homotypic PYD-PYD interactions (Fernandes-Alnemri et al., 2009; Hornung et al., 2009). Recapitulation of AIM2PYD: ASCPYD interaction yielded a binary complex with heavily skewed stoichiometry (Lu et al., 2014b). The complex is filamentous under EM examination. Immunolabeling located AIM2PYD at one end of the filaments, while ASCPYD constituted the bulk (Lu et al., 2014b). Atomic resolution structure was obtained with optimized ASCPYD filaments. The 3.8 Å model (Figure 1D) is a right-handed helical filament with three-fold symmetry (Lu et al., 2014b). ASCPYD subunits fit into a polar tube with inner and outer diameters of ~ 20 Å and ~ 90 Å, respectively. Since AIM2PYD is only found at the ends of ASCPYD filaments, it is hypothesized that AIM2 serves as the “seeds” or nucleating platforms from which ASCPYD subunits assemble into filaments. FP assays demonstrated that both AIM2PYD and AIM2FL+dsDNA accelerated spontaneous filament formation of ASCPYD (Lu et al., 2014b). EM reconstruction further strengthened this hypothesis. Negative stain EM power spectra of AIM2PYD and ASCPYD filaments are remarkably similar to each other (Morrone et al., 2015). The reconstructed AIM2PYD and ASCPYD filament structures are comparable in diameter and subunit organization (Lu et al., 2014b; Morrone et al., 2015) (Figure 1C and 1D). However, certain degree of mismatch between AIM2PYD and ASCPYD symmetry is tolerated. Subunit packing in GFP-AIM2PYD filaments is less-dense than that in ASCPYD filaments, but GFP-AIM2PYD potentiated ASCPYD polymerization at an even lower concentration than AIM2PYD (Lu et al., 2015). Another noteworthy feature of ASCPYD filament is all C-termini of PYD subunits point outward (Figure 1D), allowing ample space to accommodate the CARD domain in full-length ASC.
ASC-nucleated Casp1CARD helical filament formation
In reconstituted core AIM2 inflammasome (AIM2PYD: ASCFL: GFP-Casp1CARD), ASCFL appeared sub-stoichiometric to Casp1CARD, in the same way as the understoichiometry of AIM2PYD to ASC observed in both binary and ternary complexes (Lu et al., 2014b). Electron micrographs recaptured the similarity. In the star-shaped ternary complex, ASCFL resided in the center, while Casp1CARD were detected all along the arms of the stars (Lu et al., 2014b). Casp1CARD polymerization is nucleated by upstream adaptor ASCCARD, much like ASCPYD filament is nucleated by AIM2PYD (Lu et al., 2016). The Casp1CARD filament reconstructed at ~ 4.8 Å differs from PYD filaments in both diameter and symmetry (Figure 1E). It has an outer diameter of ~ 80 Å and an inner one < 10 Å. Casp1CARD filament is left-handed and one-start (Lu et al., 2016), unlike the right-handed PYD filaments, but shares same symmetry with MAVSCARD filament and Myddosome (Lin et al., 2010; Wu et al., 2014).
Inhibition of DNA-dependent AIM2 clustering by p202
p202 is a mouse ALR protein with two HIN domains but no PYD (Figure 1A). Like AIM2HIN, p202HIN1 binds to dsDNA through electrostatic interactions in a sequence-independent manner. Interestingly, dsDNA binding surface on p202HIN1 falls on the opposite side on HIN scaffold (Ru et al., 2013; Yin et al., 2013) (Figure 1B and 1F). p202HIN2 does not bind to dsDNA at all. Instead, it mediates the tetramerization of p202 (Yin et al., 2013) (Figure 1G). Tetramerization of p202 increases its affinity towards dsDNA for better competition with AIM2. Moreover, the interaction between p202HIN2 and AIM2HIN makes p202 a specific inhibitor of AIM2, but not other cytoplasmic DNA sensors. p202 reduces AIM2 clustering by both masking DNA sites and keeping bound AIM2 molecules away from each other.
Capping of Casp1CARD filament by INCA
Human genome encodes several PYD- or CARD-only proteins (POPs and COPs) that negatively regulate inflammasome function, including POP1/ASC2, POP2, POP3, COP1, Iceberg, and INCA (Khare et al., 2014; Le and Harton, 2013). However, their mechanisms may vary vastly. Iceberg is filamentous and promotes Casp1CARD filament formation, while INCA is monomeric and inhibits Casp1CARD polymerization (Lu et al., 2016). INCA inhibits Casp1CARD polymerization with a nanomolar Ki, three orders of magnitudes lower than Casp1CARD concentration used in the assay. It is postulated that INCA has a higher affinity towards Casp1CARD filaments rather than monomeric Casp1CARD. Indeed, in electron micrographs, INCA exclusively localized at the tips of Casp1CARD filaments (Lu et al., 2016).
The putative IFI16 inflammasome
Interferon-inducible protein 16 (IFI16), another ALR family member, forms inflammasome in both nucleus and cytoplasm (Johnson et al., 2013; Kerur et al., 2011). The HIN2 domain of IFI16 employs a surface similar to AIM2HIN to bind dsDNA (Jin et al., 2012). Binding of IFI16 to dsDNA is also cooperative and length-dependent like AIM2 (Morrone et al., 2014; Stratmann et al., 2015). However, the binding is less stable and IFI16 scans one-dimensionally on DNA through its HIN domains (Stratmann et al., 2015).
Concluding remarks
High-order structures, especially helical assemblies, are ubiquitous in AIM2 inflammasome activation. Naturally, cellular factors tightly regulate such assemblies to prevent potentially lethal AIM2 inflammasome activation (Figure 2). Nucleated helical filament formation is elucidated using AIM2 inflammasome as a model, but this mechanism is found in other inflammasomes as well. Oligomerized PYD-containing NLRP3 and NLRP6 fragments are reported to nucleate ASCPYD filament formation (Lu et al., 2015; Lu et al., 2014b). Theoretically, all ASC-dependent or PYD-containing inflammasomes can be activated in the same way. In ASC-independent NAIP-NLRC4 inflammasome, NLRC4CARD directly expedites Casp1CARD filament formation (Lu et al., 2016).
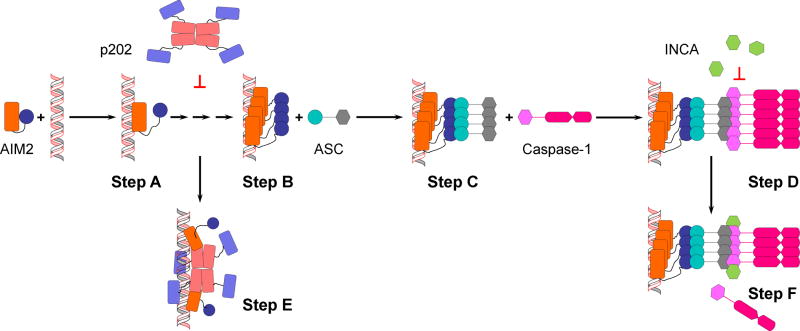
Figure 2.
A diagram of AIM2 inflammasome activation and regulation. Domains are represented in the same way as in Figure 1A. Step A: dsDNA binds to AIM2HIN. AIM2PYD is free to self-associate. Step B: AIM2 clusters on dsDNA. AIM2PYD forms filamentous structure. Step C: Filamentous AIM2PYD nucleated ASCPYD filament formation. Step D: Filamentous ASC nucleates filament formation of caspase-1. Step E: p202 blocks further access to dsDNA and separates bound AIM2. Step F: INCA caps caspase-1 filament, preventing further growth and activation.
Post-translational modifications (PTM) add another layer of complexity to inflammasome activation and regulation. Phosphorylation of NLRP3PYD disrupted interactions between two PYDs by electrostatic repulsion, therefore abrogates NLRP3 inflammasome activation (Stutz et al., 2017). Both phosphorylation and ubiquitination of ASC have been reported to promote the formation of high-order ASC structure, but the molecular mechanisms are yet unclear (Man and Kanneganti, 2015).
Although the past few years have seen much progress in understanding AIM2 inflammasome assembly and activation, some questions remain unanswered. What are the minimum activating units of upstream nucleators? Are there any oligomeric intermediates between monomers and filaments? What kind of kinetics does filament growth follow? Carefully designed experiments are required to answer these questions.
Helical filaments are a recurring theme in biology, tobacco mosaic virus capsid, microtubules, and RecA are but a few examples. In innate immunity and inflammation, RNA sensors and DD superfamily proteins often form filaments to initiate and amplify signaling (Ferrao and Wu, 2012; Sohn and Hur, 2016). Assembly of helical filaments and high-order structures in general enables signal amplification, reduces stochastic activation, and ensures rapid and robust activation upon ligand stimulation (Kagan et al., 2014).
Acknowledgments
We thank Dr. Yuan Tian for constructive discussion during manuscript preparation. This work was supported by the US National Institutes of Health grant (grant R00AI108793) and start-up funds from Florida State University to Q.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC evolutionary biology. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288:13225–13235. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Magupalli VG, Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014 doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV, Wallin MC, Gangopadhyay A, Forte E, Gottwein E, Perlman H, Reed JC, Greaves DR, Dorfleutner A, Stehlik C. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol. 2014;15:343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HT, Harton JA. Pyrin- and CARD-only Proteins as Regulators of NLR Functions. Frontiers in immunology. 2013;4:275. doi: 10.3389/fimmu.2013.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol. 2014a;426:1420–1427. doi: 10.1016/j.jmb.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman E, Wu H. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell discovery. 2015:1. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014b;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Li Y, Schmidt FI, Yin Q, Chen S, Fu TM, Tong AB, Ploegh HL, Mao Y, Wu H. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat Struct Mol Biol. 2016;23:416–425. doi: 10.1038/nsmb.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. European journal of immunology. 2016;46:269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A. 2014;111:E62–71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, Sohn J. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nature communications. 2015;6:7827. doi: 10.1038/ncomms8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Ru H, Ni X, Zhao L, Crowley C, Ding W, Hung LW, Shaw N, Cheng G, Liu ZJ. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013 doi: 10.1038/cr.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- Sohn J, Hur S. Filament assemblies in foreign nucleic acid sensors. Curr Opin Struct Biol. 2016;37:134–144. doi: 10.1016/j.sbi.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann SA, Morrone SR, van Oijen AM, Sohn J. The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex. eLife. 2015;4:e11721. doi: 10.7554/eLife.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz A, Kolbe CC, Stahl R, Horvath GL, Franklin BS, van Ray O, Brinkschulte R, Geyer M, Meissner F, Latz E. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med. 2017;214:1725–1736. doi: 10.1084/jem.20160933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, Sagulenko V, Thygesen SJ, Choubey D, Hornung V, Walz T, Stacey KJ, Wu H. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell reports. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]